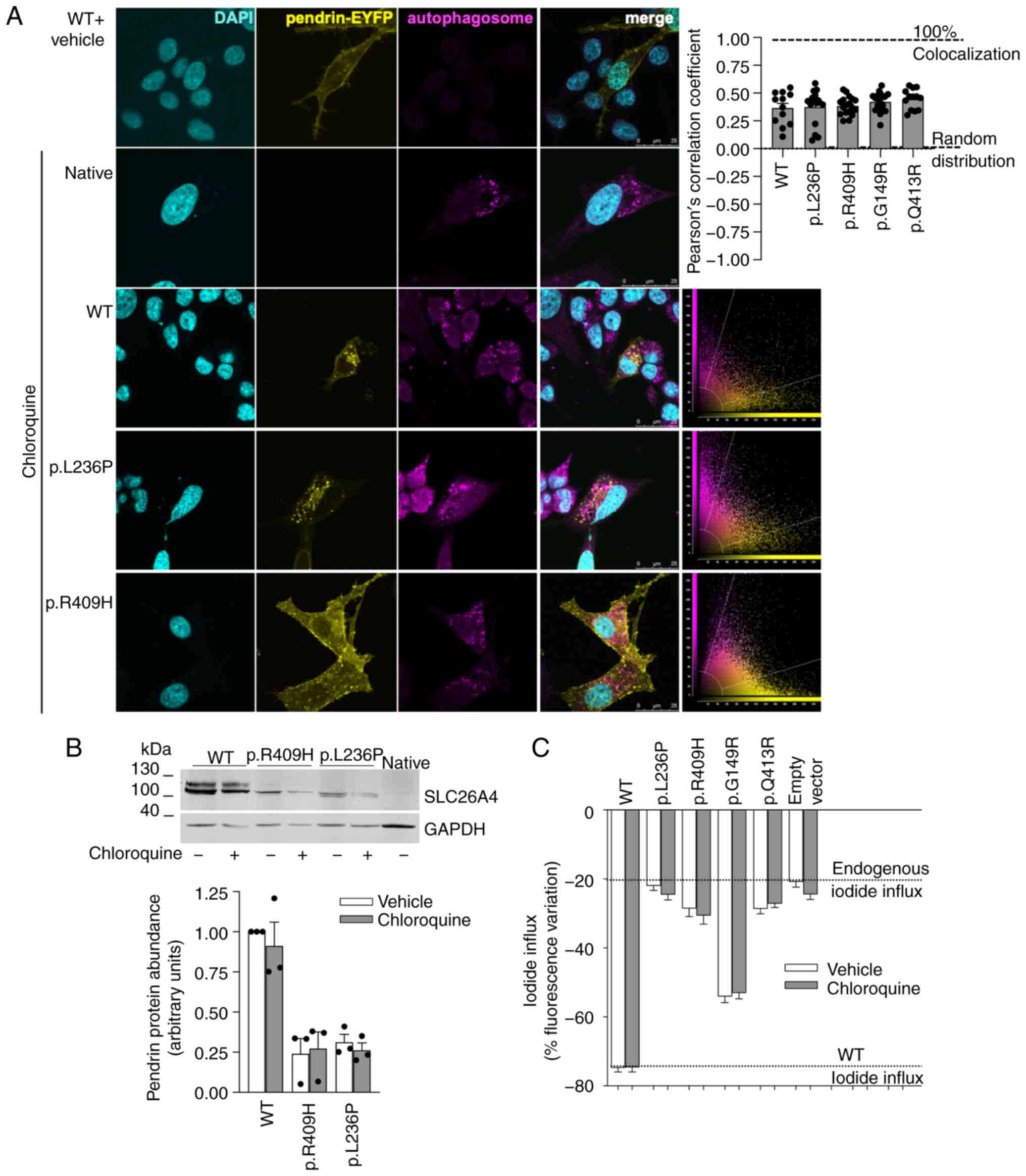
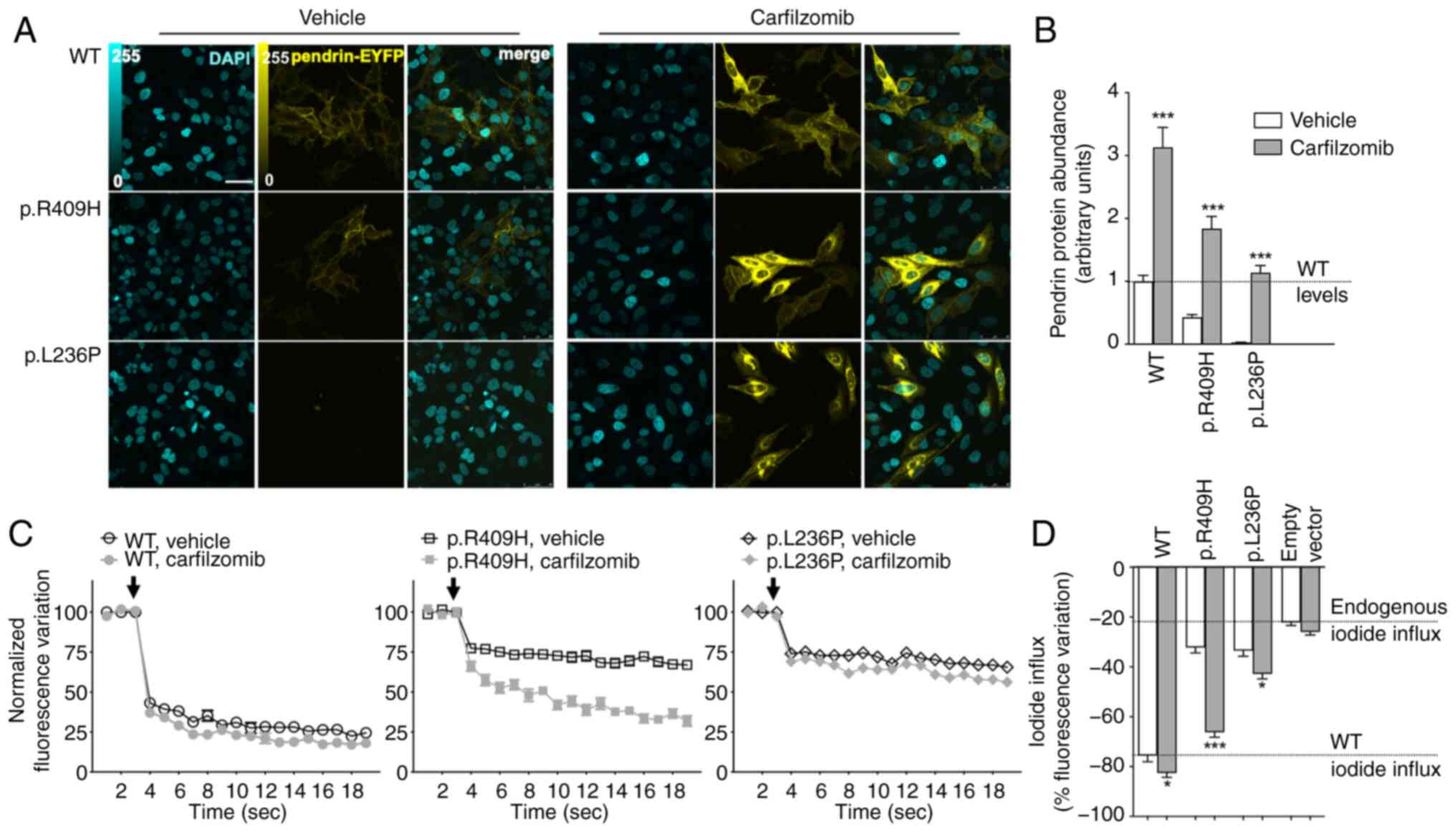
|
1
|
Everett LA, Morsli H, Wu DK and Green ED:
Expression pattern of the mouse ortholog of the Pendred's syndrome
gene (Pds) suggests a key role for pendrin in the inner ear. Proc
Natl Acad Sci USA. 96:9727–9732. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Royaux IE, Wall SM, Karniski LP, Everett
LA, Suzuki K, Knepper MA and Green ED: Pendrin, encoded by the
Pendred syndrome gene, resides in the apical region of renal
intercalated cells and mediates bicarbonate secretion. Proc Natl
Acad Sci USA. 98:4221–4226. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Royaux IE, Suzuki K, Mori A, Katoh R,
Everett LA, Kohn LD and Green ED: Pendrin, the protein encoded by
the Pendred syndrome gene (PDS), is an apical porter of iodide in
the thyroid and is regulated by thyroglobulin in FRTL-5 cells.
Endocrinology. 141:839–845. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dossena S and Paulmichl M: The role of
Pendrin in health and disease. Springer International Publishing;
Switzerland: 2017, View Article : Google Scholar
|
|
5
|
Honda K, Kim SH, Kelly MC, Burns JC,
Constance L, Li X, Zhou F, Hoa M, Kelley MW, Wangemann P, et al:
Molecular architecture underlying fluid absorption by the
developing inner ear. Elife. 6:e268512017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HM and Wangemann P: Epithelial cell
stretching and luminal acidification lead to a retarded development
of stria vascularis and deafness in mice lacking pendrin. PLoS One.
6:e179492011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wangemann P, Itza EM, Albrecht B, Wu T,
Jabba SV, Maganti RJ, Lee JH, Everett LA, Wall SM, Royaux IE, et
al: Loss of KCNJ10 protein expression abolishes endocochlear
potential and causes deafness in Pendred syndrome mouse model. BMC
Med. 2:302004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fugazzola L, Cerutti N, Mannavola D,
Vannucchi G and Beck-Peccoz P: The role of pendrin in iodide
regulation. Exp Clin Endocrinol Diabetes. 109:18–22. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Soleimani M: The multiple roles of pendrin
in the kidney. Nephrol Dial Transplant. 30:1257–1266. 2015.
View Article : Google Scholar :
|
|
10
|
Brazier F, Corniere N, Picard N, Chambrey
R and Eladari D: Pendrin: Linking acid base to blood pressure.
Pflugers Arch. 476:533–543. 2024. View Article : Google Scholar
|
|
11
|
Wall SM: Regulation of blood pressure and
salt balance by Pendrin-Positive intercalated cells: Donald seldin
lecture 2020. Hypertension. 79:706–716. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith RJH: Pendred Syndrome/Nonsyndromic
Enlarged Vestibular Aqueduct. GeneReviews®. Adam MP, Ardinger HH,
Pagon RA, et al: Seattle, WA: 1993
|
|
13
|
Fraser GR: Association of congenital
deafness with goitre (Pendred's Syndrome) a study of 207 families.
Ann Hum Genet. 28:201–249. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Griffith AJ and Wangemann P: Hearing loss
associated with enlargement of the vestibular aqueduct: Mechanistic
insights from clinical phenotypes, genotypes, and mouse models.
Hear Res. 281:11–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choi BY, Kim HM, Ito T, Lee KY, Li X,
Monahan K, Wen Y, Wilson E, Kurima K, Saunders TL, et al: Mouse
model of enlarged vestibular aqueducts defines temporal requirement
of Slc26a4 expression for hearing acquisition. J Clin Invest.
121:4516–4525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wangemann P: Mouse models for
pendrin-associated loss of cochlear and vestibular function. Cell
Physiol Biochem. 32:157–165. 2013. View Article : Google Scholar
|
|
17
|
Wangemann P and Griffith AJ: Mouse models
reveal the role of pendrin in the inner ear. The role of pendrin in
health and disease. Dossena S and Paulmichl M: Springer
International Publishing; Switzerland: pp. 7–22. 2017, View Article : Google Scholar
|
|
18
|
Wangemann P, Nakaya K, Wu T, Maganti RJ,
Itza EM, Sanneman JD, Harbidge DG, Billings S and Marcus DC: Loss
of cochlear HCO3-secretion causes deafness via endolymphatic
acidification and inhibition of Ca2+ reabsorption in a Pendred
syndrome mouse model. Am J Physiol Renal Physiol. 292:F1345–F1353.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishio A, Ito T, Cheng H, Fitzgerald TS,
Wangemann P and Griffith AJ: Slc26a4 expression prevents
fluctuation of hearing in a mouse model of large vestibular
aqueduct syndrome. Neuroscience. 329:74–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen Z, Zhu H, Li Z, Zhang S, Zhang A,
Zhang T, Fu X, Sun D, Zhang J and Gao J: A knock-in mouse model of
Pendred syndrome with Slc26a4 L236P mutation. Biochem Biophys Res
Commun. 515:359–365. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taylor JP, Metcalfe RA, Watson PF, Weetman
AP and Trembath RC: Mutations of the PDS gene, encoding pendrin,
are associated with protein mislocalization and loss of iodide
efflux: Implications for thyroid dysfunction in Pendred syndrome. J
Clin Endocrinol Metab. 87:1778–1784. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rotman-Pikielny P, Hirschberg K, Maruvada
P, Suzuki K, Royaux IE, Green ED, Kohn LD, Lippincott-Schwartz J
and Yen PM: Retention of pendrin in the endoplasmic reticulum is a
major mechanism for Pendred syndrome. Hum Mol Genet. 11:2625–2633.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dai P, Stewart AK, Chebib F, Hsu A,
Rozenfeld J, Huang D, Kang D, Lip V, Fang H, Shao H, et al:
Distinct and novel SLC26A4/Pendrin mutations in Chinese and U.S.
patients with nonsyndromic hearing loss. Physiol Genomics.
38:281–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choi BY, Stewart AK, Madeo AC, Pryor SP,
Lenhard S, Kittles R, Eisenman D, Kim HJ, Niparko J, Thomsen J, et
al: Hypo-functional SLC26A4 variants associated with nonsyndromic
hearing loss and enlargement of the vestibular aqueduct:
Genotype-phenotype correlation or coincidental polymorphisms? Hum
Mutat. 30:599–608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wasano K, Takahashi S, Rosenberg SK,
Kojima T, Mutai H, Matsunaga T, Ogawa K and Homma K: Systematic
quantification of the anion transport function of pendrin (SLC26A4)
and its disease-associated variants. Hum Mutat. 41:316–331. 2020.
View Article : Google Scholar
|
|
26
|
de Moraes VCS, Bernardinelli E, Zocal N,
Fernandez JA, Nofziger C, Castilho AM, Sartorato EL, Paulmichl M
and Dossena S: Reduction of cellular expression levels is a common
feature of functionally affected pendrin (SLC26A4) protein
variants. Mol Med. 22:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roesch S, Bernardinelli E, Nofziger C,
Tóth M, Patsch W, Rasp G, Paulmichl M and Dossena S: Functional
testing of SLC26A4 Variants-clinical and molecular analysis of a
cohort with enlarged vestibular aqueduct from Austria. Int J Mol
Sci. 19:2092018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shepshelovich J, Goldstein-Magal L,
Globerson A, Yen PM, Rotman-Pikielny P and Hirschberg K: Protein
synthesis inhibitors and the chemical chaperone TMAO reverse
endoplasmic reticulum perturbation induced by overexpression of the
iodide transporter pendrin. J Cell Sci. 118:1577–1586. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee K, Hong TJ and Hahn JS: Roles of
17-AAG-induced molecular chaperones and Rma1 E3 ubiquitin ligase in
folding and degradation of Pendrin. FEBS Lett. 586:2535–2541. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jung J, Kim J, Roh SH, Jun I, Sampson RD,
Gee HY, Choi JY and Lee MG: The HSP70 co-chaperone DNAJC14 targets
misfolded pendrin for unconventional protein secretion. Nat Commun.
7:113862016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nanami M, Pham TD, Kim YH, Yang B, Sutliff
RL, Staub O, Klein JD, Lopez-Cayuqueo KI, Chambrey R, Park AY, et
al: The role of intercalated cell Nedd4-2 in BP regulation, Ion
transport, and transporter expression. J Am Soc Nephrol.
29:1706–1719. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Galietta LJ, Haggie PM and Verkman AS:
Green fluorescent protein-based halide indicators with improved
chloride and iodide affinities. FEBS Lett. 499:220–224. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
DiCiommo DP, Duckett A, Burcescu I,
Bremner R and Gallie BL: Retinoblastoma protein purification and
transduction of retina and retinoblastoma cells using improved
alphavirus vectors. Invest Ophthalmol Vis Sci. 45:3320–3329. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Procino G, Milano S, Tamma G, Dossena S,
Barbieri C, Nicoletti MC, Ranieri M, Di Mise A, Nofziger C, Svelto
M, et al: Co-regulated pendrin and aquaporin 5 expression and
trafficking in Type-B intercalated cells under potassium depletion.
Cell Physiol Biochem. 32:184–199. 2013. View Article : Google Scholar
|
|
36
|
Pera A, Dossena S, Rodighiero S, Gandía M,
Bottà G, Meyer G, Moreno F, Nofziger C, Hernández-Chico C and
Paulmichl M: Functional assessment of allelic variants in the
SLC26A4 gene involved in Pendred syndrome and nonsyndromic EVA.
Proc Natl Acad Sci USA. 105:18608–18613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fugazzola L, Cirello V, Dossena S,
Rodighiero S, Muzza M, Castorina P, Lalatta F, Ambrosetti U,
Beck-Peccoz P, Bottà G and Paulmichl M: High phenotypic
intrafamilial variability in patients with Pendred syndrome and a
novel duplication in the SLC26A4 gene: Clinical characterization
and functional studies of the mutated SLC26A4 protein. Eur J
Endocrinol. 157:331–338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dror AA, Politi Y, Shahin H, Lenz DR,
Dossena S, Nofziger C, Fuchs H, Hrabé de Angelis M, Paulmichl M,
Weiner S and Avraham KB: Calcium oxalate stone formation in the
inner ear as a result of an Slc26a4 mutation. J Biol Chem.
285:21724–21735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dossena S, Bizhanova A, Nofziger C,
Bernardinelli E, Ramsauer J, Kopp P and Paulmichl M: Identification
of allelic variants of pendrin (SLC26A4) with loss and gain of
function. Cell Physiol Biochem. 28:467–476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dossena S, Nofziger C, Brownstein Z,
Kanaan M, Avraham KB and Paulmichl M: Functional characterization
of pendrin mutations found in the Israeli and Palestinian
populations. Cell Physiol Biochem. 28:477–484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bernardinelli E, Costa R, Nofziger C,
Paulmichl M and Dossena S: Effect of known inhibitors of ion
transport on pendrin (SLC26A4) activity in a human kidney cell
line. Cell Physiol Biochem. 38:1984–1998. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dossena S, Rodighiero S, Vezzoli V,
Bazzini C, Sironi C, Meyer G, Fürst J, Ritter M, Garavaglia ML,
Fugazzola L, et al: Fast fluorometric method for measuring pendrin
(SLC26A4) Cl-/I-transport activity. Cell Physiol Biochem. 18:67–74.
2006. View Article : Google Scholar
|
|
43
|
Dossena S, Rodighiero S, Vezzoli V,
Nofziger C, Salvioni E, Boccazzi M, Grabmayer E, Bottà G, Meyer G,
Fugazzola L, et al: Functional characterization of wild-type and
mutated pendrin (SLC26A4), the anion transporter involved in
Pendred syndrome. J Mol Endocrinol. 43:93–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tsukada K, Nishio SY, Hattori M and Usami
S: Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: Their
origin and a literature review. Ann Otol Rhinol Laryngol. 124(Suppl
1): 61S–76S. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr
M, Hijlkema KJ, Coppes RP, Engedal N, Mari M and Reggiori F:
Chloroquine inhibits autophagic flux by decreasing
autophagosome-lysosome fusion. Autophagy. 14:1435–1455. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Van Goor F, Hadida S, Grootenhuis PD,
Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J,
Olson ER, et al: Correction of the F508del-CFTR protein processing
defect in vitro by the investigational drug VX-809. Proc Natl Acad
Sci USA. 108:18843–18848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Keating D, Marigowda G, Burr L, Daines C,
Mall MA, McKone EF, Ramsey BW, Rowe SM, Sass LA, Tullis E, et al:
VX-445-Tezacaftor-ivacaftor in patients with cystic fibrosis and
one or two phe508del alleles. N Engl J Med. 379:1612–1620. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Aslam AA, Sinha IP and Southern KW:
Ataluren and similar compounds (specific therapies for premature
termination codon class I mutations) for cystic fibrosis. Cochrane
Database Syst Rev. 3:CD0120402023.PubMed/NCBI
|
|
49
|
Hosoya M, Saeki T, Saegusa C, Matsunaga T,
Okano H, Fujioka M and Ogawa K: Estimating the concentration of
therapeutic range using disease-specific iPS cells: Low-dose
rapamycin therapy for Pendred syndrome. Regen Ther. 10:54–63. 2019.
View Article : Google Scholar
|
|
50
|
Dossena S, Vezzoli V, Cerutti N, Bazzini
C, Tosco M, Sironi C, Rodighiero S, Meyer G, Fascio U, Fürst J, et
al: Functional characterization of wild-type and a mutated form of
SLC26A4 identified in a patient with Pendred syndrome. Cell Physiol
Biochem. 17:245–256. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rapid UBIquitination detection. http://old.protein.bio.unipd.it/rubi/.
2013 accessed 5 July 2024
|
|
52
|
Wang L, Hoang A, Gil-Iturbe E, Laganowsky
A, Quick M and Zhou M: Mechanism of anion exchange and
small-molecule inhibition of pendrin. Nat Commun. 15:3462024.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ward CL, Omura S and Kopito RR:
Degradation of CFTR by the ubiquitin-proteasome pathway. Cell.
83:121–127. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen YA, Peng YJ, Hu MC, Huang JJ, Chien
YC, Wu JT, Chen TY and Tang CY: The Cullin 4A/B-DDB1-Cereblon E3
ubiquitin ligase complex mediates the degradation of CLC-1 chloride
channels. Sci Rep. 5:106672015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hsu PH, Ma YT, Fang YC, Huang JJ, Gan YL,
Chang PT, Jow GM, Tang CY and Jeng CJ: Cullin 7 mediates
proteasomal and lysosomal degradations of rat Eag1 potassium
channels. Sci Rep. 7:408252017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Iwai C, Li P, Kurata Y, Morikawa K,
Maharani N, Higaki K, Sasano T, Notsu T, Ishido Y, Miake J, et al:
Hsp90 prevents interaction between CHIP and HERG proteins to
facilitate maturation of wild-type and mutant HERG proteins.
Cardiovasc Res. 100:520–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gozzetti A, Papini G, Candi V, Brambilla
CZ, Sirianni S and Bocchia M: Second generation proteasome
inhibitors in multiple myeloma. Anticancer Agents Med Chem.
17:920–926. 2017. View Article : Google Scholar
|
|
58
|
Zeniya M, Mori T, Yui N, Nomura N, Mandai
S, Isobe K, Chiga M, Sohara E, Rai T and Uchida S: The proteasome
inhibitor bortezomib attenuates renal fibrosis in mice via the
suppression of TGF-β1. Sci Rep. 7:130862017. View Article : Google Scholar
|
|
59
|
Ikeda T, Fujii H, Nose M, Kamogawa Y,
Shirai T, Shirota Y, Ishii T and Harigae H: Bortezomib treatment
induces a higher mortality rate in lupus model mice with a higher
disease activity. Arthritis Res Ther. 19:1872017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pohl C and Dikic I: Cellular quality
control by the ubiquitin-proteasome system and autophagy. Science.
366:818–822. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sheridan C: Drug makers target ubiquitin
proteasome pathway anew. Nat Biotechnol. 33:1115–1117. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sarikas A, Hartmann T and Pan ZQ: The
cullin protein family. Genome Biol. 12:2202011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bulatov E and Ciulli A: Targeting
Cullin-RING E3 ubiquitin ligases for drug discovery: Structure,
assembly and small-molecule modulation. Biochem J. 467:365–386.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Guedat P and Colland F: Patented small
molecule inhibitors in the ubiquitin proteasome system. BMC
Biochem. 8(Suppl 1): S142007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Landre V, Rotblat B, Melino S, Bernassola
F and Melino G: Screening for E3-ubiquitin ligase inhibitors:
Challenges and opportunities. Oncotarget. 5:7988–8013. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Younger JM, Chen L, Ren HY, Rosser MF,
Turnbull EL, Fan CY, Patterson C and Cyr DM: Sequential
quality-control checkpoints triage misfolded cystic fibrosis
transmembrane conductance regulator. Cell. 126:571–582. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu J, Ye J, Zou X, Xu Z, Feng Y, Zou X,
Chen Z, Li Y and Cang Y: CRL4A(CRBN) E3 ubiquitin ligase restricts
BK channel activity and prevents epileptogenesis. Nat Commun.
5:39242014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Mijnders M, Kleizen B and Braakman I:
Correcting CFTR folding defects by small-molecule correctors to
cure cystic fibrosis. Curr Opin Pharmacol. 34:83–90. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tomati V, Sondo E, Armirotti A, Caci E,
Pesce E, Marini M, Gianotti A, Jeon YJ, Cilli M, Pistorio A, et al:
Genetic inhibition of the ubiquitin ligase Rnf5 attenuates
phenotypes associated to F508del cystic fibrosis mutation. Sci Rep.
5:121382015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sondo E, Falchi F, Caci E, Ferrera L,
Giacomini E, Pesce E, Tomati V, Mandrup Bertozzi S, Goldoni L,
Armirotti A, et al: Pharmacological inhibition of the ubiquitin
ligase RNF5 rescues F508del-CFTR in cystic fibrosis airway
epithelia. Cell Chem Biol. 25:891–905.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|