Introduction
Cyclophosphamide (CY) is an effective
chemotherapeutic agent that can be used to treat various types of
cancer (1-3), such as lymphoma, breast cancer and
ovarian cancer. However, CY can also exert oxidative DNA damage on
bone marrow cells, causing myelosuppression and immunosuppression
(4). Notably, CY has been
previously reported to trigger bone marrow cell apoptosis 6 h after
administration (5). In addition,
it can cause the hematopoietic inhibition of bone marrow and reduce
the numbers of several peripheral blood cell types, such as red
blood cells (RBC), white blood cells (WBC) and platelets (PLT)
(6). Furthermore, CY frequently
causes intestinal mucositis and neutropenia, which result in
intestinal microbiota disorder (7,8).
Therefore, it is important to preserve the function of
hematopoietic stem cells (HSCs) in the bone marrow during
chemotherapy.
Ginseng (Panax ginseng C.A. Meyer) has been
used to treat diseases for thousands of years in China. Modern
pharmacological studies have previously shown that ginseng can
treat various diseases, such as cancer, inflammation, oxidative
stress, tumors, obesity and diabetes (9). The main chemical components
contained within ginseng include ginsenosides, polysaccharides,
phenolic acid and proteins (10). In the clinic, ginseng or
ginsenosides have been frequently used to reduce the side effects
induced by chemotherapy (5,11). Ginsenosides and polysaccharides
are the important active ingredients of ginseng. The total
ginsenosides (TG), ginsenoside Rb1, ginsenoside Rg1, ginsenoside
compound K (CK) or ginsenoside Rh2 have been reported to possess
dual effects, including anticancer and bone marrow-protecting
activities (12,13). The ginseng polysaccharides have a
number of pharmacological effects, such as anti-oxidative,
anti-inflammatory, immunoregulatory and intestinal microbiota
regulatory activities (14). It
has previously been shown that ginseng polysaccharides can enhance
the activity and absorption of ginsenosides by affecting intestinal
microbial metabolism via increasing β-glucosidase activity
(15).
Firmicutes (79.4%) and Bacteroidota (16.9%)
constitute the main bacterial families in the intestines of healthy
adult humans (16). Imbalance in
the intestinal microflora can weaken the resistance of the body to
pathogenic microorganisms, resulting in the loss of the protective
intestinal barrier (17).
Lactobacillus, Streptococcus and Escherichia
coli can promote goblet cell differentiation and mucus
production in the intestine to reduce the abundance of
Salmonella typhimurium and pathogenic Escherichia
coli, which preserves epithelial cell function and energy
balance (18). By contrast,
Fusobacterium have been reported to be enriched in patients
with colon cancer, which can invade intestinal epithelial cells to
enable their survival and maintenance, whereas the abundance of
Bacteroidetes and Firmicutes are reduced in patients with colon
cancer (19). Kostic et
al (20) previously reported
that Fusobacterium had a fitness advantage in the evolving
tumor microenvironment, which caused an imbalance of gut
microbiota. Notably, ginsenosides and ginseng polysaccharides have
been shown to reverse gut microbiota disorder by inhibiting the
expression of inflammatory cytokines, in turn restoring the
diversity of intestinal microflora (21-23). Ginsenosides and polysaccharides
from ginseng have shown protective effects on the imbalance of
intestinal microflora (7).
Moreover, cell apoptosis induced by CY is the main
cause of HSC damage (24), and
ginsenosides protect HSCs from damage by reducing HSC apoptosis and
inhibiting the expression of inflammation factors (25). Therefore, the present study
assessed the effects of ginseng extract (GE), TG and total
polysaccharides (TP) from ginseng on intestinal microflora
regulation and the viability of bone marrow HSCs in a mouse model
of HSC damage induced by CY via 16S ribosomal (r)RNA gene
sequencing of fecal matter and the high-throughput RNA-sequencing
of HSCs.
Materials and methods
Chemicals
CY (cat. no. S30563) was obtained from Shanghai
Yuanye Biotechnology Co., Ltd. The cell apoptosis kit (Annexin
V-FITC/PI; cat. no. 556547) was supplied by BD Pharmingen (BD
Biosciences). Blood routine reagent kits (Staining reagent kit,
cat. no. ZY4203 and Reaction reagent kit, cat. no. ZY4224) was
purchased by IDEXX Laboratories Inc. The PE-conjugated anti-TER-119
antibody (cat. no. 116207) was obtained from BioLegend, Inc.
Preparation of GE, TG and TP
Dry Panax ginseng was purchased from the
Wanliang ginseng market (Tonghua, China). The ginseng was boiled
three times to obtain the GE (6). The extracting solution of GE was
eluted with different concentrations of ethanol (80-95%), before
being centrifuged at 1,200 × g at 4°C for 20 min to obtain the
supernatant and precipitates. The supernatant was sequentially
decolorized with a decolorization resin (cat. no. D941; Tianjin
Haoju Resin Technology Co., Ltd.) and activated carbon, before
being filtered and dried at 40°C to obtain TG (26). The precipitates were sequentially
deproteinized, decolorized and dialyzed to remove components with a
molecular weight of <3,500 Da to obtain the TP (27). The composition and content of
ginsenosides and polysaccharides in the GE, TG and TP have been
reported in a previous study (6,26). The ginsenoside content of GE and
TG is 4.39 and 81.09%, respectively. By contrast, the
polysaccharide content of GE, TG and TP is 72.28, 4.68 and 89.79%,
respectively (26).
Animal model
A total of 50 male 4-week-old Kunming mice (weight,
18-20 g) were purchased from Liaoning Changsheng Biotechnology Co.,
Ltd. [animal license no. SCXK (Liao)-2015-0001]. The mice were
maintained under the conditions of controlled light (12 h
light/dark), temperature (25±1°C) and humidity (60%±5%), with ad
libitum access to food and water. After 5 days of acclimation,
mice were randomly divided into the following five groups
(n=10/group): i) The normal group (Control); ii) the group treated
with CY (Model); iii) the CY + GE group (GE, 1.0 g/kg); iv) the CY
+ TG group (TG, 0.25 g/kg); and v) the CY + TP group (TP, 1.0
g/kg).
The mice were intragastrically administered GE, TG
and TP for 28 days in the GE, TG and TP groups (0.1 ml/10 g body
weight) (7,28), whereas mice in the other groups
were administered equivalent volumes of normal saline. Mice in the
Model, GE, TG and TP groups were hypodermically injected with CY
saline solution (50 mg/kg) on days 25, 26, 27 and 28. The dose of
GE was calculated according to the human adult daily dose of
ginseng (9 g/day, 60 kg/body weight) (29), whereas the doses of TG and TP
(1.0 g/kg) were selected according to previous studies (6,7).
The fresh feces of mice were collected on day 28 and
stored at -80°C until further analysis. All mice were then
euthanized by cervical dislocation under anesthesia with 30 mg/kg
pentobarbital sodium by intraperitoneal injection after fasting for
12 h. If an animal reached the predefined humane endpoints [loss of
>15% of body weight in 1-2 days or an overall reduction of
>20% in body weight; or displaying obvious signs of suffering
(lethargy, squinted eyes, dehydration and hunched back)], they were
humanely euthanized as aforementioned. Animal death was confirmed
by cessation of respiration and heartbeat. The study protocol was
approved by the Ethics Committee of Changchun University of Chinese
Medicine (protocol no. 2023033; Changchun, China). No mice showed
abnormal signs or reached humane endpoints throughout the
experiment.
Blood cell parameters
Anesthesia with 30 mg/kg pentobarbital sodium
(concentration 5 mg/ml, dose 60 µl/10 mg body weight) was
induced by intraperitoneal injection on day 29. Subsequently, ~0.5
ml blood was collected from the retro-orbital vein (30). The mice were then sacrificed
after this blood collection. The blood cell parameters [WBC,
neutrophils, lymphocytes (LYMPH), RBC, hemoglobin (HGB),
reticulocytes (Ret) and PLT] were detected using the ProCyte DX
hematology analyzer (IDEXX Laboratories Inc.).
16S rRNA gene sequencing
A QIAamp DNA Stool Mini Kit (cat. no. 51504; Qiagen,
Inc.) was used to extract and purify the DNA of the fecal samples
(7 samples from 7 mice each group). The primers (341F: 5′-CCT AYG
GGR BGC ASC AG-3′ and 806R: 5′-GGA CTA CNN GGG TAT CTA AT-3′) were
used to amplify the 16S rRNA genes in the V3-V4 region. The library
construction (NEBNext® Ultra™ IIDNA Library Prep Kit;
cat. no. E7645; New England BioLabs, Inc.) and RNA sequencing were
performed by Novogene Bioinformatics Technology Co., Ltd. The
detailed steps of this sequencing procedure have been previously
reported (6,31). For the effective tags obtained
previously, denoise was performed with DADA2 or deblurmodule in the
QIIME2 software (version QIIME2-202006; https://library.qiime2.org) to obtain initial amplicon
sequence variants (ASVs; default: DADA2), and then ASVs with
abundance <5 were filtered out. the annotation database was the
Silva Database (https://www.arb-silva.de/). The following analyses
were performed: i) Principal co-ordinates analysis was performed
according to the weighted UniFrac distance matrices; ii)
α-diversity was calculated from 4 indexes in QIIME2, including
Chao1, Dominance, Pielou_e, and Shannon; iii) cluster analysis was
performed with principal component analysis (PCA), which was
applied to reduce the dimension of the original variables using the
ade4 package and ggplot2 package in R software (version 3.5.3); iv)
the significantly different species (P<0.05) at each taxonomic
level (phylum, class, order, family, genus and species) were
analyzed using the R software (version 3.5.3), P<0.05 was
considered to indicate a statistically significant difference; and
v) the LEfSe software (version 1.0) was used to perform LEfSe
analysis] linear discriminant analysis (LDA) score >3;
Kruskal-Wallis test, false discovery rate P<0.05] so as to
identify the biomarkers.
Apoptosis of HSCs
Bone marrow from the right femur of the mice in the
Control, Model, GE, TG and TP groups (n=10/group) was washed with
PBS into the EasySep™ buffer (cat. no. 20144; Stemcell
Technologies, Inc.) at room temperature using a syringe equipped
with a 23-gauge needle to maintain the viability of bone marrow
cells. The remaining aggregates and debris were removed by passing
the cell suspension through a 70-µm mesh nylon strainer,
before the sample was centrifuged at 300 × g for 10 min at room
temperature. The cells were then resuspended at 1×108
nucleated cells/ml in DMEM/F12 (cat. no. 11-330-032; Gibco; Thermo
Fisher Scientific, Inc.). The HSCs from bone marrow mononuclear
cells were purified using the EasySep™ Mouse Hematopoietic
Progenitor Cell Isolation Kit according to manufacturer's protocol
(cat. no. 19856; Stemcell Technologies, Inc.). HSCs were adjusted
to 1×105 cells/ml in PBS and were then labeled using
Annexin V-FITC/PI (cat. no. c1052) for 20 min at room temperature.
A total of 10,000 events were acquired to analyze cell apoptosis
using a DxFLEX flow cytometer and CytExpert Software version
2.4.0.28 (both Beckman Coulter, Inc.).
Erythroid differentiation of HSCs
HSCs (1×105) were incubated with the
PE-conjugated anti-TER-119 antibody (1:1,000) in the dark for 40
min on ice. Subsequently, TER-119 expression was detected using a
DxFLEX flow cytometer and CytExpert software version 2.4.0.28 (both
Beckman Coulter, Inc.).
RNA extraction, establishment of cDNA
library and sequencing
Total RNA was extracted from HSCs according to the
instruction manual of the TRlzol reagent (cat. no. AM9738; Thermo
Fisher Scientific, Inc.). RNA concentration and purity was measured
using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA
integrity was assessed using the RNA Nano 6000 Assay Kit (cat. no.
5067-1511) of the Agilent Bioanalyzer 2100 system (both Agilent
Technologies, Inc.). A total of 1 µg RNA per sample was used
as input material for the RNA sample preparations. Sequencing
libraries were generated using NEBNext UltraTM RNA Library Prep Kit
(cat. no. E7530S/L; New England BioLabs, Inc.) for Illumina
following the manufacturer's recommendations. To select cDNA
fragments preferentially of 240 bp in length, the library fragments
were purified with AMPure XP system (Beckman Coulter, Inc.). Then,
3 µl USER Enzyme (cat. no. E7530S/L; New England BioLabs,
Inc.) was used with size-selected, adaptor-ligated cDNA at 37°C for
15 min followed by 5 min at 95°C before PCR. PCR was then performed
using Phusion High-Fidelity DNA polymerase (cat. no. M0530S; New
England BioLabs, Inc.) with the following reaction conditions: 98°C
for 30 sec; 25 cycles of 98°C for 10 sec and 72°C for 15 sec; 72°C
for 5 min, 4°C for hold. At last, the PCR products were purified
(AMPure XP system) and the library quality was assessed on the
Agilent Bioanalyzer 2100 system. The loading concentration of the
final library was 2 nM. The clustering of the index-coded samples
was performed on a cBot Cluster Generation System using TruSeq PE
Cluster Kit v4-cBot-HS (Illumina) according to the manufacturer's
instructions. After cluster generation, the library preparations
were sequenced on an Illumina platform and paired-end reads were
generated. The libraries were constructed and sequenced by Beijing
Biomarker Technologies Co., Ltd. (www.biocloud.net).
Differentially expressed gene (DEG)
analysis
After sequencing, bioinformatics analysis was
performed using BMKCloud (www.biocloud.net) to identify the DEGs. Differential
expression analysis of two conditions/groups was performed using
DESeq2 (version 1.30.1; https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html).
An adjusted P<0.05 and a fold change (FC) of >1.5 were
considered to indicate a significant difference. Gene Ontology (GO)
enrichment analysis of DEGs was implemented by the GOseq R
packages-based Wallenius non-central hyper-geometric distribution
(32). KOBAS software (33) was used to test the statistical
enrichment of differential expression genes in Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg/). Heatmap analysis of DEGs
was conducted according to the FPKM value using the cluster
Profiler package (version 3.0.3) in the R software.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HSCs using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and cDNA was obtained using the FastKing RT Kit (Tiangen
Biotech Co., Ltd.) according to the manufacturer's protocol.
Subsequently, qPCR was performed using the CFX Connect Real-Time
PCR Detection System (Bio-Rad Laboratories, Inc.) with SuperReal
PreMix Plus (SYBR Green; Tiangen Biotech Co., Ltd.). The primer
sequences were as follows: β-actin forward, 5′-CTG TCC CTG
TAT GCC TCT G-3′ and reverse, 5′-ATG TCA CGC ACG ATT TCC-3′;
Hhex forward, 5′-CCA CCC GAG AGA AAG CGT CTG-3′ and reverse,
5′-TGC GTT GGA CAG TTT GGA CAC T-3′; Klf4 forward, 5′-GCG
GGA AGG GAG AAG ACA CTG CGT C-3′ and reverse, 5′-TAG GAG GGC CGG
GTT GTT ACT GCT-3′; Pira2 forward, 5′-ACT ACT GGA CAC CCA
GCC TT-3′ and reverse, 5′-TGA ACC TGT CAT AGC TCG GC-3′; and
Pbx1 forward, 5′-TGA AGC CTG CCT TGT TTA ATG T-3′ and
reverse, 5′-ATG TTG TCC AGT CGC ATG AGC-3′. The thermocycling
conditions were as follows: 95°C for 15 min, followed by 40 cycles
at 95°C for 10 sec, 55°C for 20 sec and 72°C for 30 sec. The
transcript levels were quantified and normalized to the internal
reference gene β-actin using the 2−ΔΔCq method
(34).
Immunohistochemical analysis
The bone marrow from the left femurs of mice in the
Control, Model, GE, TG and TP groups (n=6/group) was fixed in 10%
formalin solution for 5 days at room temperature and demineralized
with 10% EDTA for 14 days at room temperature. The samples were
then embedded in paraffin and sectioned into 4 µm slices.
Bone marrow tissue sections were dewaxed and then treated with
high-pressured 2% EDTA (pH 9.0) antigen retrieval buffer at 95°C
for 20 min for antigen retrieval. The tissue was incubated in
Endogenous Peroxidase Blocking Buffer (cat. no. P0100A; Beyotime
Institute of Biotechnology) at room temperature for 10 min to block
and eliminate the interference of endogenous peroxidase.
Subsequently, the sections were blocked with 3% BSA (cat. no.
B24726; Shanghai Angyi Biotechnology Co., Ltd.) for 1 h at room
temperature, and then incubated with primary antibodies overnight
at 4°C. The sections were then incubated with secondary antibody
[goat anti-rabbit IgG (H+L); cat. no. SA00001-2; Proteintech Group,
Inc.; diluted to 1:1,000 in 3% BSA] at room temperature for 30 min.
Finally, the binding antibody was detected by DAB (cat. no.
DAB-2031; Fuzhou Maixin Biotechnology Development Co., Ltd.)
staining at room temperature for 5 min and hematoxylin
counterstaining at room temperature for 2 min. Negative (no primary
antibody) control staining of one mouse was used in each
experiment. Images were captured using the Axioscan 7 fully
automatic digital slide scanning system (Zeiss AG). Rabbit
anti-mouse Klf4 (cat. no. 11880-1-AP; Proteintech Group, Inc.) and
rabbit anti-mouse Pbx1 (cat. no. 18204-1-AP; Proteintech Group,
Inc.) antibodies were used as primary antibodies to detect protein
expression in PBS-Tween (1%) at a concentration of 1:200.
Western blot analysis
Western blot analysis was performed as previously
reported (35). Briefly, the
proteins were extracted from the bone marrow cells of mice tibia
and femur, and then lysed in RIPA lysis buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology) containing protease inhibitors
(cat. no. 05892970001; Roche Diagnostics) at 4°C for 5 min.
Subsequently, 35 µg protein was separated by 12% SDS-PAGE
and then transferred onto PVDF membranes. The membranes were
blocked with 3% BSA for 1 h at room temperature, and then incubated
with the following primary antibodies (all from Proteintech Group,
Inc.) overnight at 4°C: β-actin (1:2,000; cat. no. BS6007M), Bcl2
(1:2,000; cat. no. 26593-1-AP) and Bax (1:2,000; cat. no.
50599-2-Ig). The membranes were then incubated with a secondary
antibody [goat anti-rabbit IgG (H+L) 1:5,000; cat. no. SA00001-2;
Proteintech Group, Inc.] for 2 h at room temperature. The bands
were visualized using an ECL luminescence reagent (cat. no.
P001-500; Hunan Hui Bai Shi Biological Technology Co., Ltd.) a
ChemiDoc™ MP imaging system (Bio-Rad Laboratories, Inc.) at room
temperature. The relative protein expression levels were normalized
to β-actin.
Statistical analysis
The DEGs were filtered according to fold change ≥1.5
and P<0.05. Quantitative data are presented as the mean ± SD.
The relative abundance of intestinal microflora was compared
between groups using one-way ANOVA followed by Dunnett's test,
whereas other data were analyzed with one-way ANOVA followed by
Tukey's test using GraphPad Prism 8.0 software (Dotmatics).
Correlation analysis was performed using Spearman's rank
correlation coefficient test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Statistical analysis of α diversity in
intestinal microflora
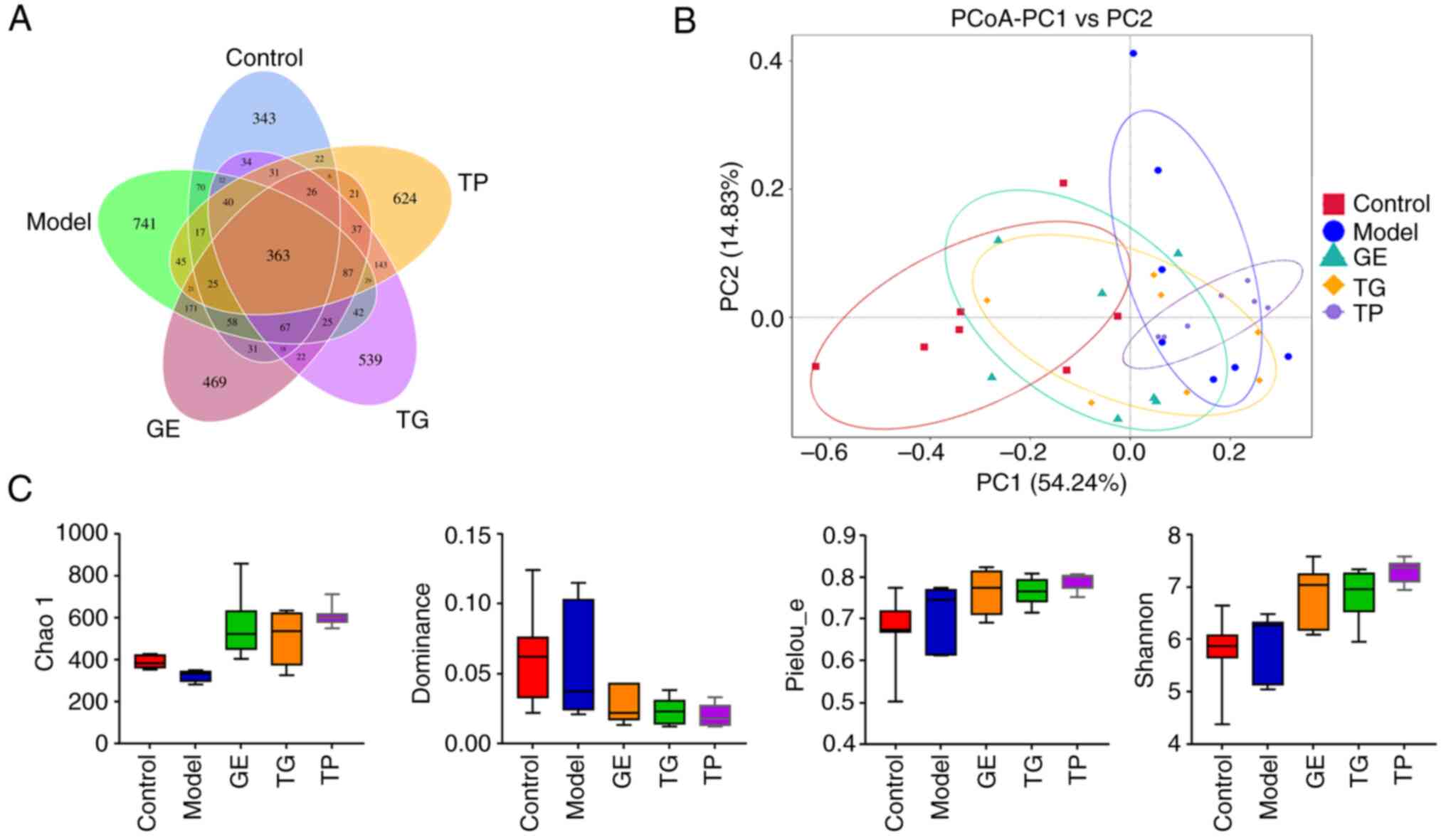
Fig. 1A shows the
number of shared and unique ASVs among the five treatment groups in
a Venn diagram. There were 343, 741, 469, 539 and 624 unique ASVs
in the Control, Model, GE, TG and TP groups, respectively. The
principal co-ordinates analysis showed that the microbial community
had some differences between the Control and Model or TP groups
(Fig. 1B). The α diversity
reflects the richness and evenness of intestinal microflora. The
Chao 1 index shows the community richness, which is positively
associated with the number of ASVs. The dominance index and
pielou_e index show the community evenness, whilst the Shannon
index shows the richness and evenness of ASVs. As shown in Fig. 1C, CY markedly decreased the Chao
1 index compared with that in the Control group. In addition, GE,
TG and TP increased the Chao 1, pielou_e and Shannon indices,
whilst reducing the dominance index compared with the Model group;
however, these differences were not statistically significant.
These results indicated that GE, TG and TP markedly increased the
community richness and evenness of ASVs in model mice. Notably, 7
samples from 7 mice in the TP group showed the best uniformity
(Fig. 1B and C).
Differences in the relative abundance of
bacteria phylum, genus and species
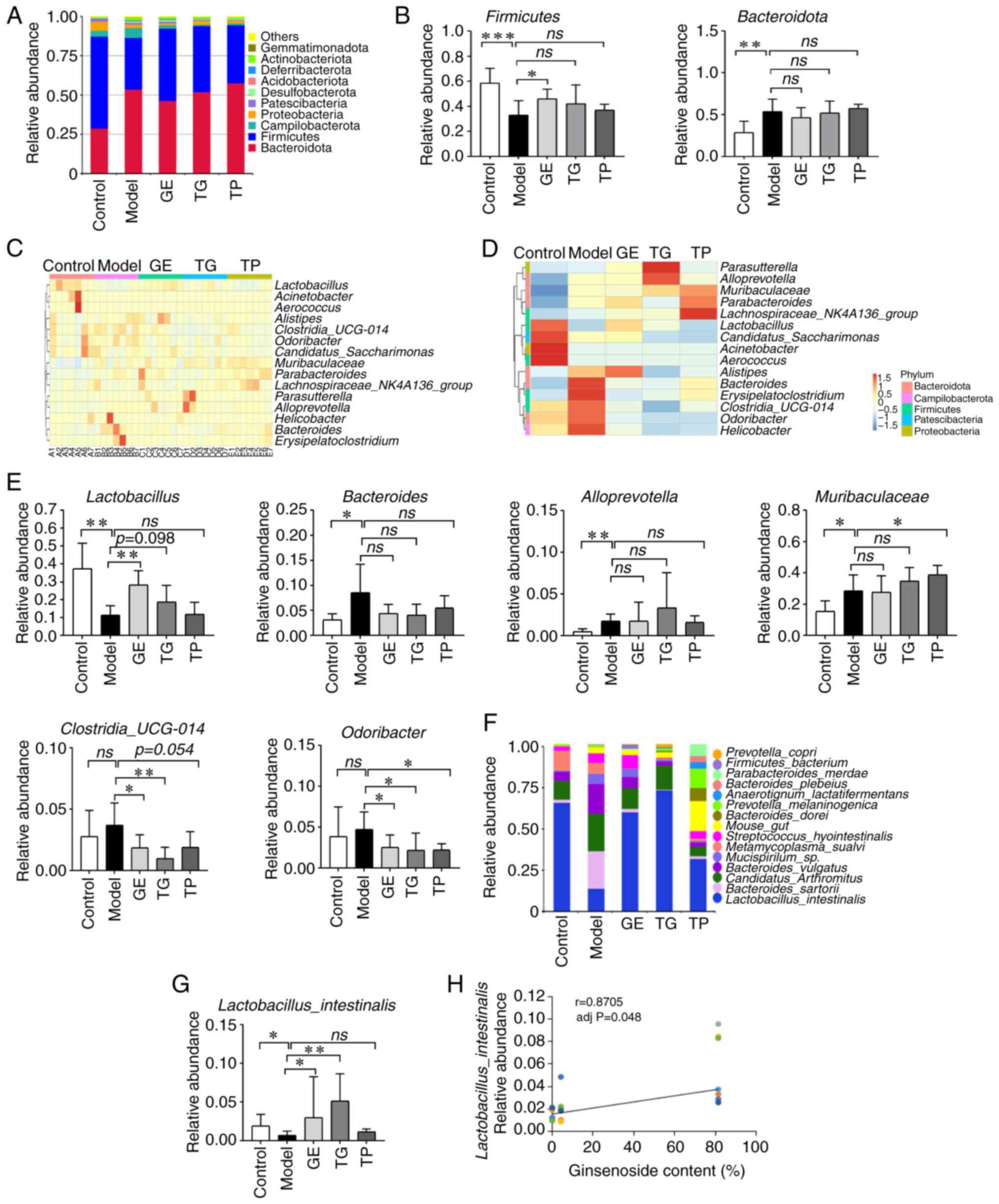
The present study next analyzed the composition and
abundance of the top 10, 15 and 15 bacteria at the phylum, genus
and species levels, respectively. CY increased the relative
abundance of Bacteroidota whilst decreasing that of Firmicutes at
the phylum level (Fig. 2A). GE
was found to significantly elevate the abundance of Firmicutes
compared with that in the Model group (Fig. 2B). TG and TP also increased the
expression of Firmicutes, whereas GE and TG also reduced the
abundance of Bacteroidota. However, the difference was not
statistically significant (Fig.
2B). Fig. 2C and D show the
top 15 genera at the genus level clustered according to the
relative abundance among the five groups (n=35 samples). CY
significantly reduced the relative abundance of
Lactobacillus, whilst increasing that of Bacteroides,
Alloprevotella and Muribaculaceae (Fig. 2E). GE significantly increased the
relative abundance of Lactobacillus. By contrast, TG also
increased the relative abundance of Lactobacillus compared
with that in the Model group, but the difference was not
significant (Fig. 2E). In
addition, GE and TG were found to reduce the relative abundance of
Clostridia_UCG-014 and Odoribacter compared with the
Model group (Fig. 2E). These
results suggest that GE at least partially reversed the abnormal
changes in some intestinal microflora, followed by TG and TP.
At the species level, CY increased the relative
abundance of Bacteroides_sartorii,
Candidatus_arthromitus, Bacteroides_vulgatus and
Mucisporollum_sp., whilst decreasing that of
Lactobacillus_ intestinalis and Metamycoplasma_sualvi
(Fig. 2F). GE, TG and TP could
reverse these aforementioned changes in the intestinal microflora.
Notably, GE and TG significantly increased the abundance of
Lactobacillus_intestinalis compared with that in model group
(Fig. 2G). In addition,
Lactobacillus_intestinalis was significantly positively
correlated with ginsenoside content (r=0.8705; Fig. 2H). Furthermore, TP increased the
abundance of low-abundance microflora compared with the Control
group, such as Mouse_gut, Bacteroides_dorei,
Prevotella_melanimogenica,
Anaerotignum_lactatifermentans, Bacteroides_plebeius
and Parabacteroides_merdae. Although CY and TP increased the
abundance of some low-abundance microflora, the bacterial species
were different. These results showed that the intestinal bacteria
of model mice treated with GE, TG and TP were markedly improved. In
general, the relative abundance and species of intestinal bacteria
recovered by GE were consistent with the Control group at the
phylum, genus and species levels.
The key microflora (biomarkers) were next identified
using LDA (Fig. 3A) and an
evolutionary branch tree (Fig.
3B). A total of 4, 9 and 48 key microflora (LDA score >3)
were detected in the GE, TG and TP groups, respectively (Fig. 3A). s_Vibrio_metschnikovii,
s_Streptococcus_hyointestinalis, s_Clostridium_
fusiformis and s_Eubacterium_plexicaudatum were key
species in the GE group. The key species in the TG group included
f_Prevotellaceae, s_Lactobacillus_ intestinalis,
g_Alloprevotella and o_Burkholderiales. By contrast, c_, p_
and o_Bacteroidales, f_Muribaculaceae and
g_Muribaculaceae were higher in the TP group compared with
that in the other groups.
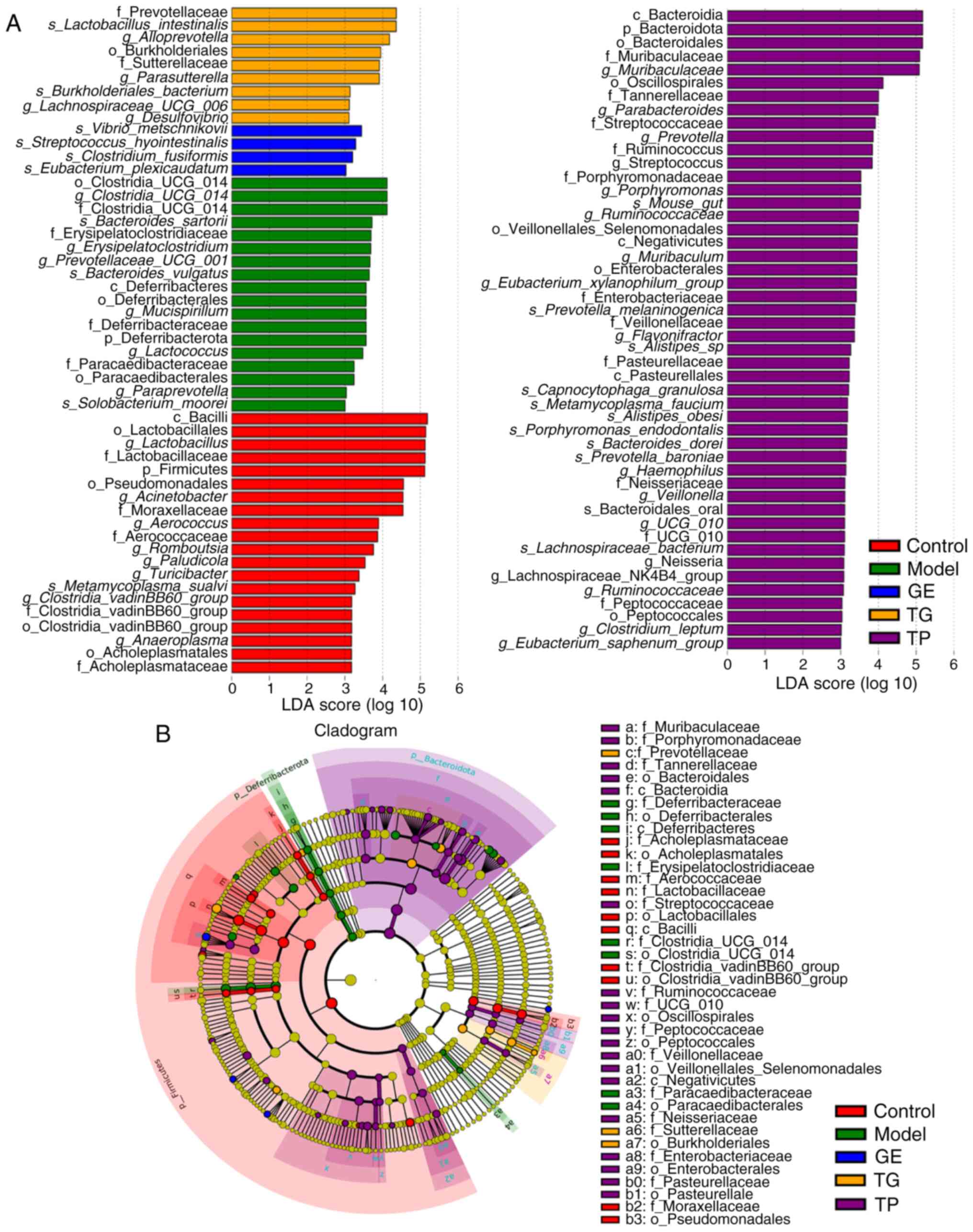 | Figure 3Linear discriminant analysis effect
size difference analysis of the relative abundance of intestinal
microflora among the GE, TG and TP groups. (A) LDA scores. (B)
Evolutionary branch diagram. (n=7). c, class; f, family; g, genus;
GE, ginseng extract; k, kingdom; LDA, linear discriminative
analysis; o, order; p, phylum; s, species; TG, total ginsenosides;
TP, total polysaccharides. |
Profiling of DEGs from HSCs using
high-throughput RNA-seq analysis
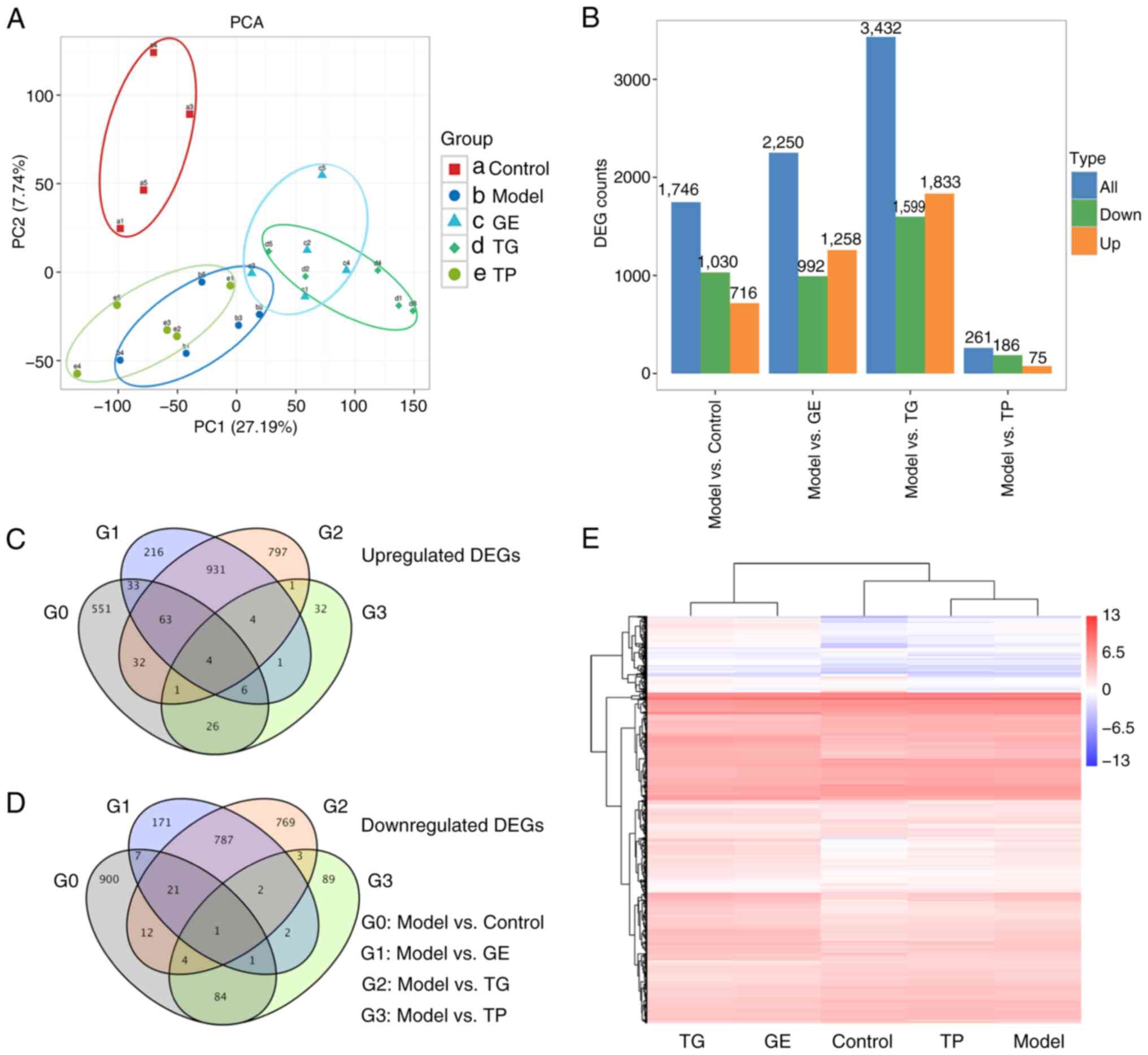
A total of 27,684 genes were annotated and used for
further analysis. There were 4,988 DEGs in all samples. PCA showed
marked differences between the Model/TP group and GE/TG groups
(Fig. 4A). Fig. 4B shows that a total of 1,746 DEGs
were obtained between the Model and Control groups, with 716
upregulated and 1,030 downregulated. There were 2,250 DEGs between
the Model and GE groups, including 1,258 upregulated and 992
downregulated DEGs. The highest number of DEGs was detected between
the Model and TG groups, including 1,833 upregulated DEGs and 1,599
down-regulated DEGs. The lowest number of DEGs was detected between
the Model and TP groups, including 75 upregulated DEGs and 186
downregulated DEGs. A Venn diagram shows the numbers of upregulated
DEGs (Fig. 4C) and downregulated
DEGs (Fig. 4D) among the groups.
There were 4 upregulated DEGs and 1 downregulated DEG in common
between the Model and Control/GE/TG/TP (Fig. 4C and D). Cluster analysis was
performed for the five groups based on 4,988 DEGs, where the
profiling of the DEGs was found to be similar between the TG and GE
groups or between the Model and TP groups (Fig. 4E).
GO analysis of the upregulated DEGs from
HSCs
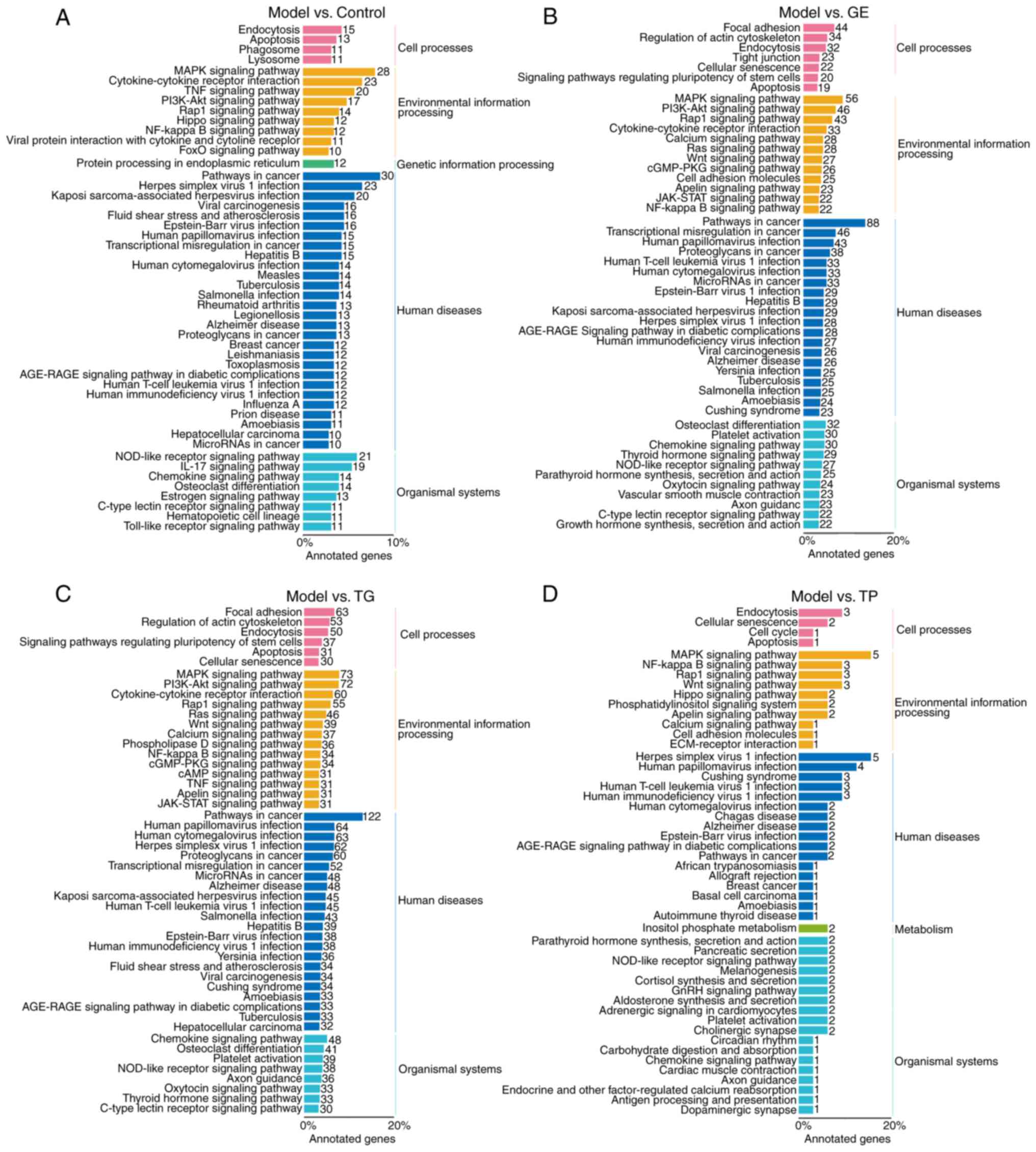
In the top 20 biological processes, upregulated
genes in the Control group compared with the Model group (Fig. 5A) were mainly enriched in immune
regulation (including 'immune response', 'neutrophil chemotaxis',
'myeloid leukocyte differentiation', 'acute inflammatory response'
and 'B cell proliferation involved in immune response') and
hemopoiesis regulation (including 'negative regulation of cell
proliferation', 'response to hypoxia', 'hemopoiesis', 'positive
regulation of nitric oxide biosynthetic process', 'cellular
response to decreased oxygen levels', 'cellular response to oxygen
levels' and 'cellular response to macrophage colony-stimulating
factor stimulus'). In the top 20 biological processes, the
upregulated genes in the GE group compared with the Model group
(Fig. 5B) were enriched in
immune regulation ('transforming growth factor beta receptor
signaling pathway' and 'negative regulation of interleukin-6
production') and hemopoiesis regulation ('small GTPase mediated
signal transduction', 'positive regulation of blood vessel
endothelial cell migration', 'regulation of small GTPase mediated
signal transduction', 'cellular response to vascular endothelial
growth factor stimulus' and 'positive regulation of nitric-oxide
synthase biosynthetic process'). Compared with the Model group, the
upregulated genes in the TG group (Fig. 5C) were related to 'inflammatory
response', 'small GTPase mediated-signal transduction' and
'positive regulation of nitric-oxide synthase biosynthetic process'
in the top 20 biological processes. Compared with the Model group,
the upregulated genes in the TP group (Fig. 5D) were enriched in immune
regulation ('negative regulation of interleukin-10 biosynthetic
process', 'cellular response to interleukin-8', 'regulation of
natural killer cell differentiation' and 'regulation of extrathymic
T cell differentiation') and 'negative regulation of
parkin-mediated stimulation of mitophagy in response to
mitochondrial depolarization' terms. These results suggest that CY
mainly affected the immune regulation and hematopoietic function of
HSCs. In addition, the upregulated genes in the GE and TG groups
showed similar enrichment results regarding biological processes
(Fig. 5B and C, pink
rectangles).
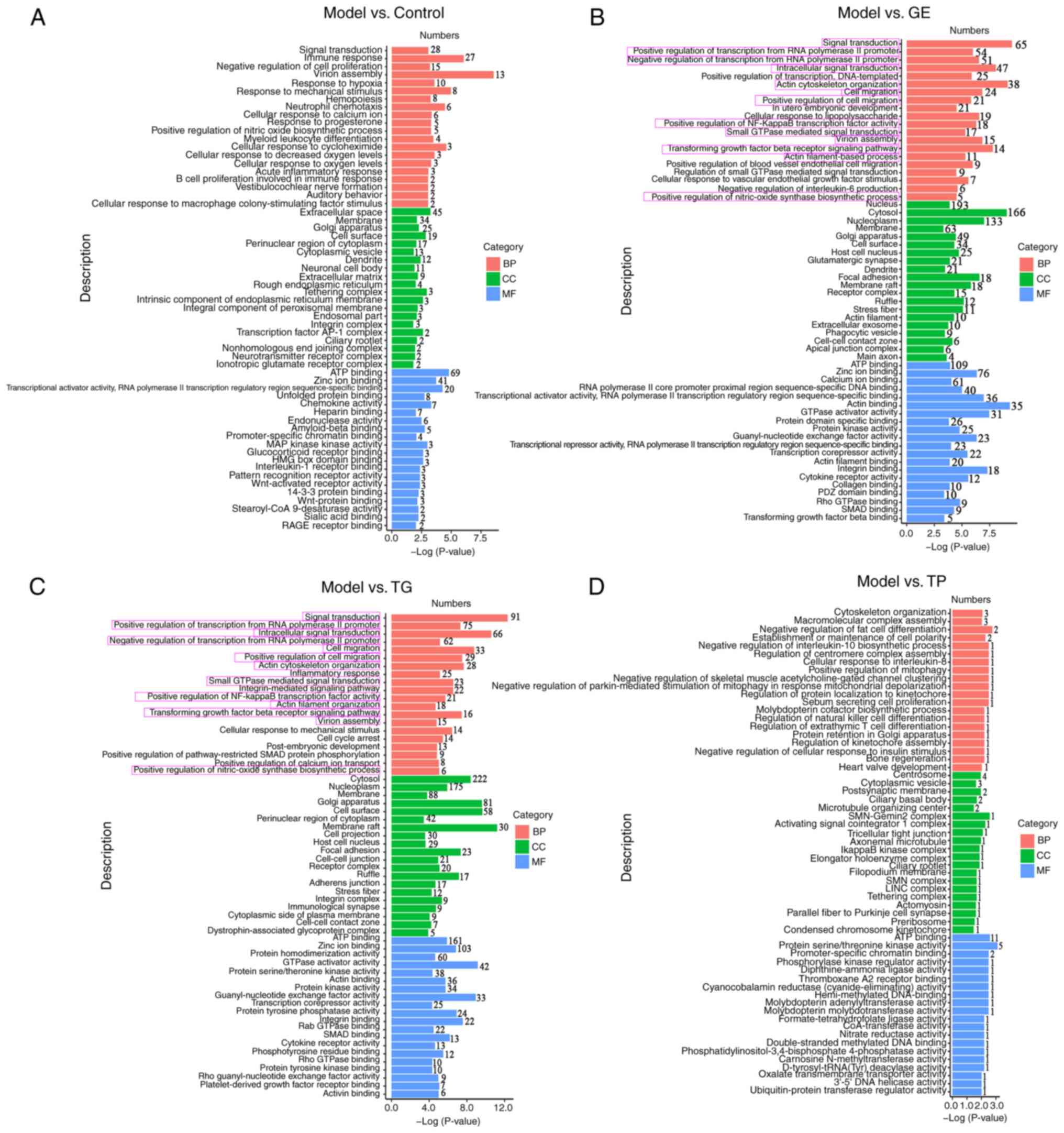 | Figure 5Top 20 GO terms associated with the
upregulated DEGs in hematopoietic stem cells between the various
indicated two treatment groups. Top 20 GO terms associated with the
DEGs between the (A) Model and Control groups, (B) Model and GE
groups, (C) Model and TG groups and (D) Model and TP groups. BP,
biological processes; CC, cellular components; DEGs, differentially
expressed genes; GE, ginseng extract; GO, Gene Ontology; MF,
molecular functions; TG, total ginsenosides; TP, total
polysaccharides. |
In the cellular component analysis, compared with
the Model group, the upregulated genes in the GE and TG groups were
mainly enriched in the 'cytosol', 'nucleoplasm', 'membrane', 'Golgi
apparatus' and 'cell surface' (Fig.
5B and C), where the upregulated genes in the GE group were
also enriched in the 'nucleus'. By contrast, the upregulated genes
in the TP group were primarily related to the 'centrosome',
'cytoplasmic vesicle' and 'postsynaptic membrane' compared with the
Model group (Fig. 5D). The main
cellular component enriched by TP were different from GE and
TG.
In the molecular function analysis, the upregulated
genes in the Control, GE, TG and TP groups were mainly enriched in
'ATP binding' (Fig. 5A-C). In
particular, the upregulated DEGs in the GE and TG groups were also
enriched in 'zinc ion binding,' 'GTPase activator activity', 'actin
binding', 'protein kinase activity', 'guanyl-nucleotide exchange
factor activity', 'transcription corepressor activity', 'integrin
binding', 'SMAD binding', 'cytokine receptor activity' and 'Rho
GTPase binding' (Fig. 5B and C).
However, TP showed obvious differences compared with the Control,
GE and TG groups regarding molecular function. The upregulated
genes in the TP group were mainly enriched in 'protein
serine/threonine kinase activity', 'promoter-specific chromatin
binding' and 'phosphorylase kinase regulator activity' (Fig. 5D). The majority of the genes
enriched in the GE and TG groups were consistent in the top 20 GO
analysis.
KEGG analysis of the upregulated DEGs
from HSCs
A total of 4,988 DEGs were involved in 297, 282, 306
and 125 KEGG pathways in the Control, GE, TG and TP groups,
respectively. The DEGs were divided mainly into four branches based
on KEGG annotations: Cellular processes, environmental information
processing, human diseases and organismal systems. In cellular
processes, the upregulated DEGs were enriched in 'endocytosis' and
'apoptosis' pathways in the Control, GE, TG and TP groups (Fig. 6A-D). Furthermore, 20 and 37 DEGs
were enriched in 'signaling pathways regulating pluripotency of
stem cells' in the GE and TG groups (Fig. 6B and C). In environmental
information processing, the DEGs in the GE and TG groups were
enriched mainly in the 'MAPK signaling pathway', 'PI3K-Akt
signaling pathway', 'Rap1 signaling pathway', 'cytokine-cytokine
receptor interaction', 'Ras signaling pathway', 'Wnt signaling
pathway', 'apelin signaling pathway', 'JAK-STAT signaling pathway'
and 'NF-kappa B signaling pathway'. The majority of these pathways
are involved in the regulation and differentiation of HSCs
(Fig. 6B and C) (36,37). The DEGs in the TP group were also
mainly enriched in the 'MAPK signaling pathway', 'NF-kappa B
signaling pathway', 'Rap1 signaling pathway', 'Wnt signaling
pathway', 'apelin signaling pathway' and 'cell adhesion molecules'.
In organismal systems, the DEGs were involved in 'osteoclast
differentiation', 'Chemokine signaling pathway', 'C-type lectin
receptor signaling pathway' and 'NOD-like receptor signaling
pathway' in the Control, GE, and TG groups (Fig. 6A-C). TP exhibited notable
differences compared with the GE and TG groups regarding organismal
systems, where the DEGs were enriched in 'parathyroid hormone
synthesis, secretion and action' and 'aldosterone synthesis and
secretion' (Fig. 6D). The DEGs
from the GE and TG groups participated mainly in regulating
HSCs-related signaling pathways, such as 'MAPK signaling pathway',
'PI3K-Akt signaling pathway', 'Rap1 signaling pathway',
'cytokine-cytokine receptor interaction', 'Ras signaling pathway',
'Wnt signaling pathway', 'JAK-STAT signaling pathway' and 'NF-kappa
B signaling pathway'. In addition, TP modulated HSCs-related
signaling pathways. However, there were clear differences between
the GE/TG and TP groups in terms of the profile of DEGs.
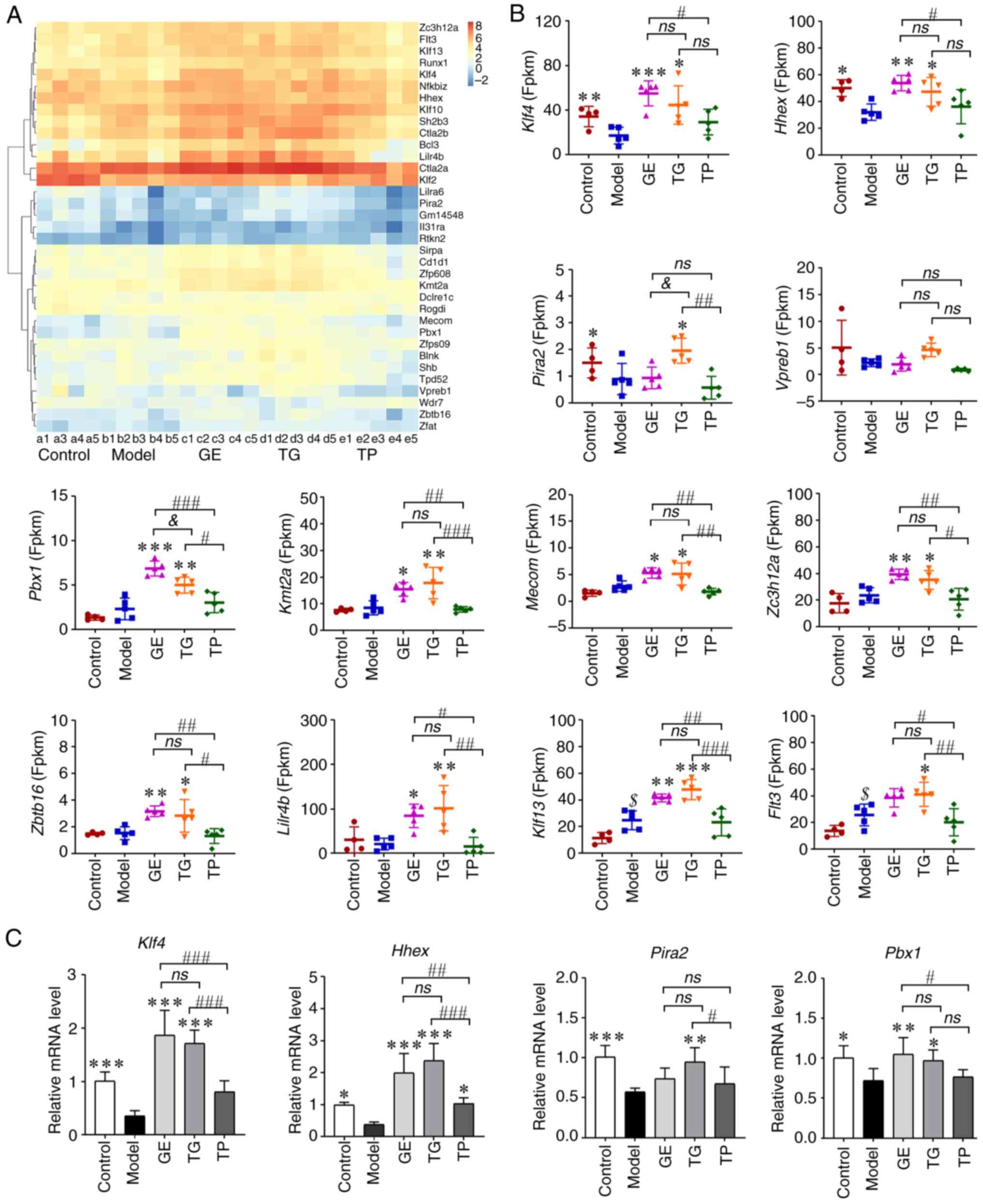
Expression analysis of the
hematopoietic-related genes of the upregulated DEGs from HSCs
According to the GO analysis, the
hematopoietic-related term of the upregulated DEGs had
'hemopoiesis', 'embryonic hemopoiesis', 'post-embryonic
hemopoiesis', 'definitive hemopoiesis', 'regulation of
hemopoiesis', 'negative regulation of hemopoiesis' and 'positive
regulation of hemopoiesis'. Compared with the Model group, 9, 19,
23 and 1 DEGs were enriched in hematopoietic-related term in the
Control, GE, TG and TP groups, respectively (Table SI). A total of 35 upregulated
hematopoietic-related genes were clustered according to FPKM
(Fig. 7A; Table SI). As shown in Fig. 7B, CY markedly decreased the
expression levels of Klf4, Hhex and Pira2,
whilst increasing those of Flt3 and Klf13, compared
with those in the Control group. These results indicated that CY
affected HSCs. GE and TG significantly increased Klf4 and
Hhex expression compared with that in the Model group,
whereas the expression levels of Hhex and Klf4 were
higher in the GE group compared with those in the TP group
(Fig. 7B). The expression levels
of Pbx1, Kmt2a, Mecom, Zc3h12a,
Zbtb16, Lilr4b, Flt3 and Klf13 were
higher in the GE and TG groups than those in the Model and TP
groups (Fig. 7B). In addition,
TG markedly increased the expression of Pira2 compared with
that in the Model, GE and TP groups (Fig. 7B). Vpreb1 expression in
the Control and TG groups was higher compared with that in the
Model group, but the difference was not statistically significant
(Fig. 7B). These results suggest
that both TG and GE activated the expression of
hematopoietic-related genes, though TP conferred almost no
activating effect on the HSCs.
The mRNA expression levels of several genes were
next detected in HSCs. As shown in Fig. 7C, CY markedly decreased the
expression levels of Klf4, Hhex, Pira2 and
Pbx1. In the drug administration groups, GE significantly
upregulated the expression levels of Klf4, Hhex and
Pbx1 compared with those in the Model and TP groups
(Fig. 7C). TG also increased
Klf4, Hhex and Pira2 expression compared with
that in the Model and TP groups (Fig. 7C). The expression of Pbx1
in HSCs treated with TG was also significantly higher compared with
that in the Model group (Fig.
7C). TP only significantly increased the expression of
Hhex compared with the Model group (Fig. 7C). Other genes measured in the TP
group exhibited slightly higher expression compared with those in
the Model group, but no statistical significance could be found. In
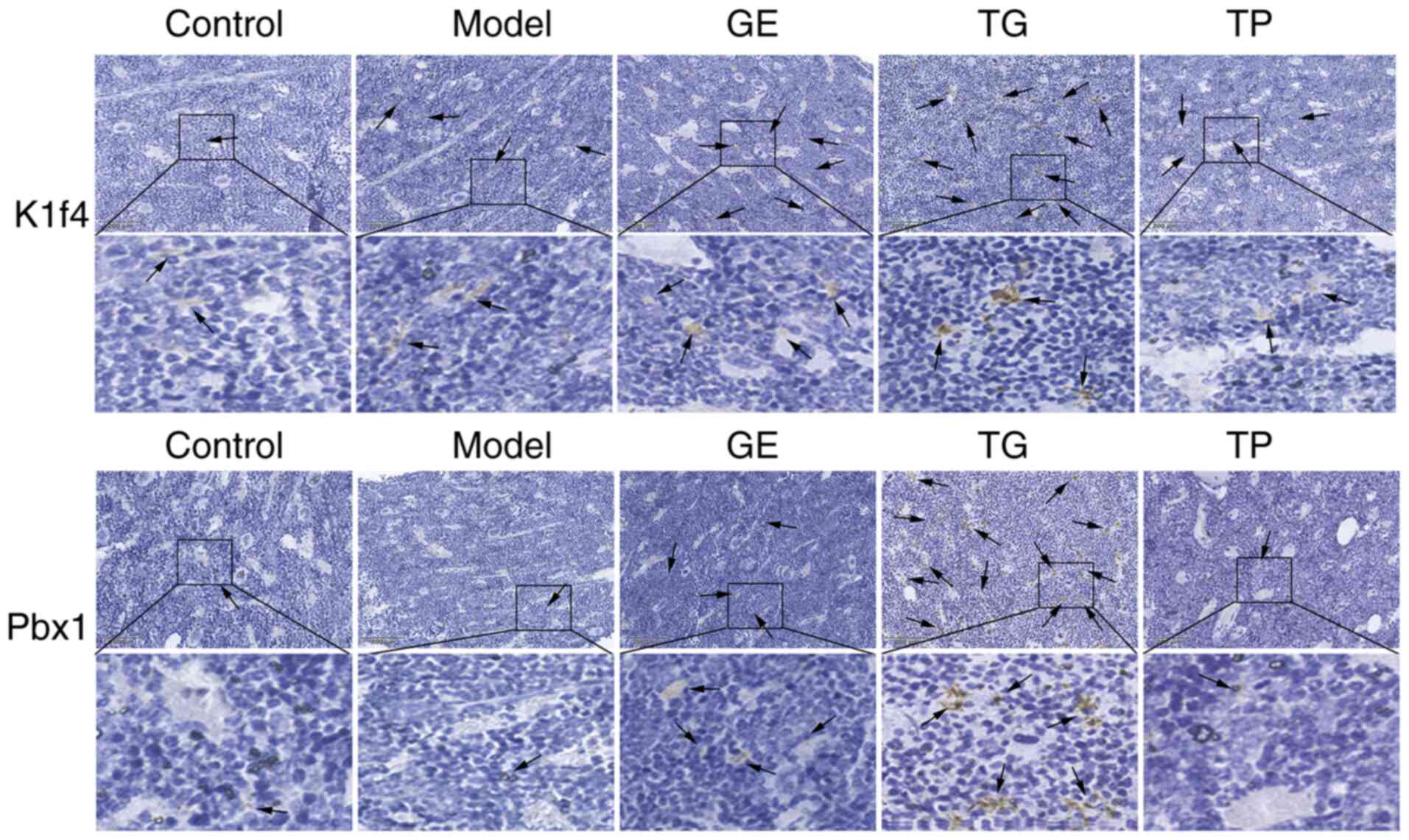
addition, the protein expression levels of Klf4 and Pbx1 were
detected by immunohistochemistry. As shown in Fig. 8A and B, positive Klf4 and Pbx1
protein expression was markedly increased in the TG and GE groups
compared with that in the Model group. The aforementioned results
appeared to be consistent with the results of RNA-seq analysis.
Effect of GE, TG and TP on the apoptosis
of HSCs
CY was found to promote the apoptosis of HSCs at
the early and late apoptosis stages, whereas GE, TG and TP could
inhibit the early apoptosis, late apoptosis and total apoptosis of
HSCs induced by CY (Fig. 9A). TG
exhibited the strongest effect on inhibiting cell apoptosis,
whereas the effect of TG and GE on inhibiting cell apoptosis was
significantly superior to TP (Fig.
9A). Furthermore, the expression levels of anti-apoptosis
(Bcl2 and Mcl1) and pro-apoptosis (Bax,
Bid and Bad) DEGs were next analyzed. The results
showed that CY significantly inhibited the expression of
Mcl1 and increased the expression of Bax (Fig. 9B). TG significantly increased the
expression levels of Bcl2 and Mcl1, whilst decreasing
those of Bax compared with the Model group. GE significantly
inhibited the expression levels of Bax and Bad
compared with those in the Model and TP groups (Fig. 9B). In addition, the expression
levels of Bid were significantly lower in the GE group
compared with those in the TP group (Fig. 9B). Notably, TP had little effect
on the expression levels of apoptotic genes compared with the Model
group. According to the western blotting analysis, TG increased the
protein expression levels of Bcl2, whereas GE and TG decreased the
protein expression levels of Bax compared with the Model group
(Fig. 9C). These results suggest
that both GE and TG inhibited the apoptosis of HSCs. However, the
inhibitory effect of TP on cell apoptosis was weaker compared with
that of GE and TG.
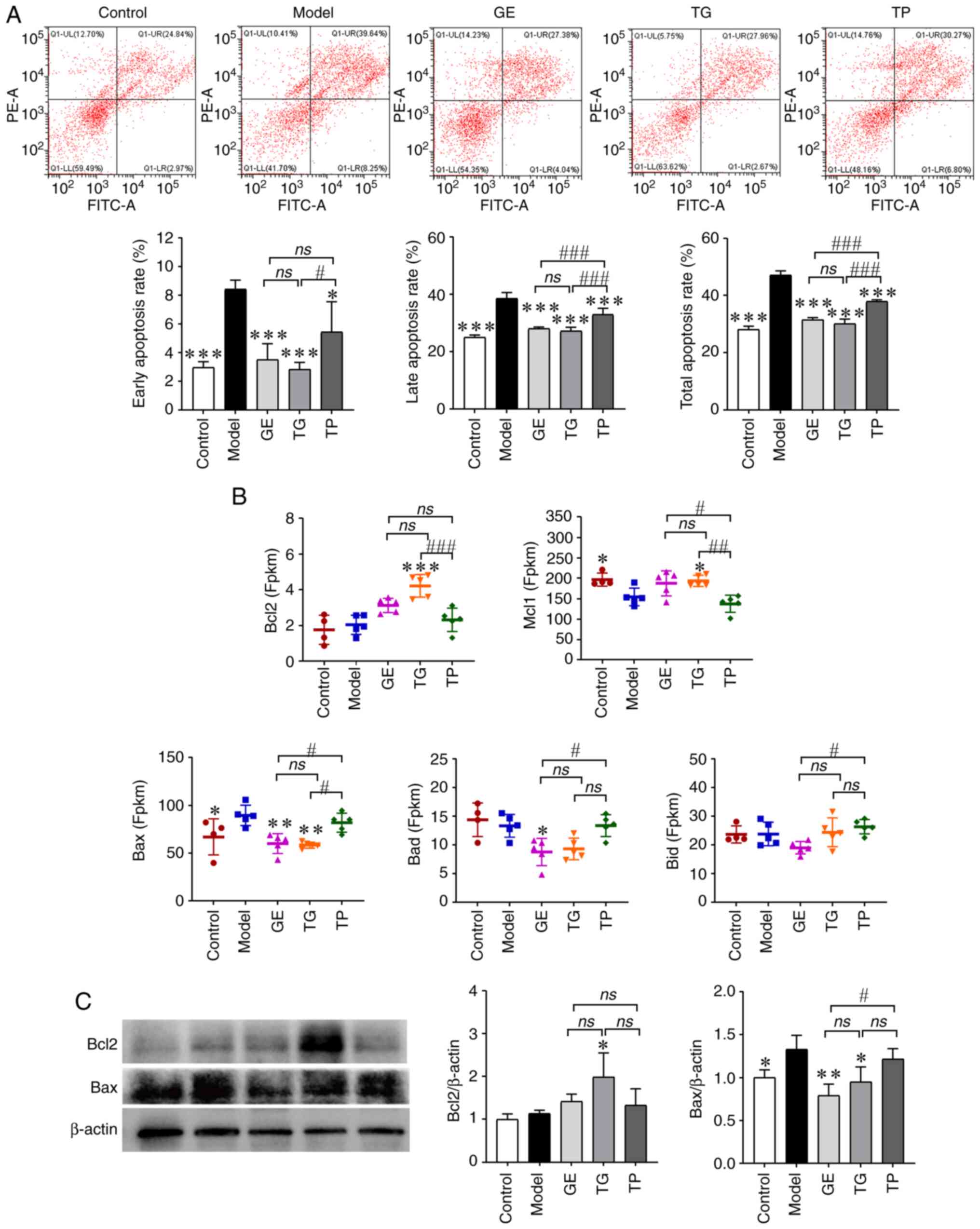 | Figure 9Effect of GE, TG and TP on the
apoptosis of HSCs. (A) Apoptosis analysis of HSCs by flow
cytometry. (B) Expression of apoptosis-associated genes among the
differentially expressed genes. (C) Protein expression of Bcl2 and
Bax. Data are presented as the mean ± SD (n=5),
*P<0.05, **P<0.01 and
***P<0.001 vs. Model group; #P<0.05,
##P<0.01 and ###P<0.001. GE, ginseng
extract; Mcl1, myeloid cell leukemia-1; HSCs, hematopoietic stem
cells; ns, no significance; TG, total ginsenosides; TP, total
polysaccharides. |
Effect of GE, TG and TP on the erythroid
differentiation of HSCs and blood cell parameters
TER-119 has been previously used as a marker of
erythroid differentiation of HSCs (38). The present study therefore
detected TER-119 expression to assess the differentiation ability
of HSCs induced by CY. The results showed that CY significantly
reduced the expression of TER-119 compared with that in the Control
group (Fig. 10A). By contrast,
GE markedly increased the expression of TER-119 compared with that
in the Model and TP groups (Fig.
10A). The expression of TER-119 in the TG and TP groups was not
statistically different compared with that in the Model group. In
the blood cell parameter analysis, the numbers of all peripheral
blood cells were found to be significantly reduced in the Model
mice (Fig. 10B), which were all
significantly reversed by GE and TG [namely WBC, neutrophils,
LYMPH, RBC, HGB, Ret and PLT; Fig.
10B]. By contrast, TP could only restore the levels of LYMPH,
RBC, HGB and PLT compared with those in the Model group (Fig. 10B). TER-119 was expressed in Ret
from HSCs (39), and this
increasing trend in TER-119, Ret and RGB is consistent among the
GE, TG, and TP groups. Notably, the improvement effect of TP on
blood cells was weaker compared with that of GE and TG.
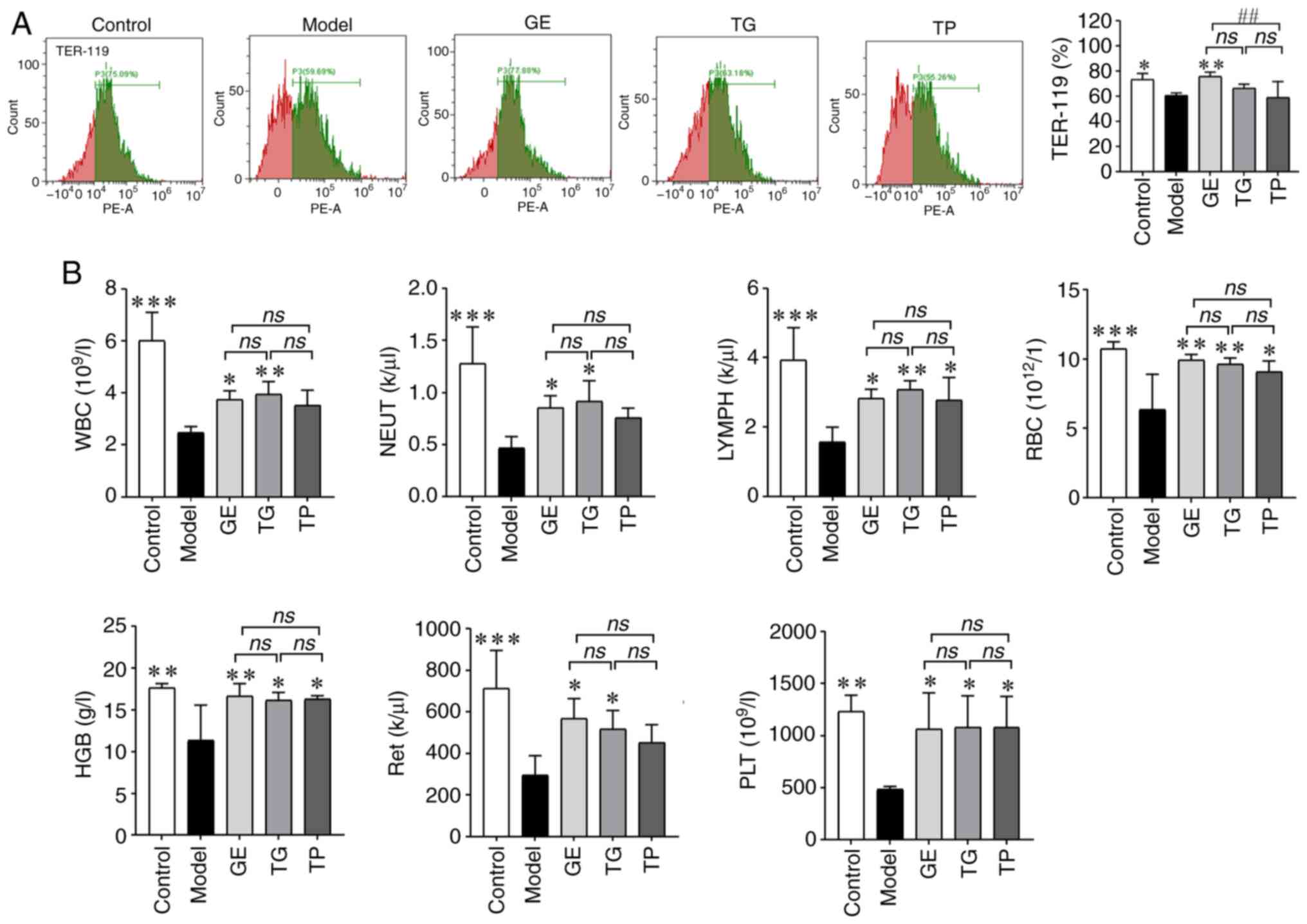 | Figure 10Effect of GE, TG and TP on the
erythroid differentiation of hematopoietic stem cells and blood
cell parameters. (A) Expression of TER-119 detected by flow
cytometry. (B) Effect of GE, TG and TP on blood cell parameters.
Data are presented as the mean ± SD (n=5). *P<0.05,
**P<0.01 and ***P<0.001 vs. Model
group; ##P<0.01. GE, ginseng extract; HGB,
hemoglobin; LYMPH, lymphocyte; NEUT, neutrophil; ns, no
significance; PLT, platelet; RBC, red blood cell; Ret,
reticulocyte; TG, total ginsenosides; TP, total polysaccharides;
WBC, white blood cell. |
Discussion
CY is a widely used chemotherapeutic drug for the
treatment of various types of cancer, such as lymphoma, breast
cancer and ovarian cancer (1,2,4).
CY can impair HSCs to affect the production of new blood cells,
further resulting in anemia, infection and bleeding (40). In addition, CY can disrupt the
balance of the intestinal microflora (7). Ginseng has previously exhibited
dual efficacy in inhibiting cancer cells and protecting the immune
system (29,41). Therefore, the present study
assessed the effect of GE, TG and TP on the intestinal microflora
and HSCs from model mice induced by CY. After drug administration
for 28 days, it was revealed that GE and TG restored the
phylum/genus/species levels of intestinal bacteria after CY
treatment. In addition, the protective effects of GE and TG on HSCs
were superior to those of TP, the latter of which was not effective
at preventing CY-induced HSC damage.
The intestinal microflora regulates host metabolism
and immune homeostasis, in turn maintaining the physiology of the
body (18,42). The α diversity estimators
revealed that CY decreased the relative abundance of some dominant
microflora, whilst increasing that of some low-abundance
microflora. Lactobacillus is a dominant and beneficial
bacteria in the intestinal microflora, which has immunoregulatory
effects by regulating the activity of epithelial cells,
macrophages, dendritic cells and regulatory T cells (43). CY significantly decreased the
relative abundance of Lactobacillus and Lactobacillus_
intestinalis at the genus and species levels. American ginseng
ginsenoside, cotreatment with American ginseng polysaccharide and
ginsenoside have all been reported to increase the abundance of
Lactobacillus in CY-induced mice (7). The present study showed that GE
exhibited the optimal improvement at the phylum and genus levels,
whereas TG showed the optimal improvement at the species level. A
previous study has shown that the ginsenoside content of TG and GE
is 4.39 and 81.09%, respectively, whilst the polysaccharide content
of GE, TG and TP is 72.28, 4.68 and 89.79%, respectively (26). Notably, ginsenoside content was
revealed to be significantly positively correlated with the
abundance of Lactobacillus_ intestinalis in the present
study. Ginseng polysaccharides have been reported to change the
intestinal microbiota and kynurenine/tryptophan ratio to exert
anti-inflammatory and immunoregulatory effects (41,44). In the present study, TP
significantly increased the abundance of low-abundance microflora,
such as Mouse_gut, Bacteroides_ dorei,
Prevotella_melanimogenica,
Anaerotignum_lactatifermentans, Bacteroides_plebeius
and Parabacteroides_merdae. Bacteroides_dorei can
reduce intestinal microbial lipopolysaccharide production (45), promote earlier IFN expression and
inhibit both local and systemic inflammatory responses (46). Park et al (47) previously reported that
Bacteroides_plebeius contains an endo-type β-agarase,
BpGH16A, which is the key enzyme that initiates the
depolymerization of agarose, where supplementary
Bacteroides_plebeius can recover disturbances of the
intestinal microbiota in chronic kidney disease (48). Prevotella_melanimogenica
has been associated with infections in humans (49), where an increase in the abundance
of Prevotella can improve glucose metabolism (50). Parabacteroides_merdae has
been shown to promote branched-chain amino acid catabolism to
alleviate obesity and hyperlipidemia (51). Therefore, TP likely participates
in the process of immune regulation and inflammation by regulating
the intestinal microflora of model mice induced by CY. The
aforementioned results indicated that GE, TG and TP can all
alleviate CY-induced abnormalities in intestinal microbiota.
However, the species of intestinal bacteria altered by TP was
different from those altered by GE and TG.
CY can induce the apoptosis of HSCs to reduce the
number of blood cells (6). In
the high-throughput RNA-seq analysis, CY was found to markedly
inhibit the expression levels of genes associated with
hematopoietic function (such as Hhex, Klf4 and
Pira2) of the bone marrow, whilst increasing those of
Flt3 (52) and
Klf13. The total extract from Korean red ginseng mainly
includes ginsenoside Rg1, ginsenoside Re, ginsenoside Rf,
ginsenoside Rg2, ginsenoside Rb1, ginsenoside Rb2, ginsenoside Rc
and ginsenoside Rd, which can promote CD34+ cell
expansion and hematopoietic colony formation, especially those of
the erythroid lineage (53).
Ginsenoside Rg1 exerts protective effects on HSCs by improving
redox homeostasis. Cao et al (35) previously reported that
ginsenoside Rg1 can reduce reactive oxygen species levels, increase
the number of mitochondria and the ratio of Bcl2/Bax, which
recovered hematopoietic function by inhibiting Bax
translocation-mediated mitochondrial apoptosis in aplastic anemia
mice. Ginsenoside Rd has been reported to promote human induced
pluripotent stem cell differentiation into myoblasts through the
Flt3 signaling pathway (54).
Flt3 can prevent stem cells and progenitors from spontaneous
apoptotic cell death by upregulating Mcl1, which is an
indispensable survival factor of hematopoiesis (55). CK has been shown to promote cell
cycle entry in bone marrow nucleated cells through the Bcl2/Bax and
MEK/ERK signaling pathways, which can also increase peripheral
blood cells of myelosuppression mice induced by CY (56).
In the present study, the DEGs in the GE and TG
groups were mainly enriched in HSCs-related signaling pathways,
such as PI3K/Akt signaling pathway, Rap1 signaling pathway,
cytokine-cytokine receptor interaction, Ras signaling pathway, Wnt
signaling pathway, TNF signaling pathway, apelin signaling pathway,
JAK-STAT signaling pathway and NF-κB signaling pathway (36,37). Tang et al (57) reported that Rg1 delayed
hematopoietic stem/progenitor cell senescence by increasing protein
expression of SIRT6, inhibiting protein expression of NF-κB and
overactivation of the Wnt/β-catenin through regulating the
SIRT6/NF-κB signaling pathway and Wnt/β-catenin signaling pathway.
Certain genes identified in the present study, such as Klf4,
Hhex, Pira2, Pbx1, Kmt2a, Mecom,
Zc3h12a, Zbtb16, Lilr4b, Flt3 and
Klf13, are involved in the self-renewal and differentiation
of HSCs (55,58-63), TG markedly increased the
expression levels of the aforementioned genes. GE significantly
enhanced the levels of Hhex, Klf4, Pbx1,
Kmt2a, Mecom, Zc3h12a, Zbtb16,
Lilr4b, Flt3 and Klf13, but TP barely
increased the expression of these genes. TG exerted the optimal
protective effect on HSCs, followed by GE and then TP, the latter
of which was not effective.
TP is mainly composed of macromolecular
polysaccharide components, which have difficulty directly acting on
target organs to improve tissue damage (64). However, TP has been reported to
ameliorate intestinal immune disorders and inflammation by
improving the intestinal microbiota (7). A previous study has demonstrated
that 12 ginsenosides are present in GE and TG, including nR1, Rg1,
Re, Rf, Ra2, Rb1, Rc, Ro, Rb2, Rb3, Rd and 20(R)-Rg3 (6). Therefore, it may be hypothesized
that the content of ginsenosides is positively associated with the
upregulated gene expression in HSCs. However, GE, TG and TP were
extracted by our laboratory and there is currently no standard
protocol for extraction, which is a limitation of the present
study. Chen et al (29)
reported that GE enhanced the immune regulation and anti-oxidant
levels of immunosuppressed mice caused by CY. In another study,
Tang et al (65) revealed
that the purity of TG was 90-93% by heating and refluxing with
petroleum ether and water-saturated n-butanol solution, which then
alleviated the CY-induced liver injury of mice by regulating the
imbalance of intestinal microflora. Furthermore, TG (purity
>70%) from the stem and leaf of Panax ginseng has been
shown to inhibit the apoptosis and genotoxicity of bone marrow and
peripheral lymphocyte cells induced by CY (5). In addition, TP (purity >90%) may
promote natural killer cell cytotoxicity in immunosuppressed mice
(66). Wan et al
(23) reported that TP, which
was extracted sequentially by 95% ethanol, hot water and 95%
ethanol, modulated tryptophan metabolism by improving the gut
microbiota, which resulted in the amelioration of impaired
intestinal barrier and the alleviation of the inflammatory
microenvironment formation of ulcerative colitis mice induced by
dextran sulfate sodium. Differential extraction methods will likely
lead to differences in the chemical composition of GE, TG and TP.
However, different components from ginseng can improve the damage
caused by CY.
Ginsenosides have low oral bioavailability and
absorption in the body. Zhou et al (67) reported that ginseng
polysaccharides can promote the exposure and absorption of
ginsenosides. Li et al (68) studied the synergistic effects of
TP and ginsenoside Rb1, before finding that the presence of TP
accelerated the microbial metabolism of Rb1 and promoted fecal
β-D-glucosidase activity, which then transformed into Rd and CK. In
particular, the biotransformation rate of CK was increased from
14.0 to 86.7%. Furthermore, TP can promote Rb1 transport across a
Caco-2 cell monolayer (15).
Wang et al (69) also
found that polysaccharides promoted the metabolism of ginsenosides.
In the present study, GE and TG protected HSCs from damage induced
by CY.
In the present study, animal models provided
valuable mechanistic insights. Namely, GE and TG were suggested to
activate the expression of HSC related-genes and proteins to
inhibit the apoptosis of HSCs of normal (non-tumorous) mice induced
by CY. However, their translation into a clinical setting has
specific challenges. HSC and progenitor cell self-renewal and
differentiation are complex processes that are important for the
production and long-term maintenance of all cell lineages and blood
cells (70). However, HSCs and
progenitor cells also directly and indirectly participate in both
tumor immune escape and the metastatic cascade (71,72). In the future, research on mouse
models of different types of cancer is required to confirm the
protective effect of ginsenosides on HSCs.
In conclusion, CY can damage HSCs, which can cause
intestinal microflora disorder and cell apoptosis, inhibit the
differentiation of HSCs and reduce the number of peripheral blood
cells. The present results showed that GE and TG can effectively
improve the imbalance of intestinal microflora and protect HSCs
from damage induced by CY. GE and TG inhibited the disordered gut
microbiota. Specifically, the relative abundances of
Lactobacillus and Lactobacillus_intestinalis were
higher in the GE and TG groups compared with those in the Model
group. However, TP mainly increased the abundance of beneficial
microflora with low-abundance. Furthermore, GE and TG could
activate the expression of hematopoietic-related genes by mediating
multiple signaling pathways of HSCs. GE blocked the apoptosis of
HSCs by inhibiting the expression of Bax and Bad, whereas TG
prevented the apoptosis of HSCs by promoting the expression of Bcl2
and Mcl1 and inhibiting the expression of Bax. Notably, GE is
similar to TG, it exhibited a protective effect on a mouse model of
HSC damage induced by CY. These findings suggest that ginsenosides
can prevent or alleviate HSC damage.
Supplementary Data
Availability of data and materials
The 16S rRNA sequencing data generated in the
present study may be found in the BioProject database under
accession number PRJNA934302 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA934302.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
ZL, XG, DZ, XL, HS and HZ contributed to the study
conception and design. ZL, TY, CJ, XG, WQ and LZ contributed to
project development and data collection. ZL, TY, XG and CJ
contributed to protocol development and manuscript writing. XG, LZ,
YY and JO contributed to data collection and analysis. HS and HZ
contributed to data analysis and manuscript editing. ZL, XG and HZ
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All animal experiments were approved and performed
by the Ethics Committee of Changchun University of Chinese Medicine
(Changchun, China; approval no. 2023033).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Science and Technology
Development Plan Project of Jilin Province (grant no.
YDZJ202401417ZYTS), the Science and Technology Major Project of
Jilin Province (grant no. 20210304001YY), the Science and
Technology Projects of Education Department of Jilin Province
(grant nos. JJKH20230990KJ and JJKH20230976KJ), the Cultivation
Project of Young Discipline Backbone Talent (grant no. 202320) and
the Innovation and Entrepreneurship Talent Funding Project of Jilin
Province (grant no. 2022ZY10).
References
|
1
|
Burke JM, Masaquel A, Wang R, Hossain F,
Li J, Zhou SQ, Ng CD and Matasar M: Cost of disease progression in
diffuse large B-cell lymphoma after frontline treatment with
rituximab plus cyclophosphamide, doxorubicin, vincristine, and
prednisone. Clin Lymphoma Myeloma Leuk. 23:e393–e404. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Velikova G, Morden JP, Haviland JS, Emery
C, Barrett-Lee P, Earl H, Bloomfield D, Brunt AM, Canney P, Coleman
R, et al: Accelerated versus standard epirubicin followed by
cyclophosphamide, methotrexate, and fluorouracil or capecitabine as
adjuvant therapy for breast cancer (UK TACT2; CRUK/05/19): Quality
of life results from a multicentre, phase 3, open-label,
randomised, controlled trial. Lancet Oncol. 24:1359–1374. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Ai L, Zhang Y, Li X, Xu S, Yang W,
Jin J, Ma Y, Hu Z, Zhang Y, et al: The EZH2-H3K27me3 axis modulates
aberrant transcription and apoptosis in cyclophosphamide-induced
ovarian granulosa cell injury. Cell Death Discov. 9:4132023.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang H, Fan W, Lei B, Tian Y and Zhang S:
The immunosuppression and immunoenhancement effects of
cyclophosphamide on normal mice. Immunol J. 34:308–312. 2018.
|
|
5
|
Zhang QH, Wu CF, Duan L and Yang JY:
Protective effects of total saponins from stem and leaf of Panax
ginseng against cyclophosphamide-induced genotoxicity and apoptosis
in mouse bone marrow cells and peripheral lymphocyte cells. Food
Chem Toxicol. 46:293–302. 2008. View Article : Google Scholar
|
|
6
|
Zhang H, Sun Y, Fan M, Zhang Y, Liang Z,
Zhang L, Gao X, He X, Li X, Zhao D, et al: Prevention effect of
total ginsenosides and ginseng extract from Panax ginseng on
cyclophosphamide-induced immunosuppression in mice. Phytother Res.
37:3583–3601. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou R, He D, Xie J, Zhou Q, Zeng H, Li H
and Huang L: The synergistic effects of polysaccharides and
ginsenosides from American ginseng (Panax quinquefolius L.)
ameliorating cyclophosphamide-induced intestinal immune disorders
and gut barrier dysfunctions based on microbiome-metabolomics
analysis. Front Immunol. 12:6659012021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Viaud S, Saccheri F, Mignot G, Yamazaki T,
Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ,
et al: The intestinal microbiota modulates the anticancer immune
effects of cyclophosphamide. Science. 342:971–976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi L, Wang C and Yuan C: Ginsenosides from
American ginseng: Chemical and pharmacological diversity.
Phytochemistry. 72:689–699. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qu D, Huo XH, Li ZM, Hua M, Lu YS, Chen
JB, Li SS, Wen LK and Sun YS: Sediment formation and analysis of
the main chemical components of aqueous extracts from different
parts of ginseng roots. Food Chem. 379:1321462022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yance DR Jr and Sagar SM: Targeting
angiogenesis with integrative cancer therapies. Integr Cancer Ther.
5:9–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wan Y, Wang J, Xu JF, Tang F, Chen L, Tan
YZ, Rao CL, Ao H and Peng C: Panax ginseng and its ginsenosides:
potential candidates for the prevention and treatment of
chemotherapy-induced side effects. J Ginseng Res. 45:617–630. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia L, Zhao Y and Liang XJ: Current
evaluation of the millennium phytomedicine-ginseng (II): Collected
chemical entities, modern pharmacology, and clinical applications
emanated from traditional Chinese medicine. Curr Med Chem.
16:2924–2942. 2009. View Article : Google Scholar
|
|
14
|
Cockburn DW and Koropatkin NM:
Polysaccharide degradation by the intestinal microbiota and its
influence on human health and disease. J Mol Biol. 428:3230–3252.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen H, Gao XJ, Li T, Jing WH, Han BL, Jia
YM, Hu N, Yan ZX, Li SL and Yan R: Ginseng polysaccharides enhanced
ginsenoside Rb1 and microbial metabolites exposure through
enhancing intestinal absorption and affecting gut microbial
metabolism. J Ethnopharmacol. 216:47–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maiuolo J, Musolino V, Gliozzi M, Carresi
C, Scarano F, Nucera S, Scicchitano M, Oppedisano F, Bosco F, Macri
R, et al: Involvement of the intestinal microbiota in the
appearance of multiple sclerosis: Aloe vera and citrus bergamia as
potential candidates for intestinal health. Nutrients. 14:27112022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li S, Han W, He Q, Zhang W and Zhang Y:
Relationship between intestinal microflora and hepatocellular
cancer based on gut-liver axis theory. Contrast Media Mol Imaging.
2022:65336282022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marchesi JR, Adams DH, Fava F, Hermes GD,
Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM,
et al: The gut microbiota and host health: A new clinical frontier.
Gut. 65:330–339. 2016. View Article : Google Scholar
|
|
19
|
Yang Z and Ji G: Fusobacterium
nucleatum-positive colorectal cancer. Oncol Lett. 18:975–982.
2019.PubMed/NCBI
|
|
20
|
Kostic AD, Gevers D, Pedamallu CS, Michaud
M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et
al: Genomic analysis identifies association of Fusobacterium with
colorectal carcinoma. Genome Res. 22:292–298. 2012. View Article : Google Scholar :
|
|
21
|
Kim YK and Yum KS: Effects of red ginseng
extract on gut microbial distribution. J Ginseng Res. 46:91–103.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding Q, Feng SW, Xu GH, Chen YY and Shi
YY: Effects of total ginsenosides from Panax ginseng stems and
leaves on gut microbiota and short-chain fatty acids metabolism in
acute lung injury mice. Zhongguo Zhong Yao Za Zhi. 48:1319–1329.
2023.In Chinese. PubMed/NCBI
|
|
23
|
Wan L, Qian C, Yang C, Peng S, Dong G,
Cheng P, Zong G, Han H, Shao M, Gong G, et al: Ginseng
polysaccharides ameliorate ulcerative colitis via regulating gut
microbiota and tryptophan metabolism. Int J Biol Macromol.
265:1308222024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazur L, Augustynek A, Deptała A, Halicka
HD and Bedner E: Effects of WR-2721 and cyclophosphamide on the
cell cycle phase specificity of apoptosis in mouse bone marrow.
Anticancer Drugs. 13:751–758. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang JB, Du MW and Zheng Y: Effect of
ginsenoside Rg1 on hematopoietic stem cells in treating aplastic
anemia in mice via MAPK pathway. World J Stem Cells. 16:591–603.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Zhang L, Zhou W, Zhang X, Su W,
Wei X and Zhang H: Protective effect of various types of ginseng
extracts on blood deficiency induced by cyclophosphamide in rat.
Lishizhen Med Mater Med Res. 33:2861–2867. 2022.In Chinese.
|
|
27
|
Duan Y, Huang J, Sun M, Jiang Y, Wang S,
Wang L, Yu N, Peng D, Wang Y, Chen W and Zhang Y: Poria cocos
polysaccharide improves intestinal barrier function and maintains
intestinal homeostasis in mice. Int J Biol Macromol.
249:1259532023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boonlert W, Benya-Aphikul H, Umka Welbat J
and Rodsiri R: Ginseng extract G115 attenuates ethanol-induced
depression in mice by increasing brain BDNF levels. Nutrients.
9:9312017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen LX, Qi YL, Qi Z, Gao K, Gong RZ, Shao
ZJ, Liu SX, Li SS and Sun YS: A comparative study on the effects of
different parts of Panax ginseng on the immune activity of
cyclophosphamide-induced immunosuppressed mice. Molecules.
24:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Parasuraman S, Raveendran R and Kesavan R:
Blood sample collection in small laboratory animals. J Pharmacol
Pharmacother. 1:87–93. 2010. View Article : Google Scholar
|
|
31
|
Zhao H, Lyu Y, Zhai R, Sun G and Ding X:
Metformin mitigates sepsis-related neuroinflammation via modulating
gut microbiota and metabolites. Front Immunol. 13:7973122022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Cao H, Wei W, Xu R and Cui X: Ginsenoside
Rg1 can restore hematopoietic function by inhibiting Bax
translocation-mediated mitochondrial apoptosis in aplastic anemia.
Sci Rep. 11:127422021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
de Roo JJD and Staal FJT: Cell signaling
pathway reporters in adult hematopoietic stem cells. Cells.
9:22642020. View Article : Google Scholar :
|
|
37
|
Montazersaheb S, Ehsani A, Fathi E and
Farahzadi R: Cellular and molecular mechanisms involved in
hematopoietic stem cell aging as a clinical prospect. Oxid Med Cell
Longev. 2022:27134832022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An X and Chen L: Flow cytometry (FCM)
analysis and fluorescence-activated cell sorting (FACS) of
erythroid cells. Methods Mol Biol. 1698:153–174. 2018. View Article : Google Scholar
|
|
39
|
Asari S, Sakamoto A, Okada S, Ohkubo Y,
Arima M, Hatano M, Kuroda Y and Tokuhisa T: Abnormal erythroid
differentiation in neonatal bcl-6-deficient mice. Exp Hematol.
33:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kurtin S: Myeloid toxicity of cancer
treatment. J Adv Pract Oncol. 3:209–224. 2012.PubMed/NCBI
|
|
41
|
Huang J, Liu D, Wang Y, Liu L, Li J, Yuan
J, Jiang Z, Jiang Z, Hsiao WW, Liu H, et al: Ginseng
polysaccharides alter the gut microbiota and kynurenine/tryptophan
ratio, potentiating the antitumour effect of antiprogrammed cell
death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1)
immunotherapy. Gut. 71:734–745. 2022. View Article : Google Scholar
|
|
42
|
O'Hara AM, O'Regan P, Fanning A, O'Mahony
C, Macsharry J, Lyons A, Bienenstock J, O'Mahony L and Shanahan F:
Functional modulation of human intestinal epithelial cell responses
by Bifidobacterium infantis and Lactobacillus salivarius.
Immunology. 118:202–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Christensen HR, Frøkiaer H and Pestka JJ:
Lactobacilli differentially modulate expression of cytokines and
maturation surface markers in murine dendritic cells. J Immunol.
168:171–178. 2002. View Article : Google Scholar
|
|
44
|
Li S, Huo X, Qi Y, Ren D, Li Z, Qu D and
Sun Y: The protective effects of Ginseng polysaccharides and their
effective subfraction against dextran sodium sulfate-induced
colitis. Foods. 11:9802022.
|
|
45
|
Yoshida N, Emoto T, Yamashita T, Watanabe
H, Hayashi T, Tabata T, Hoshi N, Hatano N, Ozawa G, Sasaki N, et
al: Bacteroides vulgatus and Bacteroides dorei reduce gut microbial
lipopolysaccharide production and inhibit atherosclerosis.
Circulation. 138:2486–2498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Song L, Huang Y, Liu G, Li X, Xiao Y, Liu
C, Zhang Y, Li J, Xu J, Lu S and Ren Z: A novel immunobiotics
Bacteroides dorei ameliorates influenza virus infection in mice.
Front Immunol. 12:8288872022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park NJ, Yu S, Kim DH, Yun EJ and Kim KH:
Characterization of BpGH16A of Bacteroides plebeius, a key enzyme
initiating the depolymerization of agarose in the human gut. Appl
Microbiol Biotechnol. 105:617–625. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pei T, Zhu D, Yang S, Hu R, Wang F, Zhang
J, Yan S, Ju L, He Z, Han Z, et al: Bacteroides plebeius improves
muscle wasting in chronic kidney disease by modulating the
gut-renal muscle axis. J Cell Mol Med. 26:6066–6078. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tett A, Pasolli E, Masetti G, Ercolini D
and Segata N: Prevotella diversity, niches and interactions with
the human host. Nat Rev Microbiol. 19:585–599. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kovatcheva-Datchary P, Nilsson A, Akrami
R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I and
Bäckhed F: Dietary fiber-induced improvement in glucose metabolism
is associated with increased abundance of Prevotella. Cell Metab.
22:971–982. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qiao S, Liu C, Sun L, Wang T, Dai H, Wang
K, Bao L, Li H, Wang W, Liu SJ and Liu H: Gut Parabacteroides
merdae protects against cardiovascular damage by enhancing
branched-chain amino acid catabolism. Nat Metab. 4:1271–1286. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
El-Serafi I, Abedi-Valugerdi M, Potácová
Z, Afsharian P, Mattsson J, Moshfegh A and Hassan M:
Cyclophosphamide alters the gene expression profile in patients
treated with high doses prior to stem cell transplantation. PLoS
One. 9:e866192014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kim SG, Lee AJ, Bae SH, Kim SM, Lee JH,
Kim MJ and Jang HB: Total extract of Korean red ginseng facilitates
human bone marrow hematopoietic colony formation in vitro. Blood
Res. 49:177–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sun C, Choi IY, Rovira Gonzalez YI,
Andersen P, Talbot CC Jr, Iyer SR, Lovering RM, Wagner KR and Lee
G: Duchenne muscular dystrophy hiPSC-derived myoblast drug screen
identifies compounds that ameliorate disease in mdx mice. JCI
Insight. 5:e1342872020.PubMed/NCBI
|
|
55
|
Kikushige Y, Yoshimoto G, Miyamoto T, Iino
T, Mori Y, Iwasaki H, Niiro H, Takenaka K, Nagafuji K, Harada M, et
al: Human Flt3 is expressed at the hematopoietic stem cell and the
granulocyte/macrophage progenitor stages to maintain cell survival.
J Immunol. 180:7358–7367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Han J, Wang Y, Cai E, Zhang L, Zhao Y, Sun
N, Zheng X and Wang S: Study of the effects and mechanisms of
ginsenoside compound K on myelosuppression. J Agric Food Chem.
67:1402–1408. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang YL, Zhou Y, Wang YP, Wang JW and Ding
JC: SIRT6/NF-κB signaling axis in ginsenoside Rg1-delayed
hematopoietic stem/progenitor cell senescence. Int J Clin Exp
Pathol. 8:5591–5596. 2015.
|
|
58
|
Jackson JT, O'Donnell K, Light A, Goh W,
Huntington ND, Tarlinton DM and McCormack MP: Hhex regulates murine
lymphoid progenitor survival independently of Stat5 and Cdkn2a. Eur
J Immunol. 50:959–971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Klaewsongkram J, Yang Y, Golech S, Katz J,
Kaestner KH and Weng NP: Krüppel-like factor 4 regulates B cell
number and activation-induced B cell proliferation. J Immunol.
179:4679–4684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Feinberg MW, Wara AK, Cao Z, Lebedeva MA,
Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, et
al: The Kruppel-like factor KLF4 is a critical regulator of
monocyte differentiation. EMBO J. 26:4138–4148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ficara F, Murphy MJ, Lin M and Cleary ML:
Pbx1 regulates self-renewal of long-term hematopoietic stem cells
by maintaining their quiescence. Cell Stem Cell. 2:484–496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Voit RA and Sankaran VG: MECOM deficiency:
From bone marrow failure to impaired B-cell development. J Clin
Immunol. 43:1052–1066. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Matsushita K, Takeuchi O, Standley DM,
Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T,
Nakamura H and Akira S: Zc3h12a is an RNase essential for
controlling immune responses by regulating mRNA decay. Nature.
458:1185–1190. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang X, Luo Y, Xiu W, Xu M and Ma Y:
Preparation and evaluation of sweet corn cob polysaccharide nano
emulsion. Sci Tech Food Ind. 43:124–133. 2022.
|
|
65
|
Tang P, Ren G, Zou H, Liu S, Zhang J, Ai
Z, Hu Y, Cui L, Nan B, Zhang Z and Wang Y: Ameliorative effect of
total ginsenosides from heat-treated fresh ginseng against
cyclophosphamide-induced liver injury in mice. Curr Res Food Sci.
8:1007342024. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sun Y, Guo M, Feng Y, Zheng H, Lei P, Ma
X, Han X, Guan H and Hou D: Effect of ginseng polysaccharides on NK
cell cytotoxicity in immunosuppressed mice. Exp Ther Med.
12:3773–3777. 2016. View Article : Google Scholar
|
|
67
|
Zhou S, Xu J, Zhu H, Wu J, Xu JD, Yan R,
Li XY, Liu HH, Duan SM, Wang Z, et al: Gut microbiota-involved
mechanisms in enhancing systemic exposure of ginsenosides by
coexisting polysaccharides in ginseng decoction. Sci Rep.
6:224742016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li J, Li R, Li N, Zheng F, Dai Y, Ge Y,
Yue H and Yu S: Mechanism of antidiabetic and synergistic effects
of ginseng polysaccharide and ginsenoside Rb1 on diabetic rat
model. J Pharm Biomed Anal. 158:451–460. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang HY, Wang C, Guo SC, Chen ZC, Peng ZT,
Duan R, Dong TTX and Tsim KWK: Polysaccharide deriving from
Ophiopogonis Radix promotes metabolism of ginsenosides in the
present of human gut microbiota based on UPLC-MS/MS assay. J Pharm
Biomed Anal. 175:1127792019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gbyli R, Song Y and Halene S: Humanized
mice as preclinical models for myeloid malignancies. Biochem
Pharmacol. 174:1137942020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cackowski FC and Taichman RS: Parallels
between hematopoietic stem cell and prostate cancer disseminated
tumor cell regulation. Bone. 119:82–86. 2019. View Article : Google Scholar
|
|
72
|
Steenbrugge J, De Jaeghere EA, Meyer E,
Denys H and De Wever O: Splenic hematopoietic and stromal cells in
cancer progression. Cancer Res. 81:27–34. 2021. View Article : Google Scholar
|