Introduction
Coronary heart disease (CHD), a leading cause of
morbidity and mortality globally, represents a significant public
health challenge (1). CHD is
primarily driven by atherosclerosis, an inflammatory process
characterized by the accumulation of lipids, inflammatory cells and
fibrous elements within the arterial wall, which leads to the
formation of atherosclerotic plaques and further compromises blood
flow and oxygen delivery to the myocardium, contributing to severe
outcomes such as myocardial infarction and heart failure (2). The underlying mechanisms of CHD are
multifactorial, involving dyslipidemia, endothelial dysfunction,
oxidative stress and chronic inflammation (3). These factors collectively
contribute to the initiation and progression of atherosclerosis,
high-lighting the need for comprehensive approaches to understand
and manage the disease. Furthermore, the heterogeneity of the
disease, coupled with the complex interplay of genetic,
environmental and lifestyle factors, complicates the identification
of precise diagnostic markers and effective therapeutic targets
(4).
Currently, circular RNAs (circRNAs) have emerged as
a pivotal class of non-coding RNAs, attracting substantial
attention in the field of cardiovascular research (5). Unlike linear RNAs, circRNAs are
characterized by their covalently closed loop structure, which
confers remarkable stability and resistance to exonuclease-mediated
degradation (6). This unique
structure not only enables circRNAs to function as microRNA (miRNA)
sponges, which contain miRNA binding sites and regulate gene
expression by sequestering miRNAs, but also allows them to interact
with RNA-binding proteins (RBPs), participating in the regulation
of gene expression (7). In CHD,
emerging evidence suggests that circRNAs play critical roles in the
regulation of vascular function, myocardial injury and inflammation
(8-10). Several studies have identified
differentially expressed circRNAs in patients with CHD, correlating
with disease severity and clinical outcomes (11,12). These findings highlight the
potential of circRNAs as non-invasive biomarkers for early
diagnosis and prognosis of CHD. Furthermore, specific circRNA has
been verified to participate in modulating key signaling pathways
associated with atherosclerosis and myocardial angiogenesis,
offering insights into novel therapeutic strategies (13,14).
This review aims to provide a comprehensive overview
of the current understanding of circRNAs in CHD. It summarizes the
molecular mechanisms by which circRNAs influence the
pathophysiology of CHD, including their roles in cardiomyocyte
death, endothelial injury, vascular dysfunction and inflammation.
In addition, it explores the potential clinical applications of
circRNAs as biomarkers and therapeutic targets, emphasizing the
translational implications of circRNA research in improving patient
outcomes.
Biogenesis and functions of circRNAs
CircRNAs are a unique class of non-coding RNAs found
in various organisms, including plants and animals, as well as in
certain plant and animal viruses, where they are linked to viral
replication and survival (15).
The stable structure contributes to their accumulation within cells
and makes them play long-term regulatory roles (16). Irrespective of the size, a
circRNA molecule may possess <100 nuclei and numerous bases,
thereby rendering each element of the reverse shearing procedure
deterministic (17). In general,
circRNAs are comprised of 2-3 exons with a median size frequently
exceeding 500 nucleotides, yet not surpassing 700 nucleotides
(18). They exhibit a high
degree of conservation across different species, with their
expression typically being specific to certain cell types, tissues
and stages of development (19).
Hence, the presence in numerous species, abundance of information,
tissue-specific nature and remarkable stability represent key
attributes of circRNAs. Given these characteristics, circRNAs have
the potential to serve as valuable indicators for specific
disorders and novel targets for therapeutic interventions.
Biogenesis
The formation of circRNAs involves specific
sequences and secondary structures within the precursor (pre)-mRNA,
which facilitate the back-splicing process (20). Intronic complementary sequences
and RBPs connect donor-splice and acceptor-splice sites by
appropriate pairing of bases or interacting with definite motifs,
playing crucial roles in circRNA biogenesis (21,22). RBPs such as Quaking and
Muscleblind bind to flanking intronic sequences to bring splice
sites into close proximity, promoting the back-splicing event
(23,24). The resulting circRNAs can
originate from three main sources: Exonic genes (exon-derived
circRNA), intronic regions (intron-derived circRNA, such as
pre-mRNA and pre-transfer RNA), or combinations of both exons and
introns, leading to three primary types: Exonic circRNAs
(EcircRNAs), circular intronic RNAs (ciRNAs) and exon-intron
circRNAs (EIciRNAs) (25-27).
The majority of identified circRNAs are derived from exonic
regions, while ciRNAs and EIciRNAs only constitute a small fraction
within this category (28).
The generation of endogenous circRNAs is
characterized by notable inefficiency, a phenomenon attributed to
the elongation process of RNA polymerase II and tightly regulated
by cis elements (29,30). Various types of circRNAs are
produced through distinct mechanisms (Fig. 1), primarily involving exon
cyclization to generate cytoplasmic circRNAs (31). The creation of exon circRNAs
predominantly occurs via two mechanisms: Lariat-driven
circularization and circularization driven by introns containing
reverse complementary sequences (32). Lariat-driven circularization
involves the direct linkage of the 5′ and 3′ ends of the pre-mRNA
to form a circular structure (33). Furthermore, this process is
associated with exon skipping, where certain exons are omitted to
facilitate circular structure formation (34). In addition, intron pairing loops
facilitate circRNA formation by promoting base complementary
pairing between introns (35).
Typically, introns excised from the pre-mRNA are debranched and
degraded by exonucleases. However, intron RNA formation is
sustained by the presence of 7 GU sequences and 11 C sequences at
the intron 5′ splice site within a lasso structure, preventing
degradation and promoting circularization (36). The generation mechanism of
exon-intron RNA mirrors that of EcircRNAs, with the distinction
that the circular structure retains the intron component (14,16,37).
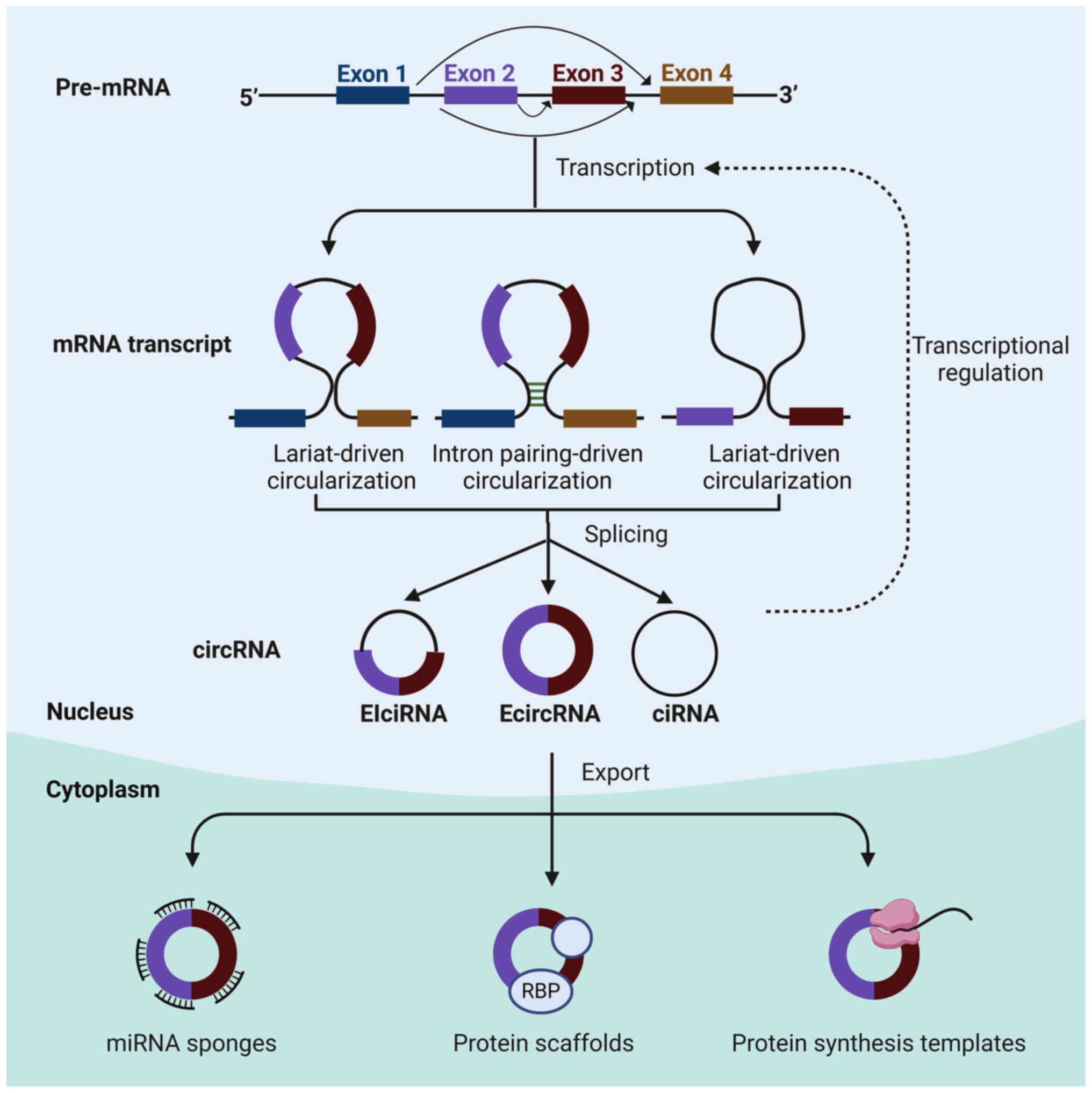 | Figure 1Biogenesis and function of circRNA.
By the lariat-driven cyclization, exon splicing generates a lariat
structure. The 5′ splice donor site of exon 2 covalently links to
the 3′ splice acceptor of exon 3. EcircRNAs are formed after the
removal of the intronic sequence. Likewise, the intron lariat forms
the ciRNAs. Besides, by intron pairing-driven circularization,
direct base-pairing of the introns flanking inverted repeats leads
to the formation of a circular structure. The introns are removed
to form EIciRNAs. CircRNAs can function as sponges to diminish
miRNA function. They also regulate gene transcription. CircRNAs can
function as decoys for proteins, like RBPs, to regulate their
functions. They can act as translatable RNAs to encode peptides.
ciRNA, circular intronic RNA; EcircRNA, circular exonic RNA;
EIciRNA, exonic circRNA; RBP, RNA-binding protein; miRNA, microRNA;
circRNA, circular RNA. |
Owing to the advancing sophistication of
contemporary biological information and RNA sequencing techniques,
a substantial volume of transcriptional data has emerged, revealing
the presence of circRNAs in various cell types within human organs
(38). In the human heart, the
percentage of expressed genes capable of generating circRNAs is 9%
(17). It is evident that as
human organs develop and certain diseases occur, the expression of
circRNA undergoes changes (39).
Research has demonstrated an elevation in circRNA expression levels
in the developing heart (40,41). While the reason for this
escalation remains elusive, several possible explanations have so
far been proposed. For instance, a study utilizing cardiomyocytes
derived from human-induced pluripotent stem cells revealed the
dynamic regulation of circRNA expression under chronic and acute
stress conditions (42). It
should be noted that N-6 methylation (m6A) has the ability to
impact circRNA biogenesis and cellular localization. Sites of m6A
position in proximity to the start and stop codons of mRNAs can
recruit the spliceosome and further facilitate back-splicing and
circRNA generation, while specific nuclear reader proteins possess
the capability to interact with m6A motifs, aiding in the export of
circRNAs from the nucleus to the cytoplasm (43,44). Furthermore, various stimuli like
high temperature or oxidative stress also affect circRNA levels
(45). Therefore, the
biogenesis, structure formation, post-transcriptional modifications
and subcellular localization of circRNAs, which are intricately
linked to their functional significance within the organism,
deserve further investigation.
Despite their stability, circRNAs can still undergo
degradation through various pathways, such as RNase L and
Argonaute-dependent and independent mechanisms (46). CircRNAs also interact with RBPs,
which can influence their stability. For instance, proteins like
transcript factor II B-related factor 1 and KH-type splicing
regulatory protein, which are regulated by signaling pathways such
as PI3K, can mediate mRNA stability and potentially affect circRNA
degradation (47). The m6A
modifications on circRNAs further alter the interaction of circRNAs
with RBPs, thereby influencing their stability and degradation
pathways (48). Understanding
the degradation pathways of circRNA can inform therapeutic
strategies, particularly in diseases where circRNAs are
dysregulated, such as CHD. Targeting specific degradation pathways
could modulate circRNA levels and their associated biological
effects.
Functions
In the last five years, investigations into circRNAs
have facilitated a more thorough comprehension of RNA's
functionality by researchers. The unique structural features of
circRNAs confer specific biological functions upon them. Current
research findings indicate that circRNAs exert a significant effect
across various developmental phases and pathophysiological states
by functioning as miRNA sponges, engaging with RBPs, serving as
modulators of transcription or translation, impacting pre-mRNA
splicing and contributing to protein translation (Fig. 1) (22,27,49-51).
One of the most well-documented functions of
circRNAs is their ability to act as miRNA sponges or competitive
endogenous RNAs (ceRNAs) (37,52). CircRNAs include a quantity of
miRNA response elements that decrease the expression abundance of
miRNA by competitive binding to miRNA, hindering its complementary
pairing with the 3′-UTR of the downstream target mRNA, thereby
alleviating the suppressive effect of miRNAs on mRNA and
subsequently enhancing mRNA expression (53,54). However, the majority of circRNAs
lack a substantial number of miRNA binding sites and their
expression levels are inferior to those of the corresponding miRNAs
(55). The first identified gene
cerebellar degeneration-related protein 1 antisense RNA (CDR1as) is
demonstrated to be responsible for the production of circRNA
molecules. Circ-CDR1as possesses >70 specific target sites for
miRNAs that effectively inhibit their functions, suggesting that
circRNAs can serve as sponges for miRNAs (37).
Apart from miRNA sponges, circRNAs can also interact
with RBPs and other regulatory proteins, regulating various
cellular processes via affecting protein localization, stability
and function (56,57). There are three prevalent manners
of interaction. Primary among these is the role of circRNAs as
sponges of proteins. Of note, it is reported that the introns
encompassing circular forms of muscleblind (circ-MBL) are rich in
MBL binding sites, which possess a specific affinity for MBL
proteins, consequently leading to a reduction in the intracellular
MBL protein levels (50).
Another mode involves circRNAs acting as protein reservoirs. For
instance, the abnormal presence of circ-forkhead box (FOX)O3 was
verified to prompt cardiac fibroblast aging through interacting
with various proteins linked to cellular stress responses, such as
DNA binding inhibitor 1, E2F transcription factor 1, hypoxia
inducible factor 1 and focal adhesion kinase, thereby sequestering
these proteins from the cytoplasm (58). Furthermore, circRNAs can serve as
scaffolds for proteins. Zeng et al (59) revealed that a circRNA derived
from angiomotin-like 1 associates with 3-phosphoinositol-dependent
protein kinase 1 (PDK1) and AKT1 to facilitate the phosphorylation
of PDK1-dependent AKT1, conferring protection to the heart. In
addition, circRNAs have the ability to attract proteins to specific
sites, which allows circRNAs to act as scaffolds or guides,
facilitating the localization of proteins to distinct regions of
the genome. For instance, circ-FECR1 recruits ten-eleven
translocation (TET)1 protein to the promoter region of its host
gene Friend leukemia integration 1, triggering the demethylation of
CpG sites and promoting active transcription (60).
Although the majority of circRNAs predominantly
localize in the cytoplasm where they are employed as protein
scaffolds or miRNA sponges, a minority of EIciRNAs are situated in
the nucleus and play crucial roles in transcriptional processes.
For instance, circRNA eukaryotic translation initiation factor 3
subunit J and circRNA poly(A)-binding protein-interacting protein 2
are confirmed to activate the promoter and modulate the
transcription of their host genes in a cis-regulatory manner
through binding to U1 small ribonucleoprotein and RNA polymerase II
transcription complexes (27).
However, the underlying mechanism of EIciRNAs regulating gene
transcription has remained largely elusive. In addition, EIciRNAs
can affect gene expression by regulating the splicing of their
linear counterparts. Furthermore, circRNAs that encompass the start
codon of the host gene function as 'mRNA traps' to hinder the
expression of the parental gene. By the circularization of an exon
that contains an ATG translation start site,
circ-homeodomain-interacting protein kinase (HIPK)2 and circ-HIPK3
are generated to repress the synthesis of the HIPK2 and HIPK3
proteins (61).
Due to the absence of essential components for
cap-dependent translation, specifically the 5′ cap and poly(A)
tail, it is widely accepted that circRNAs are incapable of being
translated into proteins. However, recent research has revealed
that circRNAs containing internal ribosome entry site fragments
(IRES) and adenine-uracil-guanine codon sites may serve as
templates for protein translation under particular conditions
(62). For instance, circ-F-box
and WD-repeat domain-containing 7 (FBXW7) has a region called the
open reading frame that starts the translation process with the
help of IRES, regardless of the 5′-cap structure, elevating the
expression of FBXW7 (63).
Additional mechanisms triggering circRNA translation include
m6A-methylation initiation of translation and rolling circle
adaptation (64,65). It is worth noting that while
circRNAs possess a certain translational capacity, their
translation efficiency is limited by the unique circular structure
of circRNAs. The roles of translation products, such as proteins
and peptides, generated from circRNAs remain elusive, necessitating
additional investigation.
In conclusion, circRNAs represent a fascinating and
rapidly evolving field of study with broad implications for
understanding gene regulation and disease mechanisms. Their unique
properties and diverse functions underscore their potential as
biomarkers and therapeutic targets. Continued research into
circRNAs will undoubtedly uncover more about their roles in various
diseases such as CHD, paving the way for innovative diagnostic and
therapeutic approaches.
Roles of circRNAs in CHD
CircRNAs have emerged as crucial players in the
pathogenesis of CHD. Research has identified numerous circRNAs that
are differentially expressed in patients with CHD compared to
healthy controls, suggesting their crucial role in disease
progression. For instance, hsa_circRNA11783-2 and hsa_circ_0000563
have been found to influence the initiation and progression of CHD,
with hsa_circ_0000563 affecting the pathological process via
targeting hub genes like ribosomal protein subunit 3 and subunit 1
(66,67). Hsa_circ_0000563 has been verified
to participate in the atherosclerotic changes in human coronary
artery segments with severe atherosclerotic stenosis (68). Furthermore, the combination of
hsa_circ_0001879 and hsa_circ_0004104 can discriminate patients
with CHD from healthy controls; in addition, upregulation of
hsa_circ_0004104 causes dysregulation of atherosclerosis-related
genes in macrophages, indicating that hsa_circ_0004104 is involved
in inflammatory response during atherosclerosis (69). Similarly, it has been
demonstrated that the combination of smoking and high
hsa_circ_0008507 expression results in the occurrence and
development of CHD (70). In
addition, the exosome-derived circRNA hsa_circ_0005540 has been
identified as a potential diagnostic biomarker for CHD, with its
expression levels being associated with disease severity and
progression (71). The ceRNA
networks involving circRNA-miRNA have been shown to regulate key
cellular and molecular biological processes, which are implicated
in CHD pathophysiology (72,73). Indeed, hsa_circ_0066439,
hsa_circ_0081241 and hsa_circ_0122984 were found to regulate
multiple signaling pathways to participate in the acute myocardial
infarction through hsa-miR-1254 and hsa-miR-328-5p (74). Furthermore, 9 circRNAs
(hsa_circ_0089378, hsa_circ_0083357, hsa_circ_0082824,
hsa_circ_0068942, hsa_circ_0057576, hsa_circ_0054537,
hsa_circ_0051172, hsa_circ_0032970 and hsa_circ_0006323) were
identified to promote the expression of transient receptor
potential melastatin-3, a calcium-permeable ion channel, by
inhibiting hsa-miR-130a-3p in patients with CHD (75). However, these circRNAs were
investigated by observational studies or bioinformatics, while
molecular mechanistic evaluations warrant further investigation.
Overall, the growing body of evidence highlights the multifaceted
roles of circRNAs in CHD, from their involvement in cell
proliferation and apoptosis to inflammation, providing new avenues
for the diagnosis and treatment of this disease.
Of note, circRNAs have distinct cell-type-specific
roles in cardiomyocytes, endothelial cells and vascular smooth
muscle cells (VSMCs), contributing to CHD progression. In
cardiomyocytes, circRNAs are involved in regulating cell apoptosis
and hypertrophic responses, both of which are critical in the
development of CHD (76).
Furthermore, circRNAs in endothelial cells are key players in
angiogenesis regulation, endothelial dysfunction and endothelial
barrier inflammation (77). In
addition to influencing the proliferation and migration of VSMCs,
circRNAs regulate their phenotypic switch between contractile and
synthetic phenotypes in response to vascular injury, affecting
plaque formation and vascular stiffening (78).
Cardiomyocyte death
Cardiomyocyte death has a pivotal role in the
progression of CHD by initiating and exacerbating a cascade of
pathological events that compromise cardiac function. Various forms
of cell death, including apoptosis, autophagy, necroptosis,
pyroptosis and ferroptosis, are implicated in the deterioration of
cardiomyocytes during CHD (79,80). The loss of cardiomyocytes weakens
the contractile power of the heart, leading to impaired cardiac
output and heart failure (81).
This cell death is often triggered by ischemic events such as
myocardial infarction, where the lack of oxygen and nutrients
causes extensive cardiomyocyte damage and death (82). The failing cardiomyocyte is
characterized by a complex interplay of abnormal signaling
pathways, oxidative stress, impaired mitochondrial function and
altered gene expression, all of which contribute to the vicious
cycle of cardiac dysfunction and cell death (83). The irreversible loss of
cardiomyocytes leads to fibrosis and scar formation, which
exacerbate heart failure and limit the heart's ability to recover
from injury (84). Thus,
understanding the mechanisms of cardiomyocyte death is essential
for developing targeted therapies to protect cardiomyocytes, reduce
heart damage and improve clinical outcomes in CHD.
CircRNAs are involved in cardiomyocyte death through
various mechanisms, primarily by acting as miRNA sponges, thereby
influencing gene expression and cellular processes. For instance,
circ-HIPK2 has been shown to interact with miR-485-5p to upregulate
the expression of autophagy-related protein 101, which facilitates
autophagy to accelerate cardiomyocyte apoptosis in
H2O2-induced myocardial oxidative injury
(8). Similarly, circ_0030235 is
upregulated in oxygen-glucose deprivation/reoxygenation
(OGD/R)-induced H9c2 cells, a widely used rat cardiomyoblast cell
line that can be induced to exhibit certain properties of cardiac
muscle cells, and aggravates mitochondrial dysfunction and
oxidative damage in cardiomyocytes by targeting miR-526b and thus
inactivating the PI3K/AKT and MEK/ERK pathways (85). These findings indicate that
circRNAs exert a promoting effect on cardiomyocyte death and
disease progression. In addition, overexpression of
circRNA-differentially expressed in normal cells and neoplasia
domain containing 4 C (DENND4C) has been observed in
OGD/R-stimulated H9c2 cells, and further mechanistic evaluation
revealed that circ-DENND4C attenuates OGD/R-induced cardiomyocyte
death by downregulation of miR-320 through activating the ERK and
mTOR pathways (86). Consistent
with this result, circ-LRP62–2 is confirmed to protect
cardiomyocytes from hypoxia-induced apoptosis. This circRNA is
downregulated in cardiomyocytes exposed to hypoxia, while its
overexpression represses cell apoptosis. Mechanically, under
hypoxia, circ-LRP62−2 recruits heterogeneous nuclear
ribonucleoprotein M and further enhances the expression of
fibroblast growth factor 9, facilitating hypoxia-adaption and
viability of cardiomyocytes (87). Thus, circRNAs also exhibit
protective effects on cardiomyocytes and delay disease progression.
Overall, the multifaceted roles of circRNAs in cardiomyocyte death,
through their interactions with miRNAs and other molecular targets,
provide a comprehensive understanding of their contribution to
cardiovascular pathophysiology and potential clinical
applications.
Therefore, circRNAs have been implicated in
regulating cardiomyocyte death by affecting processes such as cell
proliferation and autophagy, further impacting the progression of
CHD (Table I). Despite advances
in therapeutic approaches, the limited regenerative capacity of
adult cardiomyocytes poses a significant challenge in restoring
heart function after extensive cell loss (88). In this regard, targeting specific
circRNAs may improve cardiac outcomes, highlighting the potential
to mitigate cardiomyocyte death. Understanding the molecular
mechanisms by which circRNAs participate in different forms of cell
death is essential for developing effective treatments to prevent
cardiomyocyte loss and improve the prognosis for patients with
CHD.
 | Table IRole of circRNAs in the pathogenesis
of coronary heart disease. |
Table I
Role of circRNAs in the pathogenesis
of coronary heart disease.
| CircRNA | Expression | Experimental
model | Targets | Effects | Impact on CHD | (Refs.) |
|---|
| Circ-HIPK2 | Up |
H2O2-stimulated
cardiomyocytes | miR-485-5p/
ATG101 | Facilitating cell
autophagy and apoptosis | Pro-CHD | (8) |
| Circ_0030235 | Up | OGD/R-induced H9c2
cells | miR-526b, PI3K/AKT,
MEK/ERK | Promoting cell
apoptosis and reducing cell viability | Pro-CHD | (85) |
| Circ-DENND4C | Up | OGD/R-induced H9c2
cells | miR-320, ERK,
mTOR | Promoting cell
injury | Anti-CHD | (86) |
|
Hsa_circ_0007478 | Up | Ox-LDL-stimulated
macrophages | miR-765/ EFNA3 | Increasing
inflammation and foam cell formation | Pro-CHD | (10) |
|
Hsa_circ_0000280 | Down | Patients with CHD,
PDGF-BB-induced VSMCs | ELAVL1, CDKN1A
mRNA | Inhibiting cell
proliferation and neointimal hyperplasia | Anti-CHD | (109) |
| Circ-LDLR | Down | Patients with CHD,
transgenic VSMCs |
miR-26-5p/KDM6A | Inhibiting cell
proliferation and promoting cell apoptosis | Anti-CHD | (110) |
| Circ-MAP3K5 | Down | PDGF-BB-induced
VSMCs | miR-22-3p/
TET2 | Inhibiting cell
proliferation and neointima formation | Anti-CHD | (111) |
| Circ-SATB2 | Up | Transgenic
VSMCs | miR-939/ STIM1 | Inhibiting cell
proliferation and migration | Pro-CHD | (112) |
|
Hsa_circ_0031891 | Up | Patients with CAD,
PDGF-BB-induced VSMCs |
miR-579-3p/HMGB1 | Boosting cell
proliferation, migration and dedifferentiation | Pro-CHD | (113) |
| Circ_0006251 | Up | PDGF-BB-induced
VSMCs | miR-361-3p, TET3,
PPM1B | Increasing cell
proliferation | Pro-CHD | (114) |
|
Hsa_circ_0030042 | Down | Ox-LDL-stimulated
HUVECs, high-fat-diet fed ApoE−/− mice | eIF4A3, FOXO1,
Beclin-1 | Decreasing
autophagic cell death and maintaining plaque stability | Anti-CHD | (100) |
| Circ-CHFR | Up | Ox-LDL-stimulated
HUVECs |
miR-15b-5p/GADD45G | Promoting cell
apoptosis and inflammatory response | Pro-CHD | (121) |
| Circ_0004104 | Up | Patients with CAD,
ox-LDL-stimulated endothelial cells | miR-100/
TNFAIP8 | Promoting cell
apoptosis and inflammation | Pro-CHD | (122) |
|
Hsa_circ_0000284 | Down | Patients with CHD,
TNF-α and H2O2-induced EA-hy926 cells |
miR-338-3p/ETS1 | Affecting cell
proliferation and apoptosis | Pro-CHD | (9) |
|
Circ-LRP62–2 | Down | Hypoxia-treated
cardiomyocyte | hnRNPM/ FGF-9 | Inhibiting cell
apoptosis | Anti-CHD | (87) |
| Circ_0001785 | Down | Patients with CHD,
high-fat-diet fed ApoE−/− mice |
miR-513a-5p/TGFBR3 | Reducing
endothelial cell injury and delaying atherosclerosis | Anti-CHD | (99) |
| Circ-MBOAT2 | Up | Patients with CTO,
mice with hindlimb ischemia | miR-495/
NOTCH1 | Promoting
angiogenesis and improving myocardial perfusion | Anti-CHD | (101) |
|
Hsa_circ_0126672 | Up | CHD patients | miR-145-5p, NOS1,
RPS6KB1 | Affecting
atherosclerosis | Pro-CHD | (123) |
|
Hsa_circ_0092576 | Up | CHD patients | miR-145-5p, Apelin,
JAK/STAT | Enhancing cell
proliferation, inflammation and atherosclerosis | Pro-CHD | (124) |
Endothelial cell injury
Endothelial cell injury is a critical factor in the
development of CHD due to its multifaceted impacts on vascular
function and integrity. The endothelium, which lines the inner
walls of blood vessels, has a vital role in maintaining vascular
tone, regulating hemostasis and controlling inflammation and
thrombosis (89). When
endothelial cells are damaged, several pathological processes are
triggered. Firstly, endothelial injury enhances the permeability of
the vascular intima, facilitating leukocyte adhesion and
transmigration, which further exacerbates inflammation and promotes
thrombus formation (90).
Increased inflammatory mediators, such as cytokines and oxidized
lipoproteins, lead to the overexpression of adhesion molecules,
selectins and chemokines, which attract and retain leukocytes in
the subendothelial space (91).
This inflammatory cascade is compounded by oxidative stress, which
not only damages endothelial cells directly but also disrupts the
balance of pro- and anti-coagulant factors, shifting the
endothelium towards a pro-thrombotic state (92,93). Furthermore, endothelial cell
injury is linked to metabolic disturbances such as lipid metabolism
disorders, which are significant risk factors for atherosclerosis.
The damage to endothelial cells impairs their ability to regulate
lipid levels, leading to the accumulation of lipids in the arterial
walls and the formation of atherosclerotic plaques (94). Mechanical, chemical and
biological stresses, including hypertension, hyperglycemia and
infections, exacerbate endothelial dysfunction by inducing cell
senescence, autophagy dysregulation and mitochondrial stress
(95,96). Autophagy, a process crucial for
cellular homeostasis, is often impaired in endothelial cells during
aging and disease, further contributing to endothelial dysfunction
(97). The persistent
endothelial activation and injury, coupled with diminished repair
capacity, create a vicious cycle that perpetuates vascular
inflammation, thrombosis and atherosclerosis, ultimately leading to
the development and progression of CHD (98).
CircRNAs participate in regulating endothelial cell
function in CHD through complex molecular mechanisms involving the
sponging of miRNAs and subsequent modulation of mRNA targets. For
instance, hsa_circ_0000284 is downregulated in patients with CHD
and oxidative stress-induced EA-hy926 endothelial cells, and
overexpression of hsa_circ_0000284 leads to impaired cell
proliferation and increased apoptosis by sponging miR-338-3p,
thereby repressing the expression of E26 transformation-specific
sequence-1, a transcription factor involved in endothelial cell
function (9). Another circRNA,
circ_0001785, is decreased in the circulating peripheral blood of
patients with CHD but increased within atherosclerotic plaque
tissue. This circRNA is implicated in delaying atherogenesis by
alleviating aortic endothelial cell injury and the formation of
intraplaque neovascularization, via modulating the
miR-513a-5p/TGFBR3 axis (99).
In addition, hsa_circ_0030042 has been shown to suppress oxidized
low-density lipoprotein (ox-LDL)-mediated abnormal autophagy of
endothelial cells and maintain plaque stability in high-fat-diet
fed apolipoprotein E−/− mice by sponging the endogenous
eukaryotic initiation factor 4A-III, which impedes its recruitment
to beclin1 and FOXO1 mRNA (100). Furthermore, circRNA-0024103,
which is highly expressed in patients with coronary chronic total
occlusion, has been verified to accelerate tube formation and cell
migration via the miR-495/Notch1 axis in endothelial cells. This
circRNA also increased collateral formation of the ligated femoral
artery in mice after hindlimb ischemia, which is related to
myocardial perfusion improvement after revascularization (101).
Collectively, these studies highlight the
multifaceted roles of circRNAs in regulating endothelial cell
function through intricate ceRNA networks, influencing key
processes such as angiogenesis, cell proliferation and apoptosis,
thereby offering novel insights into the pathogenesis and potential
therapeutic strategies for CHD (Table I). Further studies are needed to
unravel the detailed mechanisms by which circRNAs regulate
endothelial cell function and contribute to CHD. Understanding
these mechanisms will provide insights into potential therapeutic
targets.
VSMC apoptosis
VSMC apoptosis is involved in the development and
progression of CHD by affecting vascular remodeling, plaque
stability and inflammatory responses (102). The balance between VSMC
proliferation and apoptosis is crucial in the pathogenesis of
atherosclerosis, a primary underlying cause of CHD (103). VSMCs with a synthetic phenotype
are characterized by increased migration and proliferation to
repair the damage (104). This
phenotypic switching is regulated by a network of factors,
including transcription factors, growth factors and non-coding
RNAs, which contribute to vascular aging and atherosclerosis
(104). Early atherosclerotic
lesions are characterized by intense apoptosis of VSMCs, which
decreases as the disease progresses, leading to intimal hyperplasia
and plaque formation (105).
Clonal expansion of VSMCs, initiated by a small fraction of cells
that proliferate and migrate to form oligoclonal neointima, is a
hallmark of early atherosclerotic lesions, suggesting that
selective VSMC activation drives disease progression (106). Furthermore, VSMCs can
transdifferentiate into mesenchymal and myeloid-like phenotypes,
contributing to the progression of organ fibrosis and potentially
affecting other vital organs beyond the vascular system (107). The dynamic variations in VSMC
phenotypes shape the atherosclerotic plaque microenvironment,
leading to heterogeneous clinical outcomes in CHD (108). Overall, the injury-induced
phenotypic switching and clonal expansion of VSMCs, along with the
disequilibrium between cell proliferation and apoptosis, underscore
the critical role of VSMCs in the pathogenesis of CHD, providing
potential targets for therapeutic intervention.
Dysregulated circRNAs have been implicated in the
phenotypes and function of VSMCs during CHD progression. For
instance, hsa_circ_0000280 has been shown to inhibit VSMC
proliferation and induce cell-cycle arrest by facilitating the
interaction between human antigen R and cyclin-dependent kinase
suppressor 1 mRNA, leading to cell cycle arrest at the G1/S
checkpoint and reducing neointimal hyperplasia in vivo
(109). Another circRNA,
circ-LDLR, is downregulated in CHD tissues, and its upregulation
represses proliferation and promotes apoptosis of VSMCs through the
miR-26-5p/KDM6A axis (110).
Furthermore, circ-MAP3K5 has been identified to exert
antiproliferative effects on VSMCs, where circ-MAP3K5 acts as a
master negative regulator of TET2-mediated VSMC differentiation by
sequestering miR-22-3p and thus blocking the expression of TET2
(111). Thus, these findings
indicate that circRNAs postpone CHD development via mitigating VSMC
proliferation and differentiation. Furthermore, circ-SATB2 has been
reported to promote the phenotypic switch of VSMCs from a
contractile to a synthetic state through the miR-939/stromal
interacting molecule 1 axis, which regulates VSMC phenotypic
differentiation, proliferation, apoptosis and migration (112). In addition, hsa_circ_0031891
participates in atherosclerosis by promoting VSMC proliferation,
migration and dedifferentiation through the miR-579-3p/high
mobility group box 1 axis. Silencing hsa_circ_0031891 inhibits
these processes, suggesting its role in the pathogenesis of
atherosclerosis (113).
Likewise, in platelet-derived growth factor subunit B-induced
VSMCs, circ_0006251 is upregulated to facilitate VSMC proliferation
and reduce their apoptosis by enhancing TET3 and protein
phosphatase 1B expression through sponging miR-361-3p, thereby
contributing to disease occurrence (114). Hence, circRNAs boost CHD
progression by facilitating VSMC proliferation and
differentiation.
Taken together, circRNAs are crucial regulators of
VSMC phenotype and function in CHD, regulating cell proliferation,
migration and phenotypic switching through complex molecular
pathways, making them promising targets for future therapeutic
strategies (Table I).
Inflammatory response
The inflammatory response is a complex biological
process initiated by the immune system to protect the body against
harmful stimuli such as pathogens, damaged cells or irritants
(115). It involves the
activation and recruitment of various immune cells, including
macrophages, neutrophils and T cells, which release cytokines and
other inflammatory mediators to eliminate the offending agents and
promote tissue repair (116).
However, chronic inflammation can have detrimental effects,
particularly in the context of CHD. Inflammation has a pivotal role
in the development and progression of CHD by contributing to
endothelial dysfunction, atherosclerosis and plaque instability
(117). Macrophages ingest
ox-LDL, transforming into foam cells and forming the lipid-rich
necrotic core of atheromas, while the secretion of cytokines and
proteases by these cells further exacerbates the inflammatory
process and weakens the fibrous cap, making plaques more prone to
rupture (118,119). The macrophage-mediated innate
immune response is crucial in both the initiation and resolution of
inflammation following myocardial infarction, influencing cardiac
remodeling and heart failure development (120). Consequently, the inflammatory
response is a double-edged sword in CHD, being essential for
initial defense and tissue repair but potentially harmful when it
becomes chronic.
CircRNAs have a vital role in the inflammatory
response associated with the development of CHD. For instance,
hsa_circ_0007478 has been found to be upregulated in
ox-LDL-stimulated macrophages, and its expression exacerbates lipid
metabolism imbalance and foam cell formation through the
miR-765/EFNA3 axis, along with interleukin (IL)-1β production and
NLR family pyrin domain containing 3 inflammasome activation, a key
player in cell pyroptosis and inflammation (10). Besides, in ox-LDL-stimulated
endothelial cells and patients with CHD, circ-CHFR is upregulated
to provoke atherosclerosis development by sponging miR-15b-5p,
which in turn enhances the expression of GADD45G, thus triggering
the secretion of atherosclerosis-associated cytokines, including
IL-1β, IL-6 and tumor necrosis factor (TNF)-α (121). Consistently, silencing of
circ_0004104, which is upregulated in patients with CHD, mitigates
ox-LDL-mediated inflammatory injury in endothelial cells by the
miR-100/TNFAIP8 axis, highlighting the deleterious effect of
circ_0004104 in CHD pathogenesis (122). Therefore, circRNAs can promote
endothelial cell injury and atherosclerosis progression by
enhancing inflammatory responses. Moreover, the construction of
circRNA-related ceRNA networks has revealed that hsa_circ_0126672
is involved in regulating inflammation-related pathways, such as
the JAK/ STAT and Apelin signaling pathways, which are critical for
atherosclerotic plaque progression and instability (123). By activating these signaling
pathways, hsa_circ_0092576 induces vascular inflammation via the
activation and proliferation of VSMCs, as well as mediates the
accumulation of oxidized lipids and oxidative damage to endothelial
cells, resulting in the development of atherosclerosis (124).
Therefore, circRNAs mediate the inflammatory
response in CHD by regulating miRNA and mRNA interactions,
influencing cell proliferation, oxidative stress and
atherosclerosis, thereby contributing to disease development and
progression (Table I). The
circRNA-miRNA-mRNA regulatory network is crucial in controlling the
inflammatory processes that contribute to atherosclerotic plaque
formation and instability, highlighting the therapeutic potential
of targeting these pathways. Hence, the complex interplay between
circRNAs, miRNAs and mRNAs requires further elucidation to
understand the precise mechanisms of action during CHD progression.
Furthermore, the influence of environmental factors, such as
smoking, on circRNA expression and their interaction with
inflammatory response must be considered to develop personalized
treatment strategies.
Applications of circRNAs for CHD
CircRNAs as biomarkers
CircRNAs have emerged as promising biomarkers for
the diagnosis and treatment of cardiovascular disease due to their
unique properties and regulatory roles in disease occurrence and
development (20). Compared with
linear RNAs, such as miRNAs and long non-coding RNAs, they are
covalently closed-loop structures without free 5′ and 3′ ends,
which makes them highly resistant to exonucleases and more stable
in bodily fluids, enhancing their reliability as biomarkers
(125). CircRNAs are abundant
and exhibit tissue-specific expression, making them ideal
candidates for clinical applications (126). CircRNAs have been detected in
considerable quantities in various bodily fluids, such as plasma,
serum and saliva (71,127-129). Furthermore, they exhibit a
half-life of 48 h in bodily fluids, a duration that surpasses that
of linear RNA (130). In
addition, circRNAs exhibit a broad distribution within cells, as
well as in extracellular regions, and the extracellular amount of
circRNAs is thought to reflect, at least in part, their
intracellular abundance. When cells undergo stress or pathological
conditions, transcriptional upregulation of intracellular circRNAs
may lead to an increased release of circRNAs into the extracellular
space. It is hypothesized that the expression levels of
intracellular circRNAs could impact the concentrations of
extracellular circRNAs, but the exact relationship between these
two pools of circRNAs remains to be fully elucidated (131). These distinctive attributes of
circRNA substantiate their potential suitability as biomarkers.
CircRNAs have emerged as promising biomarkers for
the early detection and prognosis of CHD due to their unique
properties and regulatory roles in gene expression. Numerous
studies have highlighted their diagnostic value, with varying
degrees of efficacy. A meta-analysis of 16 studies involving 3,962
subjects revealed that circRNAs have a pooled receiver operating
characteristic curve of 0.80, with sensitivity and specificity
values of 0.77 and 0.68, respectively, indicating their potential
as reliable biomarkers for CHD diagnosis (132). Besides, several dysregulated
circRNAs, such as circ-HECTD1, circ-ZBTB46 and hsa_circ_0001445,
have been detected in the peripheral blood of patients with CHD and
their expression level was shown to be associated with laboratory
parameters, such as hemoglobin, triglycerides and cholesterol
levels, suggesting their potential as diagnostic markers (73,133). High-throughput sequencing has
confirmed differentially expressed circRNAs in the plasma of
patients with CHD, such as hsa_circ_0069972, hsa_circ_0021509,
circ-RPRD1A and circ-HERPUD2, which show significant upregulation
and serve as new biomarkers for the diagnosis of coronary artery
disease (134,135). Specifically, exosome-derived
circRNAs have been identified as potential biomarkers for CHD, with
certain circRNAs such as hsa_circ_0001445, hsa_circ_0001360 and
hsa_ circ_0000038 showing significant downregulation in patients
with CHD (127,136). Furthermore, hsa_circ_0124644,
hsa_circ_0001946, hsa_circ_0001785, hsa_circ_0000973,
hsa_circ_0001741 and hsa_circ_0003922 in peripheral blood have
shown their ability to discriminate between patients with CHD and
healthy controls, with promising potential as diagnostic biomarkers
for CHD (137-139). In addition, bioinformatics
analyses have constructed circRNA-miRNA-mRNA regulatory networks,
revealing the involvement of circRNAs such as circ-YOD1 in lipid
metabolism and protein modification, further supporting their
diagnostic and prognostic potential (140,141). Microarray data analysis has
identified differentially expressed circRNAs in patients with CHD
with varying severity, with hsa_circ_0016868, hsa_circ_0001364,
hsa_circ_0006731 and circ-ANRIL emerging as promising biomarkers
for early CHD diagnosis (11,12,142).
Collectively, these findings suggest that circRNAs
hold substantial promise as biomarkers for CHD, offering a new
avenue for the early diagnosis, prognosis and personalized
treatment of CHD, thereby addressing unmet clinical needs such as
timely diagnosis and effective monitoring of treatment responses.
However, circRNA-based biomarkers are still undergoing experimental
evaluation in CHD. Further integration of circRNA profiling with
advanced bioinformatics, high-throughput sequencing and molecular
biology techniques will hold great promise for the identification
and validation of differentially expressed circRNAs in patients
with CHD, enabling the construction of circRNA-miRNA-mRNA
regulatory networks that provide insight into disease mechanisms
and potential therapeutic targets. However, circRNAs can be present
at low concentrations and distinguishing them from linear RNA
counterparts remains a technical challenge. Thus, developing more
sensitive, reliable and high-throughput detection techniques is
essential. Besides, circRNAs are highly heterogeneous and their
expression can vary widely across different cell types and
conditions. In this regard, identifying a panel of circRNAs rather
than relying on a single circRNA marker may enhance the diagnostic
power.
CircRNA-targeting therapeutic
strategies
Current strategies for targeting circRNAs in the
treatment of CHD are multifaceted, leveraging their unique
properties and regulatory roles in cardiovascular diseases. By
designing synthetic circRNAs or using antisense oligonucleotides to
inhibit specific circRNAs, researchers aim to modulate these
regulatory networks to achieve therapeutic effects. For instance,
targeting proatherogenic circRNAs such as circ_0002984 and
circ_0029589 could potentially mitigate atherosclerosis and its
associated risks in patients with CHD (143). However, solutions to increasing
the stability of the antisense oligonucleotide and its efficiency
should be further developed. In addition, gene therapy techniques
are being explored to either upregulate protective circRNAs or
downregulate harmful ones, thereby restoring the balance of
circRNA-miRNA-mRNA interactions crucial for cardiovascular health
(144). Physical exercise has
also been suggested as a non-pharmacological strategy to modulate
circRNA expression, offering a low-cost and accessible means to
alleviate CHD symptoms (145).
Furthermore, the development of circRNA synthesis and engineering
delivery systems holds promise for their application in
therapeutics. These systems aim to enhance the stability,
specificity and delivery efficiency of circRNA-based treatments,
potentially overcoming current challenges in circRNA therapy
(146). Thus, the integration
of circRNAs into the therapeutic landscape of CHD represents a
promising frontier, with ongoing research aimed at optimizing these
strategies for clinical application.
CircRNAs can influence patient responses to
conventional CHD treatments by modulating the molecular pathways
involved in disease mechanisms and drug efficacy. Dysregulation of
circRNAs has been linked to cholesterol homeostasis, which alters
the effectiveness of statins, leading to variable lipid-lowering
responses (147). Antiplatelet
agents like clopidogrel and ticagrelor are used to prevent
thrombosis in patients with CHD. CircRNAs can regulate platelet
activation and aggregation by controlling the expression of
proteins involved in this process (148). Upregulated circRNAs, such as
hsa_circ_0070675_CBC1, hsa-circ_13011-5_CBC1 and
hsa-circ_6406-3_CBC1, are related to clopidogrel resistance in
patients with CHD (149).
Platelet-derived circFAM13B has been implicated in platelet
aggregation processes, which affect the antiplatelet action of
ticagrelor patients with CHD (150). Hence, circRNAs represent a key
regulatory layer that can influence patient responses to
conventional CHD therapies, making them promising targets for
personalized medicine strategies in this disease.
Despite the promising potential of circRNAs as
therapeutic targets, no clinical application or ongoing clinical
trials has investigated circRNA-targeting therapeutic strategies in
CHD, which face several challenges. One significant challenge is
the accurate and sensitive detection of circRNAs, which is crucial
for understanding their biological processes. Current methodologies
for circRNA identification, including purification and sequencing
methods, have limitations in sensitivity and specificity, which can
hinder the precise characterization of circRNAs involved in CHD. In
addition, the regulatory roles of circRNAs in cardiovascular
diseases are complex, involving interactions with miRNAs and mRNAs
in regulatory networks that are not yet fully understood. This
complexity necessitates advanced technologies for identifying,
validating and analyzing circRNAs, which are still under
development. Furthermore, the involvement of circRNAs in various
pathophysiological processes, such as atherogenesis and myocardial
infarction, adds another layer of complexity. Specific circRNAs
have been identified with either atheroprotective or proatherogenic
effects, but translating these findings into therapeutic
applications requires a deeper understanding of their mechanisms
and interactions within the circRNA-miRNA-mRNA regulatory axis.
Thus, the continued exploration of circRNA functions and their
interactions within the cardiovascular system will likely yield
novel insights and more effective treatments for CHD.
Conclusions and future perspectives
CircRNAs represent a promising frontier in
cardiovascular research, offering novel insights into the molecular
mechanisms of CHD. They play crucial roles in various cellular
processes pertinent to CHD, ranging from endothelial dysfunction to
inflammation, by acting as miRNA sponges and further regulating
gene expression and signaling pathways (Fig. 2). They exhibit distinct
expression profiles in patients with CHD compared to healthy
controls, positioning them as valuable biomarkers and therapeutic
targets. However, translating circRNA research into clinical
practice requires concerted efforts to overcome current challenges.
First, advancements in high-throughput sequencing technologies and
bioinformatics tools are essential for accurate circRNA detection
and quantification. Standardized methods for circRNA isolation,
sequencing and analysis will improve reproducibility and facilitate
cross-study comparisons. Furthermore, extensive in vivo
studies are required to elucidate the precise mechanisms by which
circRNAs influence CHD pathophysiology. As numerous circRNAs serve
as miRNA sponges or regulate transcription and splicing, uncovering
their exact molecular functions in different cell types within the
cardiovascular system is critical for therapeutic targeting.
Integrating single-cell sequencing approaches could unravel
cell-type-specific roles of circRNAs, offering a more precise
understanding of their contributions to disease. In addition,
large-scale clinical studies are needed to validate the diagnostic
and prognostic utility of circRNAs identified in preliminary
research. Integrating circRNA biomarkers into existing diagnostic
frameworks could enhance early detection and risk stratification in
patients with CHD. CircRNAs could be used to monitor treatment
responses, particularly in therapies that target molecular pathways
involved in lipid metabolism, vascular remodeling and inflammation.
Furthermore, developing circRNA-based therapies involves designing
molecules that can specifically modulate circRNA activity.
Antisense oligonucleotides and small interfering RNAs targeting
circRNAs hold promise but require optimization for efficient
delivery and minimal off-target effects. Developing safe and
effective delivery systems, such as lipid nanoparticles or viral
vectors, will be crucial for translating circRNA-targeting
therapies into clinical practice. Furthermore, addressing the
potential immunogenicity of these delivery platforms is essential
to ensure safety and minimize adverse reactions. Exploring the
therapeutic potential of circRNAs in combination with existing
treatments could offer synergistic benefits, improving patient
outcomes in CHD.
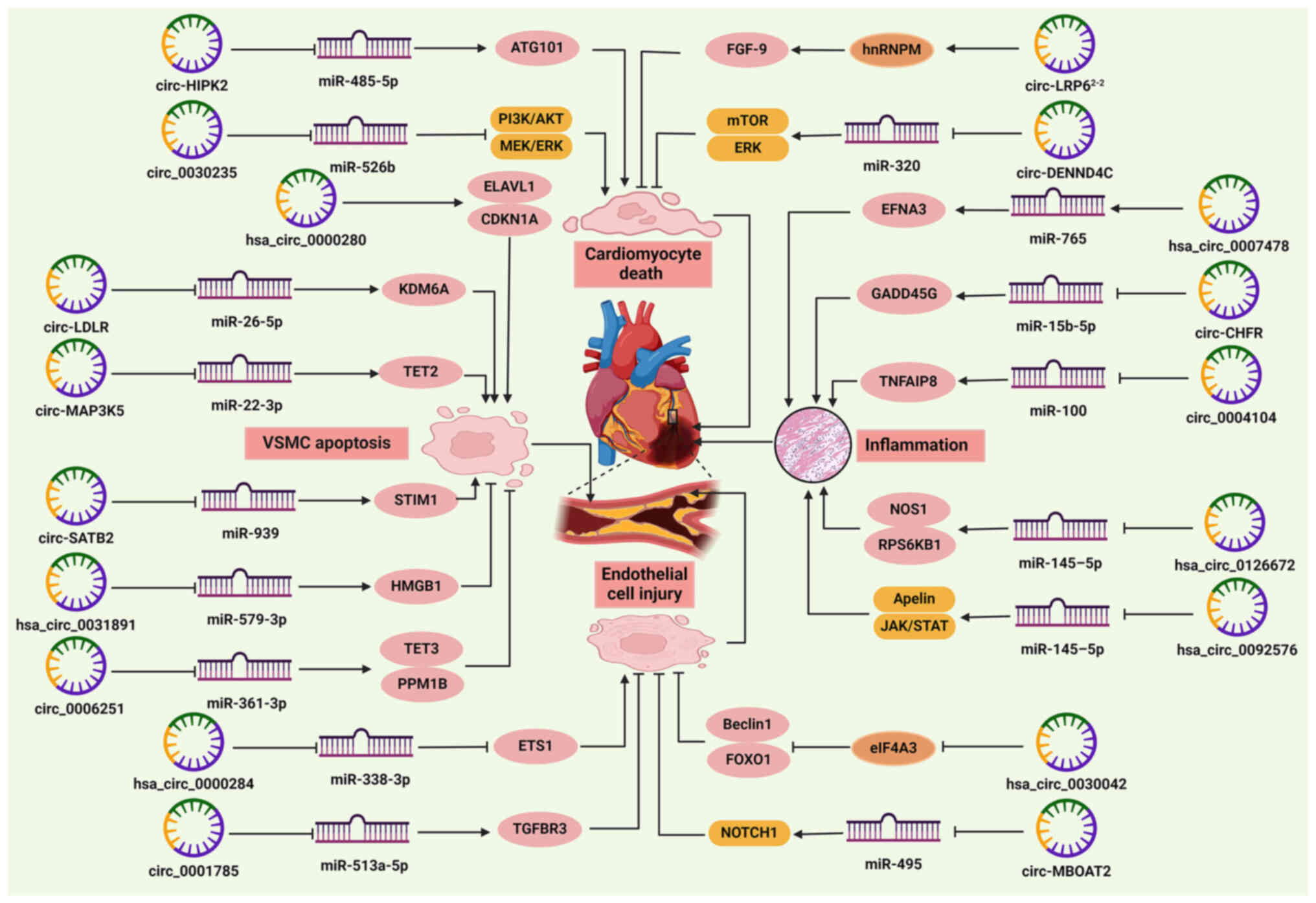 | Figure 2Role of circRNAs in the pathogenesis
of CHD. CircRNAs regulate cardiomyocyte death, endothelial cell
injury, VSMC apoptosis and cardiac inflammation by sponging miRNAs
or binding RNA-binding proteins. Circ-HIPK2 and circ_0030235
promote cardiomyocyte death, while circ-LRP62−2 and
circ-DENND4C prevent cardiomyocyte death. Hsa_circ_0000280,
circ-LDLR, circ-MAP3K5 and circ-SATB2 facilitate, but
hsa_circ_0031891 and circ_0006251 inhibit VSMC apoptosis.
Hsa_circ_0000284 promotes endothelial cell injury, whereas
circ_0001785, circ-MBOAT2, and hsa_circ_0030042 exert the opposite
effects. Hsa_circ_0007478, circ-CHFR, circ_0004104,
hsa_circ_0126672 and hsa_circ_0092576 mediate cardiac inflammation.
→ indicates a promoting effect and ⊥ indicates an inhibitory
effect. ATG, autophagy-related protein; CAD, coronary artery
disease; CDKN1A, cyclin-dependent kinase suppressor 1; CHD,
coronary heart disease; circRNA/circ, circular RNA; EFNA3,
ephrinA3; ELAVL1, human antigen R; ETS1, E26 oncogene homolog 1;
FGF9, fibroblast growth factor 9; FOXO1, forkhead box O1; GADD45G,
growth arrest and DNA damage-inducible gene γ; HMGB1, high mobility
group box 1; hnRNPM, heterogeneous nuclear ribonucleoprotein M;
KDM6A, lysine-specific demethylase 6A; miRNA/miR, microRNA; PPM1B,
protein phosphatase 1B; RPS6KB1, ribosomal protein S6 kinase β1;
STIM1, stromal interacting molecule 1; TET, ten-eleven
translocation; TGFBR3, transforming growth factor β receptor 3;
TNFAIP8, tumor necrosis factor-α-induced protein 8; VSMC, vascular
smooth muscle cell. |
Availability of data and materials
Not applicable.
Authors' contributions
ZF and YY wrote the manuscript, XY revised the
manuscript and YY designed the research. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
CDKN1A
|
cyclin-dependent kinase suppressor
1
|
|
ceRNA
|
competitive endogenous RNA
|
|
CHD
|
coronary heart disease
|
|
ciRNA
|
circular intronic RNA
|
|
circRNA
|
circular RNA
|
|
EcircRNA
|
exonic circRNA
|
|
EIciRNA
|
exon-intron circRNA
|
|
ELAVL1
|
human antigen R
|
|
FBXW7
|
F-box and WD-repeat domain-containing
7
|
|
FOXO1
|
forkhead box O1
|
|
HIPK
|
homeodomain-interacting protein
kinase
|
|
HMGB1
|
high mobility group box 1
|
|
IL
|
interleukin
|
|
IRES
|
internal ribosome entry site
fragments
|
|
LDL
|
low-density lipoprotein
|
|
miRNA
|
microRNA
|
|
m6A
|
N-6 methylation
|
|
NLRP3
|
NLR family pyrin domain containing
3
|
|
OGD/R
|
oxygen-glucose
deprivation/reoxygenation
|
|
PDGF
|
platelet-derived growth factor
|
|
PDK1
|
3-phosphoinositol-dependent protein
kinase 1
|
|
RBP
|
RNA-binding protein
|
|
STIM1
|
stromal interacting molecule 1
|
|
TET
|
ten-eleven translocation
|
|
TNF
|
tumor necrosis factor
|
|
VSMC
|
vascular smooth muscle cell
|
Acknowledgments
Not applicable.
Funding
This study is supported by the Youth Fund Project of Jiangxi
Provincial Natural Science Foundation (grant no.
20224BAB216097).
References
|
1
|
Katta N, Loethen T, Lavie CJ and Alpert
MA: Obesity and coronary heart disease: Epidemiology, Pathology,
and coronary artery imaging. Curr Probl Cardiol. 46:1006552021.
View Article : Google Scholar
|
|
2
|
Pothineni NVK, Subramany S, Kuriakose K,
Shirazi LF, Romeo F, Shah PK and Mehta JL: Infections,
atherosclerosis, and coronary heart disease. Eur Heart J.
38:3195–3201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong Y, Chen H, Gao J, Liu Y, Li J and
Wang J: Molecular machinery and interplay of apoptosis and
autophagy in coronary heart disease. J Mol Cell Cardiol. 136:27–41.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hodel F, Xu ZM, Thorball CW, de La Harpe
R, Letang-Mathieu P, Brenner N, Butt J, Bender N, Waterboer T,
Marques-Vidal PM, et al: Associations of genetic and infectious
risk factors with coronary heart disease. Elife. 12:e797422023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sygitowicz G and Sitkiewicz D: Involvement
of circRNAs in the Development of Heart Failure. Int J Mol Sci.
23:141292022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma XK, Zhai SN and Yang L: Approaches and
challenges in genome-wide circular RNA identification and
quantification. Trends Genet. 39:897–907. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwang HJ and Kim YK: Molecular mechanisms
of circular RNA translation. Exp Mol Med. 56:1272–1280. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou J, Li L, Hu H, Wu J, Chen H, Feng K
and Ma L: Circ-HIPK2 accelerates cell apoptosis and autophagy in
myocardial oxidative injury by sponging miR-485-5p and Targeting
ATG101. J Cardiovasc Pharmacol. 76:427–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dinh P, Tran C, Dinh T, Ali A and Pan S:
Hsa_circRNA_0000284 acts as a ceRNA to participate in coronary
heart disease progression by sponging miRNA-338-3p via regulating
the expression of ETS1. J Biomol Struct Dyn. 42:5114–5127. 2024.
View Article : Google Scholar
|
|
10
|
Ye B, Liang X, Zhao Y, Cai X, Wang Z, Lin
S, Wang W, Shan P, Huang W and Huang Z: Hsa_circ_0007478 aggravates
NLRP3 inflammasome activation and lipid metabolism imbalance in
ox-LDL-stimulated macrophage via miR-765/EFNA3 axis. Chem Biol
Interact. 368:1101952022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou C, Gu L, Guo Y, Zhou Y, Hua L, Chen J,
He S, Zhang S, Jia Q, Zhao C, et al: Association between circular
RNA expression content and severity of coronary atherosclerosis in
human coronary artery. J Clin Lab Anal. 34:e235522020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akan G, Nyawawa E, Nyangasa B, Turkcan MK,
Mbugi E, Janabi M and Atalar F: Severity of coronary artery disease
is associated with diminished circANRIL expression: A possible
blood based transcriptional biomarker in East Africa. J Cell Mol
Med. 28:e180932024. View Article : Google Scholar :
|
|
13
|
Cao Q, Guo Z, Du S, Ling H and Song C:
Circular RNAs in the pathogenesis of atherosclerosis. Life Sci.
255:1178372020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma X, Chen X, Mo C, Li L, Nong S and Gui
C: The role of circRNAs in the regulation of myocardial
angiogenesis in coronary heart disease. Microvasc Res.
142:1043622022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CK, Cheng R, Demeter J, Chen J,
Weingarten-Gabbay S, Jiang L, Snyder MP, Weissman JS, Segal E,
Jackson PK and Chang HY: Structured elements drive extensive
circular RNA translation. Mol Cell. 81:4300–4318.e13. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Busa VF and Leung AKL: Thrown for a (stem)
loop: How RNA structure impacts circular RNA regulation and
function. Methods. 196:56–67. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang S, Fu R, Shi J, Wu H, Mai J, Hua X,
Chen H, Liu J, Lu M and Li N: CircRNA-Mediated regulation of
angiogenesis: A new chapter in cancer biology. Front Oncol.
11:5537062021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding C and Zhou Y: Insights into circular
RNAs: Biogenesis, function and their regulatory roles in
cardiovascular disease. J Cell Mol Med. 27:1299–1314. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar
|
|
23
|
Montañés-Agudo P, van der Made I, Aufiero
S, Tijsen AJ, Pinto YM and Creemers EE: Quaking regulates circular
RNA production in cardiomyocytes. J Cell Sci. 136:sc2611202023.
View Article : Google Scholar
|
|
24
|
Pamudurti NR, Patop IL, Krishnamoorthy A,
Bartok O, Maya R, Lerner N, Ashwall-Fluss R, Konakondla JVV, Beatus
T and Kadener S: circMbl functions in cis and in trans to regulate
gene expression and physiology in a tissue-specific fashion. Cell
Rep. 39:1107402022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Monat C, Quiroga C, Laroche-Johnston F and
Cousineau B: The Ll.LtrB intron from Lactococcus lactis excises as
circles in vivo: insights into the group II intron circularization
pathway. RNA. 21:1286–1293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The Output of Protein-Coding Genes
Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is
Limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
García-Lerena JA, González-Blanco G,
Saucedo-Cárdenas O and Valdés J: Promoter-Bound Full-Length
Intronic Circular RNAs-RNA Polymerase II Complexes Regulate Gene
Expression in the Human Parasite Entamoeba histolytica. Noncoding
RNA. 8:122022.PubMed/NCBI
|
|
31
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
32
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vromman M, Anckaert J, Bortoluzzi S,
Buratin A, Chen CY, Chu Q, Chuang TJ, Dehghannasiri R, Dieterich C,
Dong X, et al: Large-scale benchmarking of circRNA detection tools
reveals large differences in sensitivity but not in precision. Nat
Methods. 20:1159–1169. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haque S and Harries LW: Circular RNAs
(circRNAs) in Health and Disease. Genes (Basel). 8:3532017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Q, Sun W, Han J, Cheng S, Yu P, Shen
L, Fan M, Tong H, Zhang H, Chen J and Chen X: The circular RNA hsa_
circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac
repair. Biochem Biophys Res Commun. 523:993–1000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Szabo L, Morey R, Palpant NJ, Wang PL,
Afari N, Jiang C, Parast MM, Murry CE, Laurent LC and Salzman J:
Statistically based splicing detection reveals neural enrichment
and tissue-specific induction of circular RNA during human fetal
development. Genome Biol. 16:1262015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Guo X, Wen Y, Huang S, Yuan X, Tang
L and Sun H: N6-Methyladenosine Modification Opens a New Chapter in
Circular RNA Biology. Front Cell Dev Biol. 9:7092992021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu T, He B, Sun H, Xiong M, Nie J, Wang S
and Pan Y: Novel insights into the interaction between
N6-methyladenosine modification and circular RNA. Mol Ther Nucleic
Acids. 27:824–837. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
46
|
Tao M, Zheng M, Xu Y, Ma S, Zhang W and Ju
S: CircRNAs and their regulatory roles in cancers. Mol Med.
27:942021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Graham JR, Hendershott MC, Terragni J and
Cooper GM: mRNA degradation plays a significant role in the program
of gene expression regulated by phosphatidylinositol 3-kinase
signaling. Mol Cell Biol. 30:5295–5305. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nisar S, Bhat AA, Singh M, Karedath T,
Rizwan A, Hashem S, Bagga P, Reddy R, Jamal F, Uddin S, et al:
Insights Into the Role of CircRNAs: Biogenesis, characterization,
functional, and clinical impact in human malignancies. Front Cell
Dev Biol. 9:6172812021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang J, Yu S, Ding L, Ma L, Chen H, Zhou
H, Zou Y, Yu M, Lin J and Cui Q: The Dual Role of Circular RNAs as
miRNA Sponges in breast cancer and colon cancer. Biomedicines.
9:15902021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration
After Myocardial Infarction in Adult Mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma B, Wang S, Wu W, Shan P, Chen Y, Meng
J, Xing L, Yun J, Hao L, Wang X, et al: Mechanisms of
circRNA/lncRNA-miRNA interactions and applications in disease and
drug research. Biomed Pharmacother. 162:1146722023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Misir S, Wu N and Yang BB: Specific
expression and functions of circular RNAs. Cell Death Differ.
29:481–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Das A, Sinha T, Shyamal S and Panda AC:
Emerging Role of Circular RNA-Protein Interactions. Noncoding RNA.
7:482021.PubMed/NCBI
|
|
57
|
Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD and
Tang JH: The Emerging Role of the Interactions between Circular
RNAs and RNA-binding Proteins in Common Human Cancers. J Cancer.
12:5206–5219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar :
|
|
59
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A Circular RNA Binds To
and Activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Conte A and Pierantoni GM: Update on the
Regulation of HIPK1, HIPK2 and HIPK3 Protein Kinases by microRNAs.
Microrna. 7:178–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 Inhibits Malignant Progression by
Sponging miR-197-3p and Encoding a 185-aa Protein in
Triple-Negative Breast Cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu C, Wu X, Gokulnath P, Li G and Xiao J:
The Functions and Mechanisms of Translatable Circular RNAs. J
Pharmacol Exp Ther. 384:52–60. 2023. View Article : Google Scholar
|
|
66
|
Li X, Zhao Z, Jian D, Li W, Tang H and Li
M: Hsa-circRNA11783-2 in peripheral blood is correlated with
coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis
Res. 14:510–515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Guo N, Zhou H, Zhang Q, Fu Y, Jia Q, Gan
X, Wang Y, He S, Li C, Tao Z, et al: Exploration and bioinformatic
prediction for profile of mRNA bound to circular RNA BTBD7_hsa_
circ_0000563 in coronary artery disease. BMC Cardiovasc Disord.
24:712024. View Article : Google Scholar
|
|
68
|
Chen JX, Hua L, Zhao CH, Jia QW, Zhang J,
Yuan JX, Zhang YJ, Jin JL, Gu MF, Mao ZY, et al: Quantitative
proteomics reveals the regulatory networks of circular RNA
BTBD7_hsa_ circ_0000563 in human coronary artery. J Clin Lab Anal.
34:e234952020. View Article : Google Scholar
|
|
69
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_ circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sun Y, Chen R, Lin S, Xie X, Ye H, Zheng
F, Lin J, Huang Q, Huang S, Ruan Q, et al: Association of circular
RNAs and environmental risk factors with coronary heart disease.
BMC Cardiovasc Disord. 19:2232019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wu WP, Pan YH, Cai MY, Cen JM, Chen C,
Zheng L, Liu X and Xiong XD: Plasma-Derived Exosomal Circular RNA
hsa_ circ_0005540 as a Novel Diagnostic Biomarker for Coronary
Artery Disease. Dis Markers. 2020:31786422020. View Article : Google Scholar
|
|
72
|
Yu F, Tie Y, Zhang Y, Wang Z, Yu L, Zhong
L and Zhang C: Circular RNA expression profiles and bioinformatic
analysis in coronary heart disease. Epigenomics. 12:439–454. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Dinh P, Peng J, Tran T, Wu D, Tran C, Dinh
T and Pan S: Identification of hsa_circ_0001445 of a novel
circRNA-miRNA-mRNA regulatory network as potential biomarker for
coronary heart disease. Front Cardiovasc Med. 10:11042232023.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yin L, Tang Y and Jiang M: Research on the
circular RNA bioinformatics in patients with acute myocardial
infarction. J Clin Lab Anal. 35:e236212021. View Article : Google Scholar :
|
|
75
|
Pan RY, Liu P, Zhou HT, Sun WX, Song J,
Shu J, Cui GJ, Yang ZJ and Jia EZ: Circular RNAs promote TRPM3
expression by inhibiting hsa-miR-130a-3p in coronary artery disease
patients. Oncotarget. 8:60280–60290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yijian L, Weihan S, Lin Y, Heng Z, Yu W,
Lin S, Shuo M, Mengyang L and Jianxun W: CircNCX1 modulates
cardiomyocyte proliferation through promoting ubiquitination of
BRG1. Cell Signal. 120:1111932024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kishore R, Garikipati VNS and Gonzalez C:
Role of Circular RNAs in Cardiovascular Disease. J Cardiovasc
Pharmacol. 76:128–137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Maguire EM and Xiao Q: Noncoding RNAs in
vascular smooth muscle cell function and neointimal hyperplasia.
FEBS J. 287:5260–5283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ma W, Wei S, Zhang B and Li W: Molecular
Mechanisms of Cardiomyocyte Death in Drug-Induced Cardiotoxicity.
Front Cell Dev Biol. 8:4342020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dorn GW II: Apoptotic and non-apoptotic
programmed cardiomyocyte death in ventricular remodelling.
Cardiovasc Res. 81:465–473. 2009. View Article : Google Scholar :
|
|
81
|
Nah J, Zablocki D and Sadoshima J: The
roles of the inhibitory autophagy regulator Rubicon in the heart: A
new therapeutic target to prevent cardiac cell death. Exp Mol Med.
53:528–536. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Martens MD, Karch J and Gordon JW: The
molecular mosaic of regulated cell death in the cardiovascular
system. Biochim Biophys Acta Mol Basis Dis. 1868:1662972022.
View Article : Google Scholar
|
|
83
|
Xiang Q, Yi X, Zhu XH, Wei X and Jiang DS:
Regulated cell death in myocardial ischemia-reperfusion injury.
Trends Endocrinol Metab. 35:219–234. 2024. View Article : Google Scholar
|
|
84
|
Parrotta EI, Lucchino V, Scaramuzzino L,
Scalise S and Cuda G: Modeling cardiac disease mechanisms using
induced pluripotent stem cell-derived cardiomyocytes: progress,
promises and challenges. Int J Mol Sci. 21:43542020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang Y, Liu S, Ding L, Wang D, Li Q and
Li D: Circ_0030235 knockdown protects H9c2 cells against
OGD/R-induced injury via regulation of miR-526b. PeerJ.
9:e114822021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang J, Zhang T, Zhang W, Zou C, Zhang Q,
Ma X and Zhu Y: Circular RNA-DENND4C in H9c2 cells relieves
OGD/R-induced injury by down regulation of microRNA-320. Cell
Cycle. 19:3074–3085. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ding W, Ding L, Lu Y, Sun W, Wang Y, Wang
J, Gao Y and Li M: Circular RNA-circLRP6 protects cardiomyocyte
from hypoxia-induced apoptosis by facilitating hnRNPM-mediated
expression of FGF-9. FEBS J. 291:1246–1263. 2024. View Article : Google Scholar
|
|
88
|
Ko T and Nomura S: Manipulating
cardiomyocyte plasticity for heart regeneration. Front Cell Dev
Biol. 10:9292562022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Theofilis P, Sagris M, Oikonomou E,
Antonopoulos AS, Siasos G, Tsioufis C and Tousoulis D: Inflammatory
mechanisms contributing to endothelial dysfunction. Biomedicines.
9:7812021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shaito A, Aramouni K, Assaf R, Parenti A,
Orekhov A, Yazbi AE, Pintus G and Eid AH: Oxidative stress-induced
endothelial dysfunction in cardiovascular diseases. Front Biosci
(Landmark Ed). 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Gallo G and Savoia C: New insights into
endothelial dysfunction in cardiometabolic diseases: Potential
mechanisms and clinical implications. Int J Mol Sci. 25:29732024.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Huang Y, Song C, He J and Li M: Research
progress in endothelial cell injury and repair. Front Pharmacol.
13:9972722022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu S, Ilyas I, Little PJ, Li H, Kamato D,
Zheng X, Luo S, Li Z, Liu P, Han J, et al: Endothelial dysfunction
in atherosclerotic cardiovascular diseases and beyond: From
mechanism to pharmacotherapies. Pharmacol Rev. 73:924–967. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang R, Wang M, Ye J, Sun G and Sun X:
Mechanism overview and target mining of atherosclerosis:
Endothelial cell injury in atherosclerosis is regulated by
glycolysis (Review). Int J Mol Med. 47:65–76. 2021. View Article : Google Scholar
|
|
95
|
Zha D, Wang S, Monaghan-Nichols P, Qian Y,
Sampath V and Fu M: Mechanisms of endothelial cell membrane repair:
Progress and Perspectives. Cells. 12:26482023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Marzoog BA: Endothelial cell autophagy in
the context of disease development. Anat Cell Biol. 56:16–24. 2023.
View Article : Google Scholar :
|
|
97
|
Wang LP, Han RM, Wu B, Luo MY, Deng YH,
Wang W, Huang C, Xie X and Luo J: Mst1 silencing alleviates
hypertensive myocardial injury associated with the augmentation of
microvascular endothelial cell autophagy. Int J Mol Med.
50:1462022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sobrevia L, Aiello EA and Contreras P:
Mechanisms of endothelial dysfunction and cardiovascular system
adaptation. Curr Vasc Pharmacol. 20:201–204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tong X, Dang X, Liu D, Wang N, Li M, Han
J, Zhao J, Wang Y, Huang M, Yang Y, et al: Exosome-derived
circ_0001785 delays atherogenesis through the ceRNA network
mechanism of miR-513a-5p/TGFBR3. J Nanobiotechnology. 21:3622023.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yu F, Zhang Y, Wang Z, Gong W and Zhang C:
Hsa_ circ_0030042 regulates abnormal autophagy and protects
atherosclerotic plaque stability by targeting eIF4A3. Theranostics.
11:5404–5417. 2021. View Article : Google Scholar :
|
|
101
|
Gao W, Li C, Yuan J, Zhang Y, Liu G, Zhang
J, Shi H, Liu H and Ge J: Circ-MBOAT2 Regulates Angiogenesis via
the miR-495/ NOTCH1 axis and associates with myocardial perfusion
in patients with coronary chronic total occlusion. Int J Mol Sci.
25:7932024. View Article : Google Scholar
|
|
102
|
Wong D, Turner AW and Miller CL: Genetic
insights into smooth muscle cell contributions to coronary artery
disease. Arterioscler Thromb Vasc Biol. 39:1006–1017. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Low EL, Baker AH and Bradshaw AC: TGFβ,
smooth muscle cells and coronary artery disease: A review. Cell
Signal. 53:90–101. 2019. View Article : Google Scholar :
|
|
104
|
Cao G, Xuan X, Hu J, Zhang R, Jin H and
Dong H: How vascular smooth muscle cell phenotype switching
contributes to vascular disease. Cell Commun Signal. 20:1802022.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Milutinović A, Šuput D and Zorc-Pleskovič
R: Pathogenesis of atherosclerosis in the tunica intima, media, and
adventitia of coronary arteries: An updated review. Bosn J Basic
Med Sci. 20:21–30. 2020.
|
|
106
|
Schnack L, Sohrabi Y, Lagache SMM, Kahles
F, Bruemmer D, Waltenberger J and Findeisen HM: Mechanisms of
trained innate immunity in oxLDL primed human coronary smooth
muscle cells. Front Immunol. 10:132019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lacolley P, Regnault V, Segers P and
Laurent S: Vascular smooth muscle cells and arterial stiffening:
Relevance in development, aging, and disease. Physiol Rev.
97:1555–1617. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang G, Liu Z, Deng J, Liu L, Li Y, Weng
S, Guo C, Zhou Z, Zhang L, Wang X, et al: Smooth muscle cell fate
decisions decipher a high-resolution heterogeneity within
atherosclerosis molecular subtypes. J Transl Med. 20:5682022.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang Z, Wang H, Guo C, Yu F, Zhang Y, Qiao
L, Zhang H and Zhang C: Role of hsa_circ_0000280 in regulating
vascular smooth muscle cell function and attenuating neointimal
hyperplasia via ELAVL1. Cell Mol Life Sci. 80:32022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dai H, Zhao N and Zheng Y: CircLDLR
modulates the proliferation and apoptosis of vascular smooth muscle
cells in coronary artery disease through miR-26-5p/KDM6A Axis. J
Cardiovasc Pharmacol. 80:132–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan
X, Liu Y, Tao J, Liu Y, Li K, et al: Circular RNA CircMAP3K5 Acts
as a MicroRNA-22-3p Sponge to Promote Resolution of Intimal
Hyperplasia Via TET2-Mediated smooth muscle cell differentiation.
Circulation. 143:354–371. 2021. View Article : Google Scholar
|
|
112
|
Mao YY, Wang JQ, Guo XX, Bi Y and Wang CX:
Circ-SATB2 upregulates STIM1 expression and regulates vascular
smooth muscle cell proliferation and differentiation through
miR-939. Biochem Biophys Res Commun. 505:119–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang L, Li H, Zheng Z and Li Y:
Hsa_circ_0031891 targets miR-579-3p to enhance HMGB1 expression and
regulate PDGF-BB-induced human aortic vascular smooth muscle cell
proliferation, migration, and dedifferentiation. Naunyn
Schmiedebergs Arch Pharmacol. 397:1093–1104. 2024. View Article : Google Scholar
|
|
114
|
Zhong W, Wang L and Xiong L: Circ_0006251
mediates the proliferation and apoptosis of vascular smooth muscle
cells in CAD via enhancing TET3 and PPM1B expression. Cell Mol Biol
(Noisy-le-grand). 69:34–39. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Medina-Leyte DJ, Zepeda-García O,
Domínguez-Pérez M, González-Garrido A, Villarreal-Molina T and
Jacobo-Albavera L: Endothelial dysfunction, inflammation and
coronary artery disease: Potential biomarkers and promising
therapeutical approaches. Int J Mol Sci. 22:38502021. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Bazoukis G, Stavrakis S and Armoundas AA:
Vagus nerve stimulation and inflammation in cardiovascular disease:
A State-of-the-Art Review. J Am Heart Assoc. 12:e0305392023.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Bhattacharya P, Kanagasooriyan R and
Subramanian M: Tackling inflammation in atherosclerosis: Are we
there yet and what lies beyond? Curr Opin Pharmacol. 66:1022832022.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Prati F, Marco V, Paoletti G and
Albertucci M: Coronary inflammation: Why searching, how to identify
and treat it. Eur Heart J Suppl. 22(Suppl E): E121–E124. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Chen R, Zhang H, Tang B, Luo Y, Yang Y,
Zhong X, Chen S, Xu X, Huang S and Liu C: Macrophages in
cardiovascular diseases: Molecular mechanisms and therapeutic
targets. Signal Transduct Target Ther. 9:1302024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Matter MA, Paneni F, Libby P, Frantz S,
Stähli BE, Templin C, Mengozzi A, Wang YJ, Kündig TM, Räber L, et
al: Inflammation in acute myocardial infarction: The good, the bad
and the ugly. Eur Heart J. 45:89–103. 2024. View Article : Google Scholar :
|
|
121
|
Li Y and Wang B: Circular RNA circCHFR
downregulation protects against oxidized low-density
lipoprotein-induced endothelial injury via regulation of
microRNA-15b-5p/growth arrest and DNA damage inducible gamma.
Bioengineered. 13:4481–4492. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ji P, Song X and Lv Z: Knockdown of
circ_0004104 alleviates oxidized low-density lipoprotein-induced
vascular endothelial cell injury by regulating miR-100/TNFAIP8
Axis. J Cardiovasc Pharmacol. 78:269–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Rafiq M, Dandare A, Javed A, Liaquat A,
Raja AA, Awan HM, Khan MJ and Naeem A: Competing Endogenous RNA
Regulatory Networks of hsa_circ_0126672 in pathophysiology of
coronary heart disease. Genes (Basel). 14:5502023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Dandare A, Rafiq M, Liaquat A, Raja AA and
Khan MJ: Identification of hsa_circ_0092576 regulatory network in
the pathogenesis of coronary heart disease. Genes Dis. 10:26–28.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Liu X, Yao X and Chen L: Expanding roles
of circRNAs in cardiovascular diseases. Noncoding RNA Res.
9:429–436. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Tang Y, Bao J, Hu J, Liu L and Xu DY:
Circular RNA in cardiovascular disease: Expression, mechanisms and
clinical prospects. J Cell Mol Med. 25:1817–1824. 2021. View Article : Google Scholar :
|
|
127
|
Vilades D, Martínez-Camblor P,
Ferrero-Gregori A, Bär C, Lu D, Xiao K, Vea À, Nasarre L, Sanchez
Vega J, Leta R, et al: Plasma circular RNA hsa_circ_0001445 and
coronary artery disease: Performance as a biomarker. FASEB J.
34:4403–4414. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Sonnenschein K, Wilczek AL, de
Gonzalo-Calvo D, Pfanne A, Derda AA, Zwadlo C, Bavendiek U,
Bauersachs J, Fiedler J and Thum T: Serum circular RNAs act as
blood-based biomarkers for hypertrophic obstructive cardiomyopathy.
Sci Rep. 9:203502019. View Article : Google Scholar
|
|
129
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar :
|
|
130
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang W, Sun L, Huang MT, Quan Y, Jiang T,
Miao Z and Zhang Q: Regulatory circular RNAs in viral diseases:
applications in diagnosis and therapy. RNA Biol. 20:847–858. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Li MZ, Zhang JN, Ren F, Yin DL, Zhao XH
and Liu K: Diagnostic value of circRNA in coronary heart disease: A
meta-analysis. Biomark Med. 17:667–677. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Fu Y, He S, Li C, Gan X, Wang Y, Zhou Y,
Jiang R, Zhang Q, Pan Y, Zhou H, et al: Detailed profiling of m6A
modified circRNAs and synergistic effects of circRNA and
environmental risk factors for coronary artery disease. Eur J
Pharmacol. 951:1757612023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
He S, Fu Y, Li C, Wang Y, Zhou H, Jiang R,
Zhang Q, Jia Q, Chen X and Jia EZ: Interaction between the
expression of hsa_circRPRD1A and hsa_circHERPUD2 and classical
coronary risk factors promotes the development of coronary artery
disease. BMC Med Genomics. 16:1312023. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhong Q, Jin S, Zhang Z, Qian H, Xie Y,
Yan P, He W and Zhang L: Identification and verification of circRNA
biomarkers for coronary artery disease based on WGCNA and the LASSO
algorithm. BMC Cardiovasc Disord. 24:3052024. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang W, Cui J, Li L, Zhu T and Guo Z:
Identification of Plasma Exosomes hsa_circ_0001360 and
hsa_circ_0000038 as key biomarkers of coronary heart disease.
Cardiol Res Pract. 2024:55571432024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Huang S, Zeng Z, Sun Y, Cai Y, Xu X, Li H
and Wu S: Association study of hsa_circ_0001946, hsa-miR-7-5p and
PARP1 in coronary atherosclerotic heart disease. Int J Cardiol.
328:1–7. 2021. View Article : Google Scholar
|
|
139
|
Tong X, Zhao X, Dang X, Kou Y and Kou J:
circRNA, a novel diagnostic biomarker for coronary heart disease.
Front Cardiovasc Med. 10:10706162023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ji WF, Chen JX, He S, Zhou YQ, Hua L, Hou
C, Zhang S, Gan XK, Wang YJ, Zhou HX, et al: Characteristics of
circular RNAs expression of peripheral blood mononuclear cells in
humans with coronary artery disease. Physiol Genomics. 53:349–357.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Miao L, Yin RX, Zhang QH, Liao PJ, Wang Y,
Nie RJ and Li H: A novel circRNA-miRNA-mRNA network identifies
circ-YOD1 as a biomarker for coronary artery disease. Sci Rep.
9:183142019. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Ward Z, Schmeier S, Pearson J, Cameron VA,
Frampton CM, Troughton RW, Doughty RN, Richards AM and Pilbrow AP:
Identifying candidate circulating RNA markers for coronary artery
disease by deep RNA-Sequencing in human plasma. Cells. 11:31912022.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Dergunova LV, Vinogradina MA, Filippenkov
IB, Limborska SA and Dergunov AD: Circular RNAs variously
participate in coronary atherogenesis. Curr Issues Mol Biol.
45:6682–6700. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wang L, Xu GE, Spanos M, Li G, Lei Z,
Sluijter JPG and Xiao J: Circular RNAs in cardiovascular diseases:
Regulation and therapeutic applications. Research (Wash D C).
6:00382023.PubMed/NCBI
|
|
145
|
Goina CA, Goina DM, Farcas SS and
Andreescu NI: The role of circular RNA for early diagnosis and
improved management of patients with cardiovascular diseases. Int J
Mol Sci. 25:29862024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Long Q, Lv B, Jiang S and Lin J: The
landscape of circular RNAs in cardiovascular diseases. Int J Mol
Sci. 24:45712023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chen W, Xu J, Wu Y, Liang B, Yan M, Sun C,
Wang D, Hu X, Liu L, Hu W, et al: The potential role and mechanism
of circRNA/miRNA axis in cholesterol synthesis. Int J Biol Sci.
19:2879–2896. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Neu CT, Gutschner T and Haemmerle M:
Post-Transcriptional expression control in platelet biogenesis and
function. Int J Mol Sci. 21:76142020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yu R, Yu Q, Li Z, Li J, Yang J, Hu Y,
Zheng N, Li X, Song Y, Li J, et al: Transcriptome-wide map of
N6-methyladenosine (m6A) profiling in coronary artery disease (CAD)
with clopidogrel resistance. Clin Epigenetics. 15:1942023.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zou Y, Wang Y, Yao Y, Wu Y, Lv C and Yin
T: Platelet-derived circFAM13B associated with anti-platelet
responsiveness of ticagrelor in patients with acute coronary
syndrome. Thromb J. 22:532024. View Article : Google Scholar : PubMed/NCBI
|
















