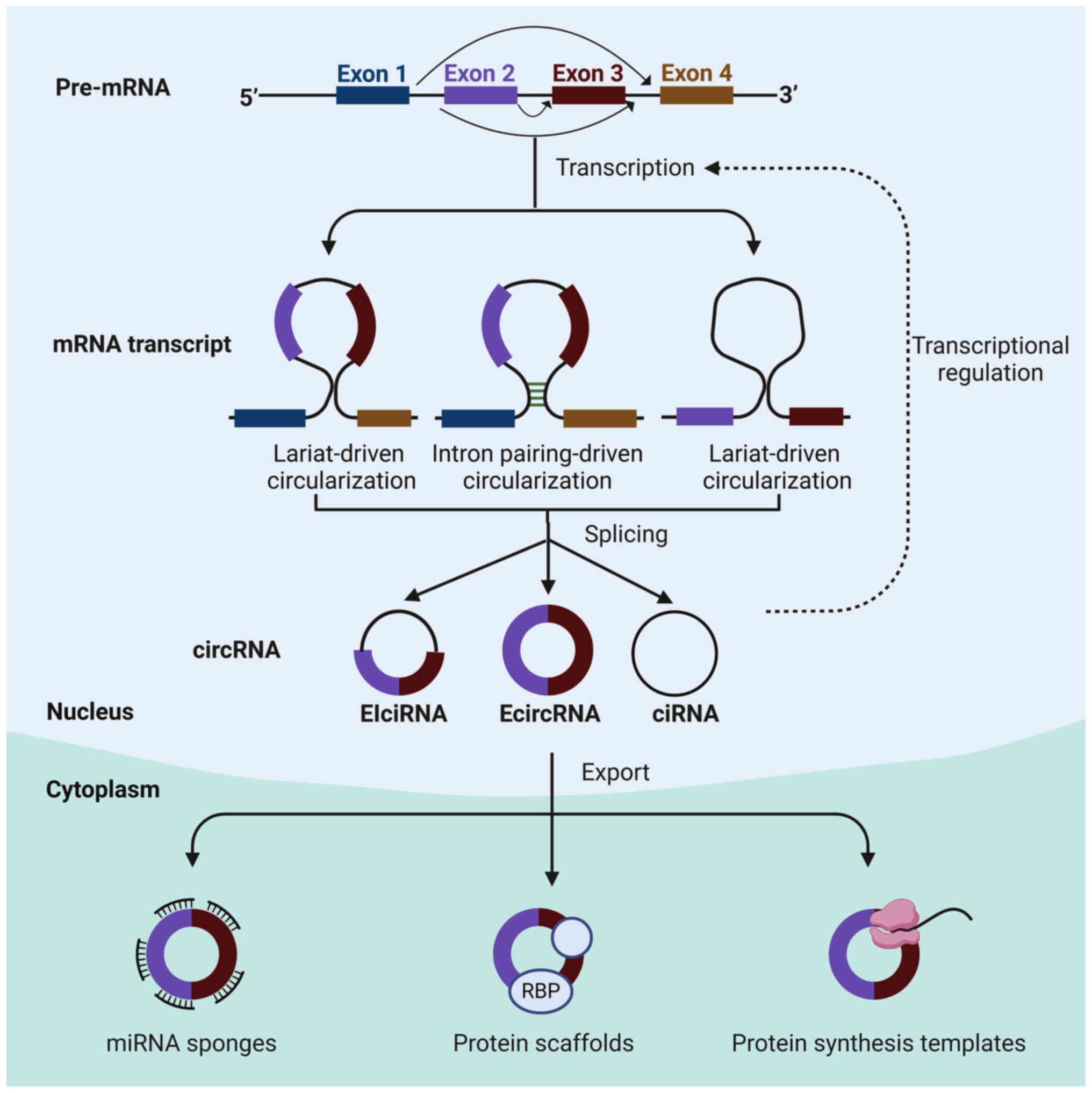
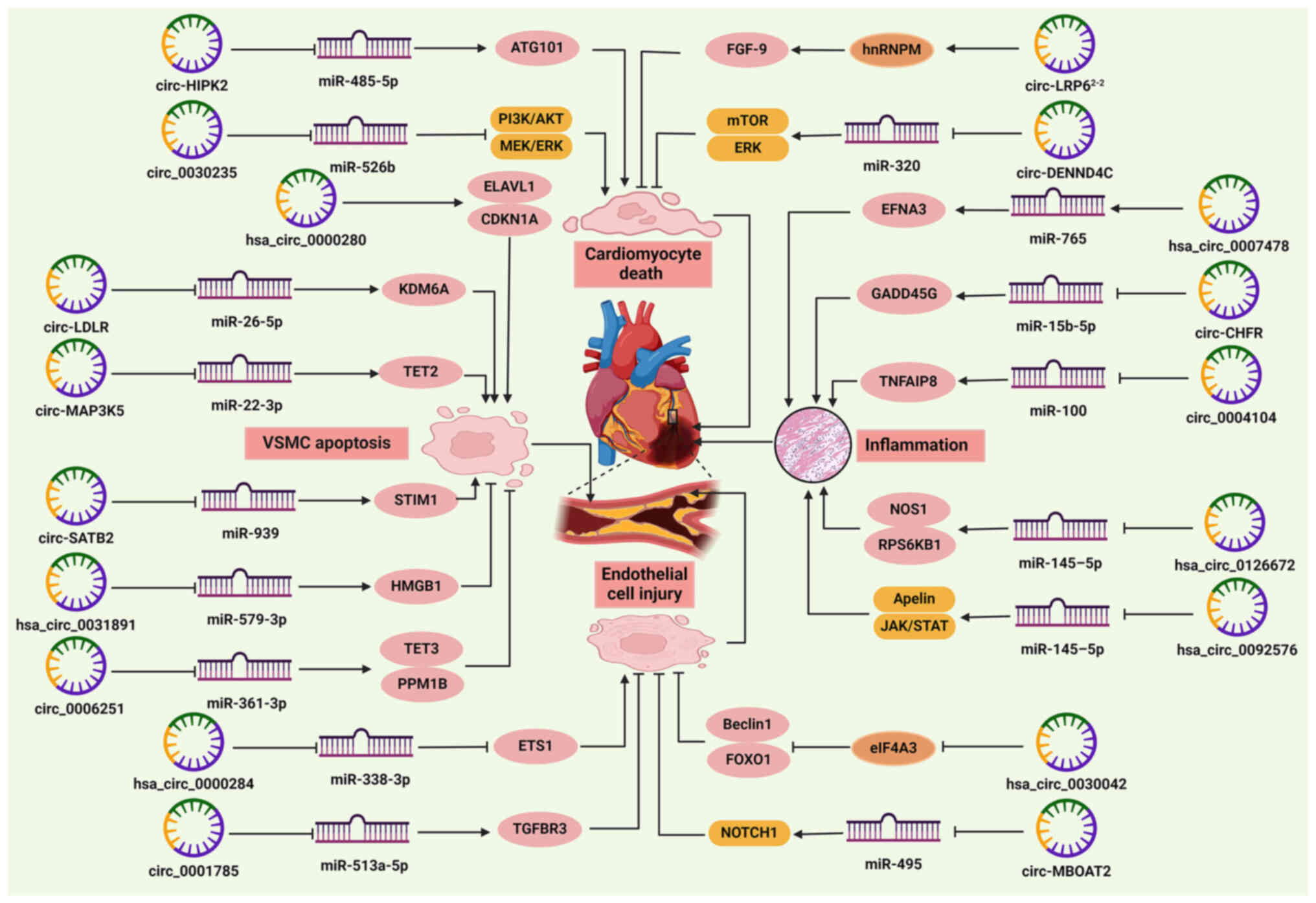
|
1
|
Katta N, Loethen T, Lavie CJ and Alpert
MA: Obesity and coronary heart disease: Epidemiology, Pathology,
and coronary artery imaging. Curr Probl Cardiol. 46:1006552021.
View Article : Google Scholar
|
|
2
|
Pothineni NVK, Subramany S, Kuriakose K,
Shirazi LF, Romeo F, Shah PK and Mehta JL: Infections,
atherosclerosis, and coronary heart disease. Eur Heart J.
38:3195–3201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong Y, Chen H, Gao J, Liu Y, Li J and
Wang J: Molecular machinery and interplay of apoptosis and
autophagy in coronary heart disease. J Mol Cell Cardiol. 136:27–41.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hodel F, Xu ZM, Thorball CW, de La Harpe
R, Letang-Mathieu P, Brenner N, Butt J, Bender N, Waterboer T,
Marques-Vidal PM, et al: Associations of genetic and infectious
risk factors with coronary heart disease. Elife. 12:e797422023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sygitowicz G and Sitkiewicz D: Involvement
of circRNAs in the Development of Heart Failure. Int J Mol Sci.
23:141292022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma XK, Zhai SN and Yang L: Approaches and
challenges in genome-wide circular RNA identification and
quantification. Trends Genet. 39:897–907. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang HJ and Kim YK: Molecular mechanisms
of circular RNA translation. Exp Mol Med. 56:1272–1280. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou J, Li L, Hu H, Wu J, Chen H, Feng K
and Ma L: Circ-HIPK2 accelerates cell apoptosis and autophagy in
myocardial oxidative injury by sponging miR-485-5p and Targeting
ATG101. J Cardiovasc Pharmacol. 76:427–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dinh P, Tran C, Dinh T, Ali A and Pan S:
Hsa_circRNA_0000284 acts as a ceRNA to participate in coronary
heart disease progression by sponging miRNA-338-3p via regulating
the expression of ETS1. J Biomol Struct Dyn. 42:5114–5127. 2024.
View Article : Google Scholar
|
|
10
|
Ye B, Liang X, Zhao Y, Cai X, Wang Z, Lin
S, Wang W, Shan P, Huang W and Huang Z: Hsa_circ_0007478 aggravates
NLRP3 inflammasome activation and lipid metabolism imbalance in
ox-LDL-stimulated macrophage via miR-765/EFNA3 axis. Chem Biol
Interact. 368:1101952022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou C, Gu L, Guo Y, Zhou Y, Hua L, Chen J,
He S, Zhang S, Jia Q, Zhao C, et al: Association between circular
RNA expression content and severity of coronary atherosclerosis in
human coronary artery. J Clin Lab Anal. 34:e235522020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akan G, Nyawawa E, Nyangasa B, Turkcan MK,
Mbugi E, Janabi M and Atalar F: Severity of coronary artery disease
is associated with diminished circANRIL expression: A possible
blood based transcriptional biomarker in East Africa. J Cell Mol
Med. 28:e180932024. View Article : Google Scholar :
|
|
13
|
Cao Q, Guo Z, Du S, Ling H and Song C:
Circular RNAs in the pathogenesis of atherosclerosis. Life Sci.
255:1178372020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma X, Chen X, Mo C, Li L, Nong S and Gui
C: The role of circRNAs in the regulation of myocardial
angiogenesis in coronary heart disease. Microvasc Res.
142:1043622022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CK, Cheng R, Demeter J, Chen J,
Weingarten-Gabbay S, Jiang L, Snyder MP, Weissman JS, Segal E,
Jackson PK and Chang HY: Structured elements drive extensive
circular RNA translation. Mol Cell. 81:4300–4318.e13. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Busa VF and Leung AKL: Thrown for a (stem)
loop: How RNA structure impacts circular RNA regulation and
function. Methods. 196:56–67. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang S, Fu R, Shi J, Wu H, Mai J, Hua X,
Chen H, Liu J, Lu M and Li N: CircRNA-Mediated regulation of
angiogenesis: A new chapter in cancer biology. Front Oncol.
11:5537062021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding C and Zhou Y: Insights into circular
RNAs: Biogenesis, function and their regulatory roles in
cardiovascular disease. J Cell Mol Med. 27:1299–1314. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar
|
|
23
|
Montañés-Agudo P, van der Made I, Aufiero
S, Tijsen AJ, Pinto YM and Creemers EE: Quaking regulates circular
RNA production in cardiomyocytes. J Cell Sci. 136:sc2611202023.
View Article : Google Scholar
|
|
24
|
Pamudurti NR, Patop IL, Krishnamoorthy A,
Bartok O, Maya R, Lerner N, Ashwall-Fluss R, Konakondla JVV, Beatus
T and Kadener S: circMbl functions in cis and in trans to regulate
gene expression and physiology in a tissue-specific fashion. Cell
Rep. 39:1107402022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Monat C, Quiroga C, Laroche-Johnston F and
Cousineau B: The Ll.LtrB intron from Lactococcus lactis excises as
circles in vivo: insights into the group II intron circularization
pathway. RNA. 21:1286–1293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The Output of Protein-Coding Genes
Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is
Limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
García-Lerena JA, González-Blanco G,
Saucedo-Cárdenas O and Valdés J: Promoter-Bound Full-Length
Intronic Circular RNAs-RNA Polymerase II Complexes Regulate Gene
Expression in the Human Parasite Entamoeba histolytica. Noncoding
RNA. 8:122022.PubMed/NCBI
|
|
31
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
32
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vromman M, Anckaert J, Bortoluzzi S,
Buratin A, Chen CY, Chu Q, Chuang TJ, Dehghannasiri R, Dieterich C,
Dong X, et al: Large-scale benchmarking of circRNA detection tools
reveals large differences in sensitivity but not in precision. Nat
Methods. 20:1159–1169. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haque S and Harries LW: Circular RNAs
(circRNAs) in Health and Disease. Genes (Basel). 8:3532017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Q, Sun W, Han J, Cheng S, Yu P, Shen
L, Fan M, Tong H, Zhang H, Chen J and Chen X: The circular RNA hsa_
circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac
repair. Biochem Biophys Res Commun. 523:993–1000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Szabo L, Morey R, Palpant NJ, Wang PL,
Afari N, Jiang C, Parast MM, Murry CE, Laurent LC and Salzman J:
Statistically based splicing detection reveals neural enrichment
and tissue-specific induction of circular RNA during human fetal
development. Genome Biol. 16:1262015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Guo X, Wen Y, Huang S, Yuan X, Tang
L and Sun H: N6-Methyladenosine Modification Opens a New Chapter in
Circular RNA Biology. Front Cell Dev Biol. 9:7092992021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu T, He B, Sun H, Xiong M, Nie J, Wang S
and Pan Y: Novel insights into the interaction between
N6-methyladenosine modification and circular RNA. Mol Ther Nucleic
Acids. 27:824–837. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
46
|
Tao M, Zheng M, Xu Y, Ma S, Zhang W and Ju
S: CircRNAs and their regulatory roles in cancers. Mol Med.
27:942021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Graham JR, Hendershott MC, Terragni J and
Cooper GM: mRNA degradation plays a significant role in the program
of gene expression regulated by phosphatidylinositol 3-kinase
signaling. Mol Cell Biol. 30:5295–5305. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nisar S, Bhat AA, Singh M, Karedath T,
Rizwan A, Hashem S, Bagga P, Reddy R, Jamal F, Uddin S, et al:
Insights Into the Role of CircRNAs: Biogenesis, characterization,
functional, and clinical impact in human malignancies. Front Cell
Dev Biol. 9:6172812021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang J, Yu S, Ding L, Ma L, Chen H, Zhou
H, Zou Y, Yu M, Lin J and Cui Q: The Dual Role of Circular RNAs as
miRNA Sponges in breast cancer and colon cancer. Biomedicines.
9:15902021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration
After Myocardial Infarction in Adult Mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma B, Wang S, Wu W, Shan P, Chen Y, Meng
J, Xing L, Yun J, Hao L, Wang X, et al: Mechanisms of
circRNA/lncRNA-miRNA interactions and applications in disease and
drug research. Biomed Pharmacother. 162:1146722023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Misir S, Wu N and Yang BB: Specific
expression and functions of circular RNAs. Cell Death Differ.
29:481–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Das A, Sinha T, Shyamal S and Panda AC:
Emerging Role of Circular RNA-Protein Interactions. Noncoding RNA.
7:482021.PubMed/NCBI
|
|
57
|
Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD and
Tang JH: The Emerging Role of the Interactions between Circular
RNAs and RNA-binding Proteins in Common Human Cancers. J Cancer.
12:5206–5219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar :
|
|
59
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A Circular RNA Binds To
and Activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Conte A and Pierantoni GM: Update on the
Regulation of HIPK1, HIPK2 and HIPK3 Protein Kinases by microRNAs.
Microrna. 7:178–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 Inhibits Malignant Progression by
Sponging miR-197-3p and Encoding a 185-aa Protein in
Triple-Negative Breast Cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu C, Wu X, Gokulnath P, Li G and Xiao J:
The Functions and Mechanisms of Translatable Circular RNAs. J
Pharmacol Exp Ther. 384:52–60. 2023. View Article : Google Scholar
|
|
66
|
Li X, Zhao Z, Jian D, Li W, Tang H and Li
M: Hsa-circRNA11783-2 in peripheral blood is correlated with
coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis
Res. 14:510–515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Guo N, Zhou H, Zhang Q, Fu Y, Jia Q, Gan
X, Wang Y, He S, Li C, Tao Z, et al: Exploration and bioinformatic
prediction for profile of mRNA bound to circular RNA BTBD7_hsa_
circ_0000563 in coronary artery disease. BMC Cardiovasc Disord.
24:712024. View Article : Google Scholar
|
|
68
|
Chen JX, Hua L, Zhao CH, Jia QW, Zhang J,
Yuan JX, Zhang YJ, Jin JL, Gu MF, Mao ZY, et al: Quantitative
proteomics reveals the regulatory networks of circular RNA
BTBD7_hsa_ circ_0000563 in human coronary artery. J Clin Lab Anal.
34:e234952020. View Article : Google Scholar
|
|
69
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_ circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sun Y, Chen R, Lin S, Xie X, Ye H, Zheng
F, Lin J, Huang Q, Huang S, Ruan Q, et al: Association of circular
RNAs and environmental risk factors with coronary heart disease.
BMC Cardiovasc Disord. 19:2232019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wu WP, Pan YH, Cai MY, Cen JM, Chen C,
Zheng L, Liu X and Xiong XD: Plasma-Derived Exosomal Circular RNA
hsa_ circ_0005540 as a Novel Diagnostic Biomarker for Coronary
Artery Disease. Dis Markers. 2020:31786422020. View Article : Google Scholar
|
|
72
|
Yu F, Tie Y, Zhang Y, Wang Z, Yu L, Zhong
L and Zhang C: Circular RNA expression profiles and bioinformatic
analysis in coronary heart disease. Epigenomics. 12:439–454. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Dinh P, Peng J, Tran T, Wu D, Tran C, Dinh
T and Pan S: Identification of hsa_circ_0001445 of a novel
circRNA-miRNA-mRNA regulatory network as potential biomarker for
coronary heart disease. Front Cardiovasc Med. 10:11042232023.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yin L, Tang Y and Jiang M: Research on the
circular RNA bioinformatics in patients with acute myocardial
infarction. J Clin Lab Anal. 35:e236212021. View Article : Google Scholar :
|
|
75
|
Pan RY, Liu P, Zhou HT, Sun WX, Song J,
Shu J, Cui GJ, Yang ZJ and Jia EZ: Circular RNAs promote TRPM3
expression by inhibiting hsa-miR-130a-3p in coronary artery disease
patients. Oncotarget. 8:60280–60290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yijian L, Weihan S, Lin Y, Heng Z, Yu W,
Lin S, Shuo M, Mengyang L and Jianxun W: CircNCX1 modulates
cardiomyocyte proliferation through promoting ubiquitination of
BRG1. Cell Signal. 120:1111932024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kishore R, Garikipati VNS and Gonzalez C:
Role of Circular RNAs in Cardiovascular Disease. J Cardiovasc
Pharmacol. 76:128–137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Maguire EM and Xiao Q: Noncoding RNAs in
vascular smooth muscle cell function and neointimal hyperplasia.
FEBS J. 287:5260–5283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ma W, Wei S, Zhang B and Li W: Molecular
Mechanisms of Cardiomyocyte Death in Drug-Induced Cardiotoxicity.
Front Cell Dev Biol. 8:4342020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dorn GW II: Apoptotic and non-apoptotic
programmed cardiomyocyte death in ventricular remodelling.
Cardiovasc Res. 81:465–473. 2009. View Article : Google Scholar :
|
|
81
|
Nah J, Zablocki D and Sadoshima J: The
roles of the inhibitory autophagy regulator Rubicon in the heart: A
new therapeutic target to prevent cardiac cell death. Exp Mol Med.
53:528–536. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Martens MD, Karch J and Gordon JW: The
molecular mosaic of regulated cell death in the cardiovascular
system. Biochim Biophys Acta Mol Basis Dis. 1868:1662972022.
View Article : Google Scholar
|
|
83
|
Xiang Q, Yi X, Zhu XH, Wei X and Jiang DS:
Regulated cell death in myocardial ischemia-reperfusion injury.
Trends Endocrinol Metab. 35:219–234. 2024. View Article : Google Scholar
|
|
84
|
Parrotta EI, Lucchino V, Scaramuzzino L,
Scalise S and Cuda G: Modeling cardiac disease mechanisms using
induced pluripotent stem cell-derived cardiomyocytes: progress,
promises and challenges. Int J Mol Sci. 21:43542020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang Y, Liu S, Ding L, Wang D, Li Q and
Li D: Circ_0030235 knockdown protects H9c2 cells against
OGD/R-induced injury via regulation of miR-526b. PeerJ.
9:e114822021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang J, Zhang T, Zhang W, Zou C, Zhang Q,
Ma X and Zhu Y: Circular RNA-DENND4C in H9c2 cells relieves
OGD/R-induced injury by down regulation of microRNA-320. Cell
Cycle. 19:3074–3085. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ding W, Ding L, Lu Y, Sun W, Wang Y, Wang
J, Gao Y and Li M: Circular RNA-circLRP6 protects cardiomyocyte
from hypoxia-induced apoptosis by facilitating hnRNPM-mediated
expression of FGF-9. FEBS J. 291:1246–1263. 2024. View Article : Google Scholar
|
|
88
|
Ko T and Nomura S: Manipulating
cardiomyocyte plasticity for heart regeneration. Front Cell Dev
Biol. 10:9292562022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Theofilis P, Sagris M, Oikonomou E,
Antonopoulos AS, Siasos G, Tsioufis C and Tousoulis D: Inflammatory
mechanisms contributing to endothelial dysfunction. Biomedicines.
9:7812021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shaito A, Aramouni K, Assaf R, Parenti A,
Orekhov A, Yazbi AE, Pintus G and Eid AH: Oxidative stress-induced
endothelial dysfunction in cardiovascular diseases. Front Biosci
(Landmark Ed). 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Gallo G and Savoia C: New insights into
endothelial dysfunction in cardiometabolic diseases: Potential
mechanisms and clinical implications. Int J Mol Sci. 25:29732024.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Huang Y, Song C, He J and Li M: Research
progress in endothelial cell injury and repair. Front Pharmacol.
13:9972722022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu S, Ilyas I, Little PJ, Li H, Kamato D,
Zheng X, Luo S, Li Z, Liu P, Han J, et al: Endothelial dysfunction
in atherosclerotic cardiovascular diseases and beyond: From
mechanism to pharmacotherapies. Pharmacol Rev. 73:924–967. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang R, Wang M, Ye J, Sun G and Sun X:
Mechanism overview and target mining of atherosclerosis:
Endothelial cell injury in atherosclerosis is regulated by
glycolysis (Review). Int J Mol Med. 47:65–76. 2021. View Article : Google Scholar
|
|
95
|
Zha D, Wang S, Monaghan-Nichols P, Qian Y,
Sampath V and Fu M: Mechanisms of endothelial cell membrane repair:
Progress and Perspectives. Cells. 12:26482023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Marzoog BA: Endothelial cell autophagy in
the context of disease development. Anat Cell Biol. 56:16–24. 2023.
View Article : Google Scholar :
|
|
97
|
Wang LP, Han RM, Wu B, Luo MY, Deng YH,
Wang W, Huang C, Xie X and Luo J: Mst1 silencing alleviates
hypertensive myocardial injury associated with the augmentation of
microvascular endothelial cell autophagy. Int J Mol Med.
50:1462022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sobrevia L, Aiello EA and Contreras P:
Mechanisms of endothelial dysfunction and cardiovascular system
adaptation. Curr Vasc Pharmacol. 20:201–204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tong X, Dang X, Liu D, Wang N, Li M, Han
J, Zhao J, Wang Y, Huang M, Yang Y, et al: Exosome-derived
circ_0001785 delays atherogenesis through the ceRNA network
mechanism of miR-513a-5p/TGFBR3. J Nanobiotechnology. 21:3622023.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yu F, Zhang Y, Wang Z, Gong W and Zhang C:
Hsa_ circ_0030042 regulates abnormal autophagy and protects
atherosclerotic plaque stability by targeting eIF4A3. Theranostics.
11:5404–5417. 2021. View Article : Google Scholar :
|
|
101
|
Gao W, Li C, Yuan J, Zhang Y, Liu G, Zhang
J, Shi H, Liu H and Ge J: Circ-MBOAT2 Regulates Angiogenesis via
the miR-495/ NOTCH1 axis and associates with myocardial perfusion
in patients with coronary chronic total occlusion. Int J Mol Sci.
25:7932024. View Article : Google Scholar
|
|
102
|
Wong D, Turner AW and Miller CL: Genetic
insights into smooth muscle cell contributions to coronary artery
disease. Arterioscler Thromb Vasc Biol. 39:1006–1017. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Low EL, Baker AH and Bradshaw AC: TGFβ,
smooth muscle cells and coronary artery disease: A review. Cell
Signal. 53:90–101. 2019. View Article : Google Scholar :
|
|
104
|
Cao G, Xuan X, Hu J, Zhang R, Jin H and
Dong H: How vascular smooth muscle cell phenotype switching
contributes to vascular disease. Cell Commun Signal. 20:1802022.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Milutinović A, Šuput D and Zorc-Pleskovič
R: Pathogenesis of atherosclerosis in the tunica intima, media, and
adventitia of coronary arteries: An updated review. Bosn J Basic
Med Sci. 20:21–30. 2020.
|
|
106
|
Schnack L, Sohrabi Y, Lagache SMM, Kahles
F, Bruemmer D, Waltenberger J and Findeisen HM: Mechanisms of
trained innate immunity in oxLDL primed human coronary smooth
muscle cells. Front Immunol. 10:132019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lacolley P, Regnault V, Segers P and
Laurent S: Vascular smooth muscle cells and arterial stiffening:
Relevance in development, aging, and disease. Physiol Rev.
97:1555–1617. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang G, Liu Z, Deng J, Liu L, Li Y, Weng
S, Guo C, Zhou Z, Zhang L, Wang X, et al: Smooth muscle cell fate
decisions decipher a high-resolution heterogeneity within
atherosclerosis molecular subtypes. J Transl Med. 20:5682022.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang Z, Wang H, Guo C, Yu F, Zhang Y, Qiao
L, Zhang H and Zhang C: Role of hsa_circ_0000280 in regulating
vascular smooth muscle cell function and attenuating neointimal
hyperplasia via ELAVL1. Cell Mol Life Sci. 80:32022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dai H, Zhao N and Zheng Y: CircLDLR
modulates the proliferation and apoptosis of vascular smooth muscle
cells in coronary artery disease through miR-26-5p/KDM6A Axis. J
Cardiovasc Pharmacol. 80:132–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan
X, Liu Y, Tao J, Liu Y, Li K, et al: Circular RNA CircMAP3K5 Acts
as a MicroRNA-22-3p Sponge to Promote Resolution of Intimal
Hyperplasia Via TET2-Mediated smooth muscle cell differentiation.
Circulation. 143:354–371. 2021. View Article : Google Scholar
|
|
112
|
Mao YY, Wang JQ, Guo XX, Bi Y and Wang CX:
Circ-SATB2 upregulates STIM1 expression and regulates vascular
smooth muscle cell proliferation and differentiation through
miR-939. Biochem Biophys Res Commun. 505:119–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang L, Li H, Zheng Z and Li Y:
Hsa_circ_0031891 targets miR-579-3p to enhance HMGB1 expression and
regulate PDGF-BB-induced human aortic vascular smooth muscle cell
proliferation, migration, and dedifferentiation. Naunyn
Schmiedebergs Arch Pharmacol. 397:1093–1104. 2024. View Article : Google Scholar
|
|
114
|
Zhong W, Wang L and Xiong L: Circ_0006251
mediates the proliferation and apoptosis of vascular smooth muscle
cells in CAD via enhancing TET3 and PPM1B expression. Cell Mol Biol
(Noisy-le-grand). 69:34–39. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Medina-Leyte DJ, Zepeda-García O,
Domínguez-Pérez M, González-Garrido A, Villarreal-Molina T and
Jacobo-Albavera L: Endothelial dysfunction, inflammation and
coronary artery disease: Potential biomarkers and promising
therapeutical approaches. Int J Mol Sci. 22:38502021. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Bazoukis G, Stavrakis S and Armoundas AA:
Vagus nerve stimulation and inflammation in cardiovascular disease:
A State-of-the-Art Review. J Am Heart Assoc. 12:e0305392023.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Bhattacharya P, Kanagasooriyan R and
Subramanian M: Tackling inflammation in atherosclerosis: Are we
there yet and what lies beyond? Curr Opin Pharmacol. 66:1022832022.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Prati F, Marco V, Paoletti G and
Albertucci M: Coronary inflammation: Why searching, how to identify
and treat it. Eur Heart J Suppl. 22(Suppl E): E121–E124. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Chen R, Zhang H, Tang B, Luo Y, Yang Y,
Zhong X, Chen S, Xu X, Huang S and Liu C: Macrophages in
cardiovascular diseases: Molecular mechanisms and therapeutic
targets. Signal Transduct Target Ther. 9:1302024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Matter MA, Paneni F, Libby P, Frantz S,
Stähli BE, Templin C, Mengozzi A, Wang YJ, Kündig TM, Räber L, et
al: Inflammation in acute myocardial infarction: The good, the bad
and the ugly. Eur Heart J. 45:89–103. 2024. View Article : Google Scholar :
|
|
121
|
Li Y and Wang B: Circular RNA circCHFR
downregulation protects against oxidized low-density
lipoprotein-induced endothelial injury via regulation of
microRNA-15b-5p/growth arrest and DNA damage inducible gamma.
Bioengineered. 13:4481–4492. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ji P, Song X and Lv Z: Knockdown of
circ_0004104 alleviates oxidized low-density lipoprotein-induced
vascular endothelial cell injury by regulating miR-100/TNFAIP8
Axis. J Cardiovasc Pharmacol. 78:269–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Rafiq M, Dandare A, Javed A, Liaquat A,
Raja AA, Awan HM, Khan MJ and Naeem A: Competing Endogenous RNA
Regulatory Networks of hsa_circ_0126672 in pathophysiology of
coronary heart disease. Genes (Basel). 14:5502023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Dandare A, Rafiq M, Liaquat A, Raja AA and
Khan MJ: Identification of hsa_circ_0092576 regulatory network in
the pathogenesis of coronary heart disease. Genes Dis. 10:26–28.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Liu X, Yao X and Chen L: Expanding roles
of circRNAs in cardiovascular diseases. Noncoding RNA Res.
9:429–436. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Tang Y, Bao J, Hu J, Liu L and Xu DY:
Circular RNA in cardiovascular disease: Expression, mechanisms and
clinical prospects. J Cell Mol Med. 25:1817–1824. 2021. View Article : Google Scholar :
|
|
127
|
Vilades D, Martínez-Camblor P,
Ferrero-Gregori A, Bär C, Lu D, Xiao K, Vea À, Nasarre L, Sanchez
Vega J, Leta R, et al: Plasma circular RNA hsa_circ_0001445 and
coronary artery disease: Performance as a biomarker. FASEB J.
34:4403–4414. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Sonnenschein K, Wilczek AL, de
Gonzalo-Calvo D, Pfanne A, Derda AA, Zwadlo C, Bavendiek U,
Bauersachs J, Fiedler J and Thum T: Serum circular RNAs act as
blood-based biomarkers for hypertrophic obstructive cardiomyopathy.
Sci Rep. 9:203502019. View Article : Google Scholar
|
|
129
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar :
|
|
130
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang W, Sun L, Huang MT, Quan Y, Jiang T,
Miao Z and Zhang Q: Regulatory circular RNAs in viral diseases:
applications in diagnosis and therapy. RNA Biol. 20:847–858. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Li MZ, Zhang JN, Ren F, Yin DL, Zhao XH
and Liu K: Diagnostic value of circRNA in coronary heart disease: A
meta-analysis. Biomark Med. 17:667–677. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Fu Y, He S, Li C, Gan X, Wang Y, Zhou Y,
Jiang R, Zhang Q, Pan Y, Zhou H, et al: Detailed profiling of m6A
modified circRNAs and synergistic effects of circRNA and
environmental risk factors for coronary artery disease. Eur J
Pharmacol. 951:1757612023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
He S, Fu Y, Li C, Wang Y, Zhou H, Jiang R,
Zhang Q, Jia Q, Chen X and Jia EZ: Interaction between the
expression of hsa_circRPRD1A and hsa_circHERPUD2 and classical
coronary risk factors promotes the development of coronary artery
disease. BMC Med Genomics. 16:1312023. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhong Q, Jin S, Zhang Z, Qian H, Xie Y,
Yan P, He W and Zhang L: Identification and verification of circRNA
biomarkers for coronary artery disease based on WGCNA and the LASSO
algorithm. BMC Cardiovasc Disord. 24:3052024. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang W, Cui J, Li L, Zhu T and Guo Z:
Identification of Plasma Exosomes hsa_circ_0001360 and
hsa_circ_0000038 as key biomarkers of coronary heart disease.
Cardiol Res Pract. 2024:55571432024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Huang S, Zeng Z, Sun Y, Cai Y, Xu X, Li H
and Wu S: Association study of hsa_circ_0001946, hsa-miR-7-5p and
PARP1 in coronary atherosclerotic heart disease. Int J Cardiol.
328:1–7. 2021. View Article : Google Scholar
|
|
139
|
Tong X, Zhao X, Dang X, Kou Y and Kou J:
circRNA, a novel diagnostic biomarker for coronary heart disease.
Front Cardiovasc Med. 10:10706162023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ji WF, Chen JX, He S, Zhou YQ, Hua L, Hou
C, Zhang S, Gan XK, Wang YJ, Zhou HX, et al: Characteristics of
circular RNAs expression of peripheral blood mononuclear cells in
humans with coronary artery disease. Physiol Genomics. 53:349–357.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Miao L, Yin RX, Zhang QH, Liao PJ, Wang Y,
Nie RJ and Li H: A novel circRNA-miRNA-mRNA network identifies
circ-YOD1 as a biomarker for coronary artery disease. Sci Rep.
9:183142019. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Ward Z, Schmeier S, Pearson J, Cameron VA,
Frampton CM, Troughton RW, Doughty RN, Richards AM and Pilbrow AP:
Identifying candidate circulating RNA markers for coronary artery
disease by deep RNA-Sequencing in human plasma. Cells. 11:31912022.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Dergunova LV, Vinogradina MA, Filippenkov
IB, Limborska SA and Dergunov AD: Circular RNAs variously
participate in coronary atherogenesis. Curr Issues Mol Biol.
45:6682–6700. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wang L, Xu GE, Spanos M, Li G, Lei Z,
Sluijter JPG and Xiao J: Circular RNAs in cardiovascular diseases:
Regulation and therapeutic applications. Research (Wash D C).
6:00382023.PubMed/NCBI
|
|
145
|
Goina CA, Goina DM, Farcas SS and
Andreescu NI: The role of circular RNA for early diagnosis and
improved management of patients with cardiovascular diseases. Int J
Mol Sci. 25:29862024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Long Q, Lv B, Jiang S and Lin J: The
landscape of circular RNAs in cardiovascular diseases. Int J Mol
Sci. 24:45712023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chen W, Xu J, Wu Y, Liang B, Yan M, Sun C,
Wang D, Hu X, Liu L, Hu W, et al: The potential role and mechanism
of circRNA/miRNA axis in cholesterol synthesis. Int J Biol Sci.
19:2879–2896. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Neu CT, Gutschner T and Haemmerle M:
Post-Transcriptional expression control in platelet biogenesis and
function. Int J Mol Sci. 21:76142020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yu R, Yu Q, Li Z, Li J, Yang J, Hu Y,
Zheng N, Li X, Song Y, Li J, et al: Transcriptome-wide map of
N6-methyladenosine (m6A) profiling in coronary artery disease (CAD)
with clopidogrel resistance. Clin Epigenetics. 15:1942023.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zou Y, Wang Y, Yao Y, Wu Y, Lv C and Yin
T: Platelet-derived circFAM13B associated with anti-platelet
responsiveness of ticagrelor in patients with acute coronary
syndrome. Thromb J. 22:532024. View Article : Google Scholar : PubMed/NCBI
|