Introduction
The incidence of melanoma worldwide has been
increasing more rapidly than any other type of cancer over the past
few decades (1). It mainly affects
young populations and has become the fifth most common type of
cancer in females. Compared to other skin cancers, such as basal
cell carcinoma and squamous cell carcinoma, melanoma has a higher
metastatic potential. Therefore, early detection is essential,
since complete surgical removal is the only treatment with a high
probability of complete recovery for early-stage melanoma. Patients
with regional and distant metastatic disease have a much worse
prognosis and current treatments, even novel therapeutic options,
have limited effects on overall survival. This emphasizes the need
to continue the research for more effective treatment
modalities.
The development and progression of malignant
melanoma has been associated with genetic changes, deregulated
signaling pathways and changes in the tumor microenvironment
(2,3). Apart from the identification of
affected genes, transcription factors and epigenetic changes in
regulatory genes, the detailed molecular analysis of melanoma cell
lines and tumor samples at different stages has also assigned a
role for microRNAs (miRNAs) in the development, progression and
invasiveness of melanoma. Recently, several profiling experiments
have identified numerous differentially regulated miRNAs that are
associated with the early and late progression of malignant
melanoma, providing a series of candidate miRNAs to be analyzed for
their potential as diagnostic markers or therapeutic targets
(4–6). One of these, miR-145, seems to be
associated with the early progression of melanoma, since its
expression is upregulated in primary melanoma cell lines in
contrast to normal human epidermal melanocytes (NHEMs) (5). A previous study demonstrated that
miR-145 was upregulated in two primary melanoma tissue samples, as
compared to NHEMs, and two melanoma cell lines with different
metastatic potential (7). By
profiling a large panel of melanoma tissue samples, Segura et
al(8) defined a melanoma miRNA
signature capable of predicting post-recurrence survival in
metastatic melanoma. Of note, this study revealed that a higher
expression of miR-145 was associated with longer patient survival.
This finding is consistent with previous reports on lung, colon,
breast and prostate cancer, suggesting a tumor suppressor role for
miR-145 (9,10).
The exact function or mechanism of miR-145 in
malignant melanoma is unclear. To date, only one study has
investigated the effect of the ectopic expression of miR-145 in
melanoma, using two canine and two human melanoma cell lines
(11). Depending on the cell lines
under study, the overexpression of miR-145 inhibited cell growth
and/or cell migration, suggesting that miR-145 acts as a tumor
suppressor in both canine and human malignant melanomas. In the
present study, we aimed to collect additional evidence to sustain
the hypothesis that miR-145 is a melanoma tumor suppressor by
investigating its possible involvement, not only in cell
proliferation and migration, but also in cell invasiveness. miR-145
was overexpressed in three human melanoma cell lines, and its
effect on cell growth, migration and invasion, as well as on the
expression of a number of target genes was evaluated.
Materials and methods
Cell culture
In this study, we chose three different melanoma
cell lines with a decreasing metastatic potential (BLM, FM3P and
WM793). The highly metastatic melanoma cell line, BLM, was
originally obtained from Dr Leon Van Kempen (Department of
Biochemistry, Nijmegen, The Netherlands) and cultured in Dulbecco’s
modified Eagle’s medium (DMEM) (Life Technologies Europe B.V.,
Ghent, Belgium) supplemented with 10% fetal calf serum (FCS), 200
μM L-glutamine, 50 μg/ml streptomycin, 50 U/ml
penicillin and Fungizone. The metastatic melanoma cell line, FM3P
(12), was cultured in RPMI-1640
medium (Life Technologies Europe B.V.) supplemented with 10% FCS,
200 μM L-glutamine, 50 μg/ml streptomycin, 50 U/ml
penicillin and Fungizone. The melanoma cell line, WM793, was kindly
provided by Dr Carola Berkling (Department of Dermatology, Ludwig
Maximilian University of Munich, Munich, Germany). This cell line
was derived from a primary melanoma (stage I) in the sternal area
and cultured in MCDB153:Leibovitz L-15 (4:1) medium supplemented
with 2% FCS, 1.68 mM CaCl2, 5 μg/ml insulin, 50
μg/ml streptomycin, 50 U/ml penicillin and Fungizone
(information is also available at http://ccr.coriell.org, catalog ID WC00062). Human
primary epidermal melanocyte cultures were established as
previously described (13,14). All cells were incubated at a
temperature of 37°C, 99% humidity and 10% CO2.
RNA isolation and real-time quantitative
PCR
Total RNA, including miRNAs, was extracted from the
melanoma cell lines and melanocytes (pooled from three different
donors) using the miRNeasy Mini kit (Qiagen, Venlo, The
Netherlands) according to the manufacturer’s recommendations. A
DNase treatment was performed and first-strand cDNA was generated
by reverse transcription using the iScript cDNA synthesis kit
according to the manufacturer’s instructions (Bio-Rad, Eke,
Belgium). Relative gene expression levels were determined using a
SYBR-Green I reverse transcription-PCR assay as described by
Vandesompele et al(15) and
the comparative Cq method was used for quantification. PCR
reactions were performed by using SYBR®-Green I master
mix (Eurogentec, Ougrée Seraing, Belgium) and were run on a MyiQ™
iCycler (Bio-Rad). To correct for differences in RNA quantities and
cDNA synthesis efficiency, relative gene expression levels were
normalized using the geometric mean of three reference genes
(RPL13A, UBC and SDHA), as described
previously by Vandesompele et al(16).
Pre-miR/short interfering RNA (siRNA)
transfections and miRNA quantification
To achieve the upregulation of miR-145, chemically
modified double-stranded (ds) nucleic acids were transfected with
minimal cellular stress to mimic endogenous mature miRNAs
(pre-miR-145, PM11480; Life Technologies Europe B.V.). This enabled
the detailed analysis of the biological effects of miRNAs via
gain-of-function experiments. Pre-miR negative control 2 (AM17111;
Life Technologies Europe B.V.) is a random-sequence pre-miR
molecule that has been extensively tested in cell lines and tissues
and validated to not produce identifiable effects on known miRNA
function. To induce the knockdown of fascin homolog 1 (FSCN1),
validated synthetic siRNA molecules were used, obtained from
Qiagen, Hilden, Germany (Hs_FSCN1_3 Flexitube siRNA, SI00421806). A
scrambled sequence (5′-AUUAUCUAGGAGAUAUCAC-3′), showing no homology
to any known gene, was used as the negative control (Eurogentec).
For qPCR and western blot analysis, melanoma cells were plated into
60-mm dishes (or 6-well plates) at a density of 500,000 per
dish/well in their specific medium, depending on the cell line
used. Twenty-four hours later the medium was replaced with 2.4 ml
of fresh medium. The final pre-miR-145 or siRNA concentration was
50 nM following the addition of 18 μl HiPerFect Transfection
Reagent (Qiagen) to 100 μl of serum-free culture medium.
This solution was mixed by vortexing and incubated for 10 min at
room temperature to allow the formation of transfection complexes.
These complexes were added in a dropwise manner onto the cells. The
medium was replaced 24 h later and gene silencing was monitored at
the desired time points by harvesting the cells for either RNA
isolation or for preparation of whole-cell lysates (see below). All
transfections were performed in triplicate.
Quantification of miR-145 by TaqMan Real-Time PCR
was carried out as described by the manufacturer (Life Technologies
Europe B.V.). Briefly, 800 ng of template RNA was
reverse-transcribed using the TaqMan MicroRNA Reverse Transcription
kit and the multiplex RT primer pools containing miRNA-specific
stem-loop primers. An RT-product diluted five times was introduced
into a 5-μl PCR reaction. Reactions were run in 384-well
plates on a 7900HT RT-qPCR system (Applied Biosystems Europe,
Halle, Belgium) at 95°C for 10 min, followed by 40 cycles at 95°C
for 15 sec and 60°C for 1 min. miR-145 expression was normalized
between different samples based on the geometric means of the
expression values of three small nucleolar RNAs (U43, U48 and U49)
(Life Technologies Europe B.V.).
Western blot analysis
Lysates of melanoma cells or pooled melanocytes
(n=3) were produced and immunoblotting was performed as previously
described by Van Gele et al(14). Primary antibodies included the
purified rabbit anti-myosin-Va (MYO5A) exon F (1/10,000) (14), a rabbit polyclonal antibody against
RAB27A (H-60) (1/200, sc-22756, Santa Cruz, Heidelberg, Germany), a
polyclonal antibody against FSCN1 (IM20) (1/200, ab49815, Abcam,
Cambridge, UK), a rabbit polyclonal antibody against GAPDH (G9545)
(1/10,000, Sigma-Aldrich, Bornem, Belgium) and a mouse monoclonal
antibody anti-α-tubulin clone B-5-1-2 (T5168) (1/8,000,
Sigma-Aldrich). The latter was used to control loading. The blots
were incubated with the appropriate HRP-conjugated secondary
antibody (1/3,000, Amersham Biosciences Ltd., Buckinghamshire, UK).
Detection was performed with an ECL detection system kit (Amersham
Biosciences, Ltd.).
Cell growth, wound healing and invasion
assays
All three melanoma cell lines were transfected with
pre-miR-145 or siFSCN1 and their respective negative controls, as
described above, in order to examine the effect on cell
proliferation, cell migration and invasion. All experiments were
performed in triplicate. Cell growth was determined using the
trypan blue exclusion test of cell viability. Viable cells were
counted 48 h after transfection, while non-viable cells containing
a blue cytoplasm were excluded. Cell migration activity was
evaluated by wound healing migration assays. Cells were plated in
six-well tissue culture dishes and grown to confluence. The
confluent cell monolayer was scraped with a plastic pipette tip of
1 mm diameter which was followed by the addition of fresh medium,
specific for each cell line. On the exterior bottom side of each
dish, a mark was made at six arbitrary places where the width of
the wound was measured with an inverted microscope (objective ×4)
at the time of wound induction (initiation, 0 h) and after 12 h of
incubation at 37°C. Migration was expressed as the mean ± SEM of
the difference between the measurement at wound initiation and at
12 h. The results of the control-transfected cells were rescaled to
100%.
Cell invasion assays were performed by the use of
collagen type I matrices, as described by De Wever et
al(17). Briefly, six-well
tissue culture dishes were filled with 1.35 ml of neutralized type
I collagen (BD Biosciences, Erembodegem, Belgium) and incubated
overnight at 37°C to allow gelling. Cells were harvested using PBS
buffer and trypsin/EDTA and seeded on top of the collagen gels.
Cultures transfected with specific siRNAs or miRNAs were incubated
for 24 h at 37°C. The cell invasion index (cells with invasive
extensions versus the total number of cells) was calculated by
manually counting the number of invading and non-invading cells
present in ten independent microscopic fields. The invasion index
of the control-transfected cells was rescaled to 100%. Statistical
significance was determined by the Mann-Whitney test by use of SPSS
version 19.0 software (IBM).
Results
miR-145 expression in melanoma cell lines
and human primary melanocytes
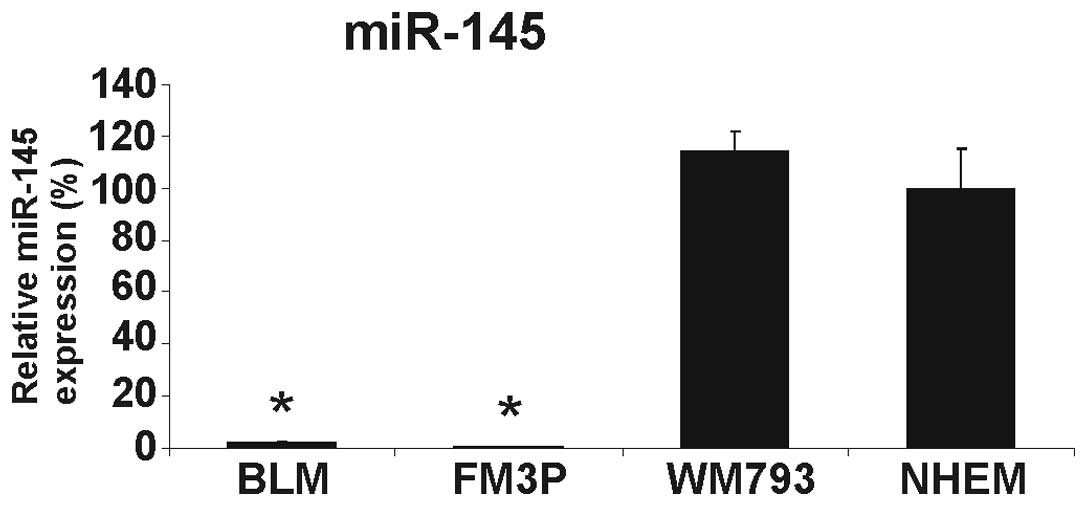
We first determined the expression level of miR-145
in three melanoma cell lines and in pooled NHEMs from three donors.
The expression of miR-145 was extremely low in the two metastatic
melanoma cell lines (BLM and FM3P) and slightly, but not
significantly, increased in the primary melanoma cell line, WM793,
compared to the NHEMs (Fig. 1).
The downregulation of miR-145 in the metastatic cancer cell lines
was in accordance with the role of miR-145 as a tumor suppressor
miRNA in different types of cancer, including melanoma (8,11).
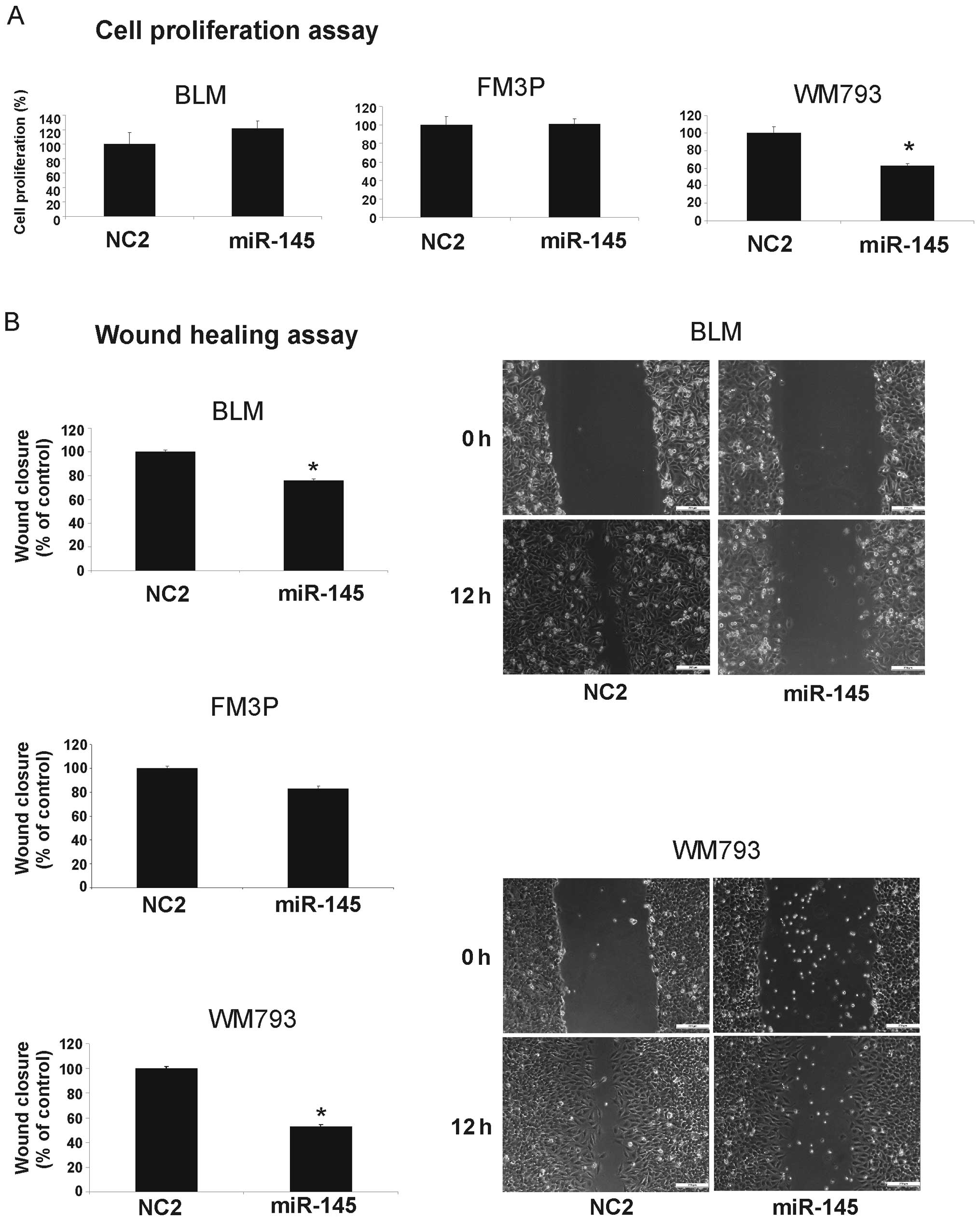
Effect of miR-145 expression on cell
growth, invasion and migration activity in melanoma cell lines
To investigate the functional role of miR-145 in the
melanoma cell lines, we performed gain-of-function experiments by
transfecting miR-145 mimics into each melanoma cell line. The
overexpression of miR-145 resulted in decreased cell growth
compared to the negative control-transfected cells (NC2) only in
the WM793 cell line (Fig. 2A).
Wound healing assays showed significant cell migration inhibition
in the miR-145 transfectants compared to the controls in the BLM
and WM793 cells (% inhibition: BLM, 24±1.4% SEM and WM793, 47±1.6%
SEM) (P<0.001), whereas in the FM3P cells, only a slight
suppression of migration was observed (Fig. 2B).
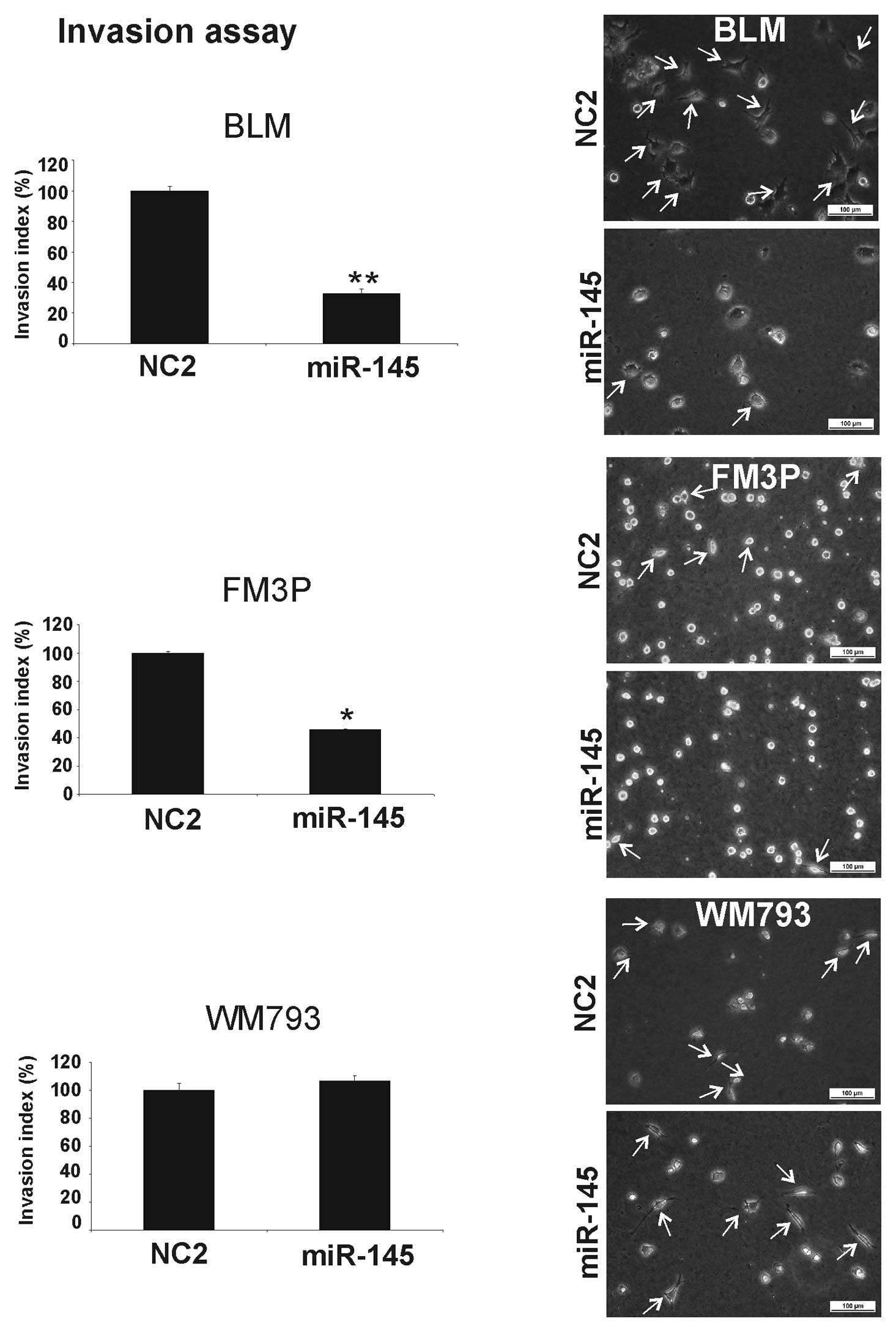
The collagen I invasion assay also showed a
significant inhibition of cell invasion when miR-145 was
overexpressed in the BLM (68±4% SEM) (P<0.001) and FM3P (55±1%
SEM) (P<0.01) cell lines (Fig.
3). No inhibitory effect was observed in the miR-145
transfectants in the primary WM793 cell line.
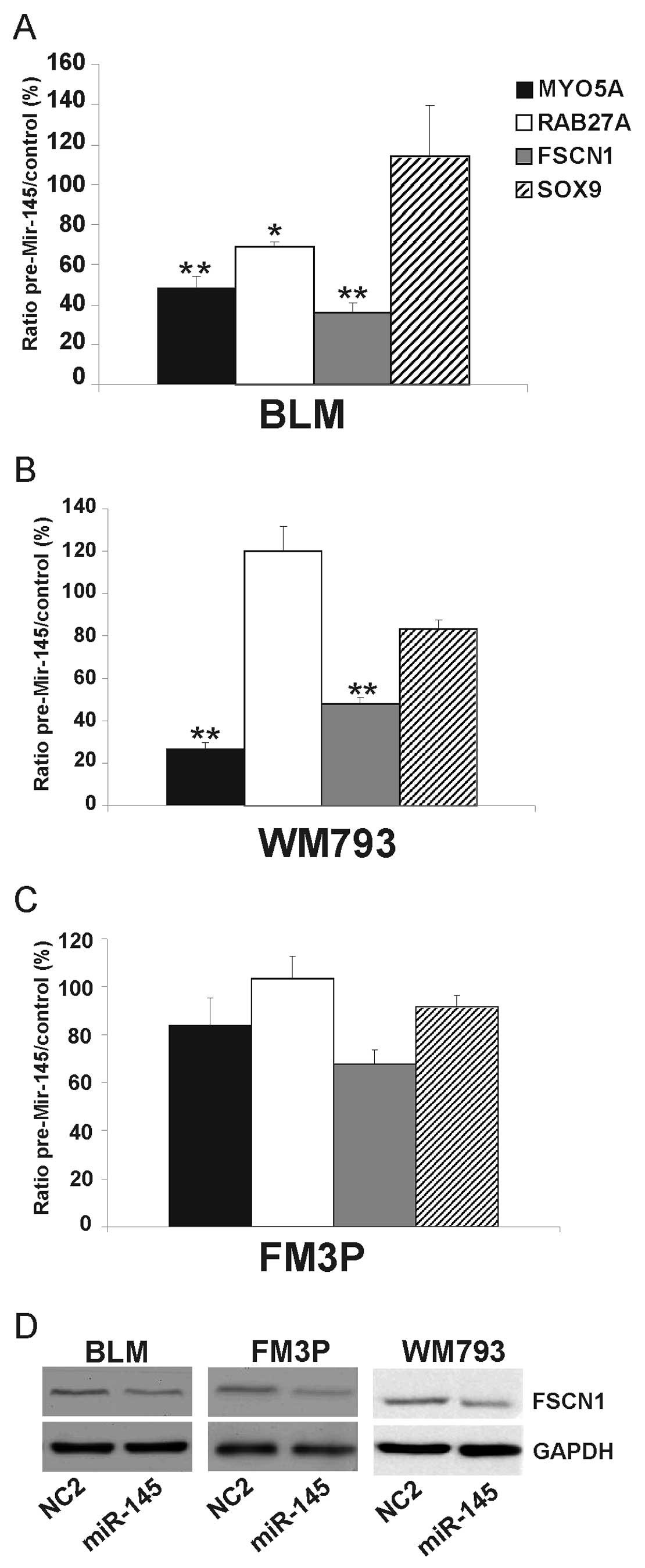
Effect of miR-145 overexpression on
(potential) target genes in melanoma cell lines
We overexpressed miR-145 in all three cell lines and
measured the effect of miR-145 on a number of (potential) target
genes (FSCN1, MYO5A and SOX9) and on an
indirect target (RAB27A) (18). Transfection experiments were
performed in triplicate with miR-145 mimic ds oligonucleotides
(pre-miR-145) and a negative control miRNA. A significant decrease
was observed for the target genes, MYO5A and FSCN1,
in the BLM cells (52±5% SEM and 64±5% SEM, respectively)
(P<0.001) and in the WM793 cells (73±2% SEM and 52±3% SEM,
respectively) (P<0.001) (Fig. 4A
and B). The expression levels of MYO5A and FSCN1
were also decreased in the FM3P cells, although not to a
significant extent (Fig. 4C). The
expression of RAB27A was significantly reduced in the BLM
cells (P<0.05) but not in the WM793 and FM3P cells. There were
no significant differences observed in SOX9 expression among the
three examined cell lines.
Since FSCN1 is a well-known target gene of miR-145
in many different types of cancer (19–21),
we examined via western blot analysis whether the ectopic
expression of miR-145 reduced FSCN1 protein levels. A reduction in
FSCN1 expression was observed following the overexpression of
miR-145, compared to the negative control conditions in all three
examined cell lines (Fig. 4D).
Effect of FSCN1 knockdown on cell growth,
invasion and migration activity in melanoma cell lines
To further examine the mechanism by which miR-145
exerts its invasion suppressor role in our studied melanoma cell
lines, we decided to knock down FSCN1 and study this effect
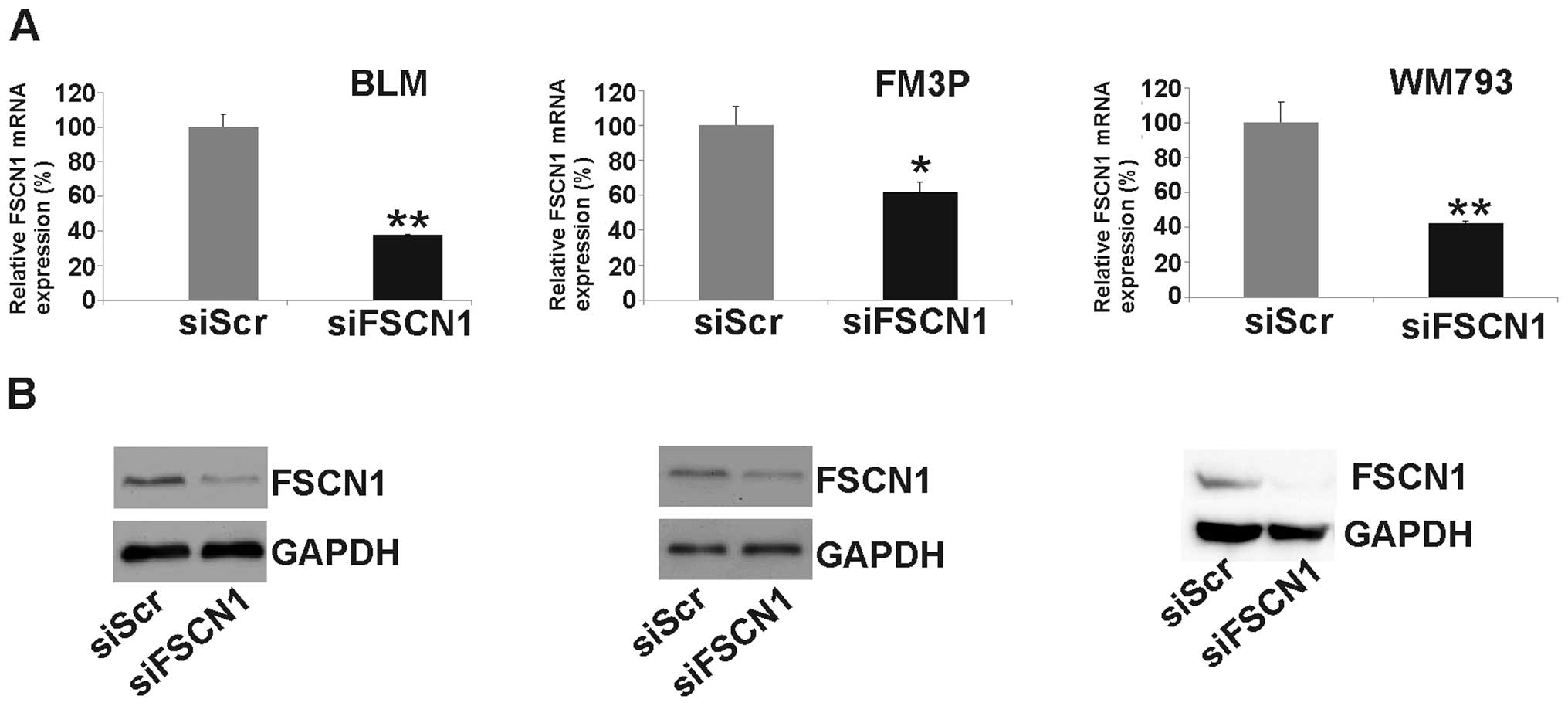
by functional assays. After the transfection of siFSCN1 into
the melanoma cell lines we determined its expression at the mRNA
and protein levels. Real-time qPCR revealed a significant reduction
in FSCN1 expression at the mRNA level [62±0.3% SEM
(P<0.001), 39±6% SEM (P<0.01) and 58±1% SEM (P<0.001), in
BLM, FM3P and WM793 cells, respectively] (Fig. 5A). The silencing of FSCN1 was also
confirmed at the protein level by western blot analysis (Fig. 5B). The knockdown of FSCN1
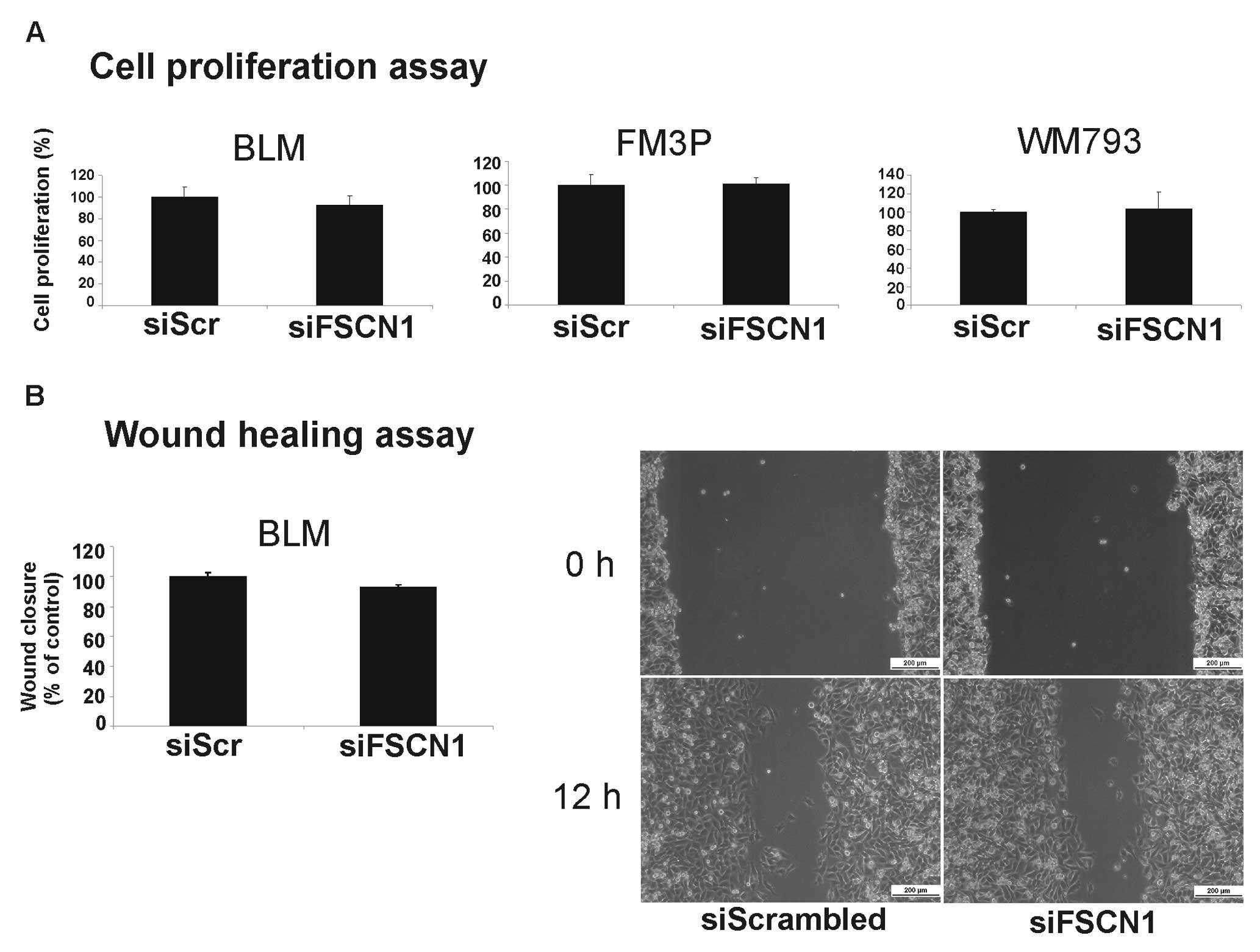
did not result in a significantly reduced cell growth (Fig. 6A) or the inhibition of migration in
any of the transfected cell lines. Fig. 6B depicts the cell migration results
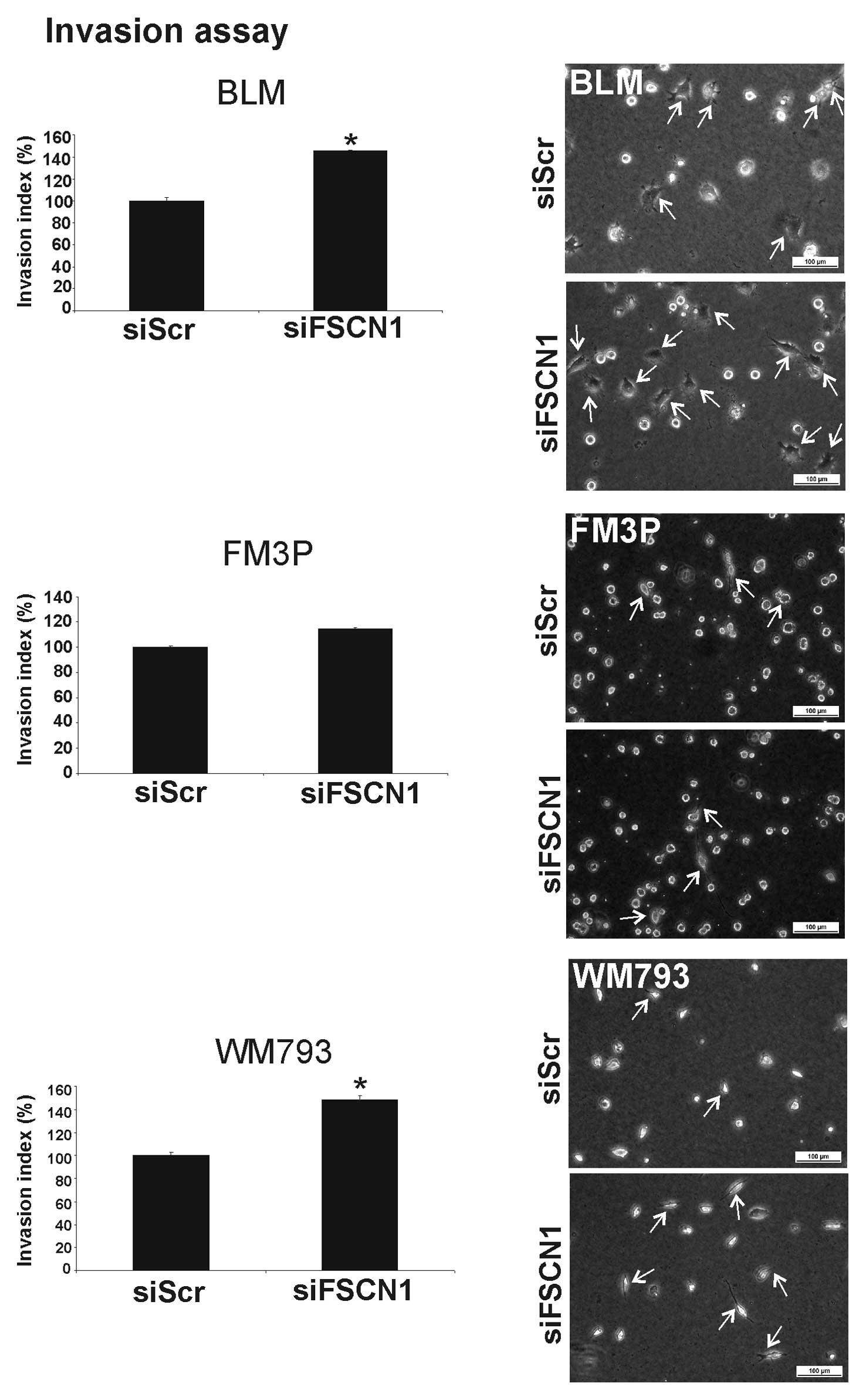
for the BLM cell line. Surprisingly, the silencing of FSCN1
resulted in a significant increase in cell invasion in two out of
the three studied melanoma cell lines (BLM, 45±1% SEM increase; and
WM793, 40±3% SEM increase) (P<0.001) (Fig. 7). These results led to the
conclusion that the invasion suppressor function of miR-145 is
possibly caused by other target gene(s) or pathways.
MYO5A may be another possible target gene,
since its mRNA expression was also reduced after the overexpression
of miR-145 in our melanoma cell lines (Fig. 4A–C). However, when examining the
protein expression levels of MYO5A and RAB27A by western blot
analysis in the BLM, FM3P and WM793 cells, we detected only a
moderate protein expression of MYO5A and a very low expression
level of RAB27A in the FM3P cells as compared to NHEMs, while
neither protein was expressed in the other two cell lines, even
following long exposure times (data not shown). Due to the low or
absent expression of MYO5A in our studied melanoma cell lines we
could not further examine the effect of MYO5A (or RAB27A) knockdown
by functional assays. Based on these results, however, we assumed
that miR-145 does not exert its invasion suppressor activity
through the direct targeting of MYO5A in our melanoma cell
lines.
Discussion
Based on miRNA profiling studies from malignant
melanoma tissues it is assumed that miR-145 plays a tumor
suppressor role in melanoma (8,22).
Functional data unraveling the role of miR-145 in melanoma are
limited. In this study, we used three human melanoma cell lines in
which we examined the expression of miR-145. Our experiments
suggest that miR-145 expression is biphasic, in that its expression
is slightly increased in primary, non-metastatic melanoma cells
compared to normal melanocytes and is significantly reduced as the
cancer progresses from non-metastatic to metastatic. Furthermore,
our data suggest diverse functions of miR-145 among different
melanoma cell lines, depending on the invasive capacity. This
hypothesis was supported by our in vitro experiments using
miR-145 mimics. In primary, non-invasive melanoma cells, miR-145
mimics exerted an anti-proliferative effect (tumor suppressor
response), whereas in metastatic melanoma cells, migration and
matrix invasion were suppressed by miR-145 overexpression (invasion
suppressor response). A similar phenomenon was observed by Sachdeva
et al while studying the effect of miR-145 on cell growth in
non-metastatic MCF-7 breast cancer cells versus two other
metastatic breast cancer cell lines (23,24).
The observed miR-145 effects may be regulated
through the modulation of the expression of genes essential for the
formation of an invasive front. To determine the mechanims by which
miR-145 exerts its invasion suppressor function, we investigated
the expression of several target genes (FSCN1, MYO5A
and SOX9) and an indirect target (RAB27A) following
the overexpression of miR-145 in the melanoma cell lines. The
expression of SOX9, recently reported as a miR-145 target in
mouse mesenchymal stem cells (25), was not significantly reduced at the
mRNA level following the ectopic expression of miR-145. Therefore,
we did not focus any further on this gene. The mRNA expression of
MYO5A, however, was reduced following the overexpression of
miR-145 in our melanoma cell lines. We recently demonstrated that
MYO5A was a direct target of miR-145 by luciferase assays
and functional studies in pigment cells (18). In addition, we reported that the
downregulation of MYO5A following miR-145 overexpression also
resulted in the reduced expression of RAB27A in normal primary
melanocytes (18). Both proteins
are part of a tripartite complex (MYO5A-RAB27A-MLPH) involved in
the intracellular transport of melanosomes in pigment cells. Loss
of one of the members of this tripartite complex results in its
destabilization (14). To our
knowledge, the presence of a functional MYO5A-RAB27A(-MLPH) complex
in melanoma has not been shown to exist thus far. Due to the low or
absent protein expression of MYO5A and RAB27A in our studied
melanoma cell lines, compared to normal human melanocytes, we were
unable to perform further functional assays to examine the effects
of MYOVA (or RAB27A) knockdown in melanoma cells. Consequently,
this led to the assumption that miR-145 does not exert its invasion
suppressor function through the direct targeting of MYO5A, which is
mainly involved in melanogenesis, or the disruption of the
MYO5A-RAB27A(-MLPH) melanosomal pathway, which is probably not
present or functional in this set of studied melanoma cell
lines.
Of note, mRNA and protein expression levels of FSCN1
were reduced in all three studied melanoma cell lines following the
overexpression of miR-145. FSCN1 is an actin-binding protein
required for the formation of actin-based cell-surface protrusions
that mediate interactions between cells and the extracellular
matrix, cell-to-cell interactions and cell migration; it is
required for the formation of cytoplasmic bundles of microfilaments
that contribute to cellular architecture and movement (26,27).
Several studies have already reported that miR-145 targets FSCN1 in
different human cancer types. In these cancers, the loss of miR-145
has been shown to promote the upregulation of FSCN1 expression,
contributing to oncogenesis and tumor progression (19,20).
Based on this knowledge, as well as on our obtained expression
data, we further focused on the role of FSCN1 in our melanoma cell
lines. The knockdown of FSCN1 did not affect cell growth in
these cell lines. These observations are in accordance with those
reported by Noguchi et al(11) and point to the fact that FSCN1 does
not affect cell proliferation. In contrast to the latter study, the
silencing of FSCN1 did not result in reduced cell migration
in our studied melanoma cell lines, further suggesting that miR-145
exerts its invasion suppressor role independently of FSCN1,
even when the expression level of FSCN1 was reduced
following the overexpression of miR-145. In this study, the effect
of FSCN1 knockdown on cell invasion was studied for the
first time in melanoma cell lines. Surprisingly, we found that the
silencing of FSCN1 resulted in an increase in cell
invasiveness, and thus metastatic potential, in two out of the
three studied melanoma cell lines, namely BLM and WM793. As already
mentioned, FSCN1 expression is often highly upregulated in tumors
compared to normal matching tissues which exhibit no FSCN1
expression. The high expression of FSCN1 is associated with
metastasis and poor prognosis in these types of cancer. In
melanoma, however, FSCN1 seems to follow a different expression
pattern. Yildiz et al(28)
demonstrated by immunohistochemical analysis that FSCN1 was less
frequently expressed in malignant melanoma (including metastatic
melanoma) compared to benign and dysplastic nevi. These results are
in accordance with those reported by Goncharuk et
al(29), who performed
immunohistochemical staining for FSCN1 in different skin
neoplasias. They concluded that almost all melanocytic nevi
expressed FSCN1, while no or weak FSCN1 expression was present in
melanomas with pagetoid intra-epidermal spread and in invasive
tumors with a high metastatic risk. The expression of FSCN1
decreases as formation and progression stage of melanoma advances.
Both studies concluded that the loss of FSCN1 contributes to the
development of invasive and metastatic phenotypes of melanoma. The
downregulation or loss of the actin-bundling properties of FSCN1,
which is probably associated with the disorganization of cell-cell
and cell-matrix interactions, would stimulate cell motility and be
an important step in the progression from locally invasive to
widely disseminated melanoma. Our in vitro data point out
that the loss of FSCN1 stimulates invasiveness and correspond with
the conclusions obtained from these in vivo studies. It is
clear from these data that FSCN1 plays a different role in melanoma
than in other tumor types. Instead of exerting an oncogenic
function it may be a metastasis suppressor gene involved in
reducing invasive potential in melanoma. We hypothesize that its
expression in melanoma is possibly regulated by miRNAs different
from miR-145, as a low expression of FSCN1 in metastatic melanoma
would correspond with a high expression of miR-145, which is not
the case based on previous reports (11,22)
and on our in vitro data. Further research, aimed towards
determining the expression levels of miR-145 and FSCN1 in different
stages of melanoma, is warranted to support this hypothesis.
In conclusion, our results provide additional
evidence that miR-145 acts as an invasion suppressor in malignant
melanoma; when lost, tumor progression and metastatic potential are
stimulated. Reintroduction of miR-145 could possibly reduce the
invasiveness or metastatic potential of melanomas, making miR-145
potentially useful in the therapy of melanoma, either alone or in
combination with other existing treatment regimens. Future research
is warranted, using in vivo models with suitable (topical)
delivery systems. The mechanism of action of miR-145 or its
regulatory effect on target genes and pathways in melanoma remain
unclear. SOX9 and MYO5A were not involved in the
biological effects caused by miR-145 mimics. In addition, we
illustrated that miR-145 does not necessarily act via the target
FSCN1 in melanoma, in contrast to other tumor types, which
indicates that other miR-145 target genes, such as MUC1,
MMP-11, FLI1 and JAM-A(24,30,31)
or pathways independent of FSCN1 are involved in cell
migration and invasion. Further research is required to clarify
these observations.
Acknowledgements
We thank Martine De Mil for her
assistance with cell culturing and Marie-Chantal Herteleer for her
technical assistance. We thank Wendy De Rycke for her assistance
with the collagen I invasion assays.
References
|
1
|
Garbe C and Leiter U: Melanoma
epidemiology and trends. Clin Dermatol. 27:3–9. 2009. View Article : Google Scholar
|
|
2
|
Chin L: The genetics of malignant
melanoma: lessons from mouse and man. Nat Rev Cancer. 3:559–570.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin K, Baritaki S, Militello L, Malaponte
G, Bevelacqua Y and Bonavida B: The role of B-RAF mutations in
melanoma and the induction of EMT via dysregulation of the
NF-κB/Snail/RKIP/PTEN circuit. Genes Cancer. 1:409–420.
2010.PubMed/NCBI
|
|
4
|
Howell PM Jr, Li X, Riker AI and Xi Y:
MicroRNA in melanoma. Ochsner J. 10:83–92. 2010.PubMed/NCBI
|
|
5
|
Mueller DW and Bosserhoff AK: Role of
miRNAs in the progression of malignant melanoma. Br J Cancer.
101:551–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mueller DW and Bosserhoff AK: The evolving
concept of ‘melano-miRs’-microRNAs in melanomagenesis. Pigment Cell
Melanoma Res. 23:620–626. 2010.
|
|
7
|
Molnár V, Tamási V, Bakos B, Wiener Z and
Falus A: Changes in miRNA expression in solid tumors: an miRNA
profiling in melanomas. Semin Cancer Biol. 18:111–122.
2008.PubMed/NCBI
|
|
8
|
Segura MF, Belitskaya-Lévy I, Rose AE, et
al: Melanoma microRNA signature predicts post-recurrence survival.
Clin Cancer Res. 16:1577–1586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
10
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Noguchi S, Mori T, Hoshino Y, et al:
Comparative study of anti-oncogenic microRNA-145 in canine and
human malignant melanoma. J Vet Med Sci. 74:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kirkin AF, Petersen TR, Olsen AC, Li L,
thor Straten P and Zeuthen J: Generation of human-melanoma-specific
T lymphocyte clones defining novel cytolytic targets with panels of
newly established melanoma cell lines. Cancer Immunol Immunother.
41:71–81. 1995. View Article : Google Scholar
|
|
13
|
Naeyaert JM, Eller M, Gordon PR, Park HY
and Gilchrest BA: Pigment content of cultured human melanocytes
does not correlate with tyrosinase message level. Br J Dermatol.
125:297–303. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Van Gele M, Geusens B, Schmitt AM, Aguilar
L and Lambert J: Knockdown of myosin Va isoforms by RNAi as a tool
to block melanosome transport in primary human melanocytes. J
Invest Dermatol. 128:2474–2484. 2008.PubMed/NCBI
|
|
15
|
Vandesompele J, De Paepe A and Speleman F:
Elimination of primer-dimer artifacts and genomic coamplification
using a two-step SYBR green I real-time RT-PCR. Anal Biochem.
303:95–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vandesompele J, De Preter K, Pattyn F, et
al: Accurate normalization of real-time quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome
Biol. 3:Research00342002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
De Wever O, Hendrix A, De Boeck A, et al:
Modeling and quantification of cancer cell invasion through
collagen type I matrices. Int J Dev Biol. 54:887–896.
2010.PubMed/NCBI
|
|
18
|
Dynoodt P, Mestdagh P, Van Peer G, et al:
Identification of miR-145 as a key regulator of the pigmentary
process. J Invest Dermatol. August 16–2012.(E-pub ahead of print).
View Article : Google Scholar
|
|
19
|
Chiyomaru T, Enokida H, Tatarano S, et al:
miR-145 and miR-133a function as tumour suppressors and directly
regulate FSCN1 expression in bladder cancer. Br J Cancer.
102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fuse M, Nohata N, Kojima S, et al:
Restoration of miR-145 expression suppresses cell proliferation,
migration and invasion in prostate cancer by targeting FSCN1. Int J
Oncol. 38:1093–1101. 2011.PubMed/NCBI
|
|
21
|
Kim SJ, Oh JS, Shin JY, et al: Development
of microRNA-145 for therapeutic application in breast cancer. J
Control Release. 155:427–434. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mascellani N, Tagliavini L, Gamberoni G,
et al: Using miRNA expression data for the study of human cancer.
Minerva Biotec. 20:23–30. 2008.
|
|
23
|
Sachdeva M, Zhu S, Wu F, et al: p53
represses c-Myc through induction of the tumor suppressor miR-145.
Proc Natl Acad Sci USA. 106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang B, Guo H, Zhang Y, Chen L, Ying D and
Dong S: MicroRNA-145 regulates chondrogenic differentiation of
mesenchymal stem cells by targeting Sox9. PLoS One. 6:e216792011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kureishy N, Sapountzi V, Prag S, Anilkumar
N and Adams JC: Fascins, and their roles in cell structure and
function. Bioessays. 24:350–361. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hashimoto Y, Skacel M and Adams JC: Roles
of fascin in human carcinoma motility and signaling: prospects for
a novel biomarker? Int J Biochem Cell Biol. 37:1787–1804. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yildiz L, Kefeli M, Aydin O and Kandemir
B: Fascin expression in melanocytic lesions of the skin. Eur J
Dermatol. 19:445–450. 2009.PubMed/NCBI
|
|
29
|
Goncharuk VN, Ross JS and Carlson JA:
Actin-binding protein fascin expression in skin neoplasia. J Cutan
Pathol. 29:430–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Guo H, Zhang H, et al: Putative
tumor suppressor miR-145 inhibits colon cancer cell growth by
targeting oncogene Friend leukemia virus integration 1 gene.
Cancer. 117:86–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Götte M, Mohr C, Koo CY, et al:
miR-145-dependent targeting of junctional adhesion molecule A and
modulation of fascin expression are associated with reduced breast
cancer cell motility and invasiveness. Oncogene. 29:6569–6580.
2010.PubMed/NCBI
|