|
1.
|
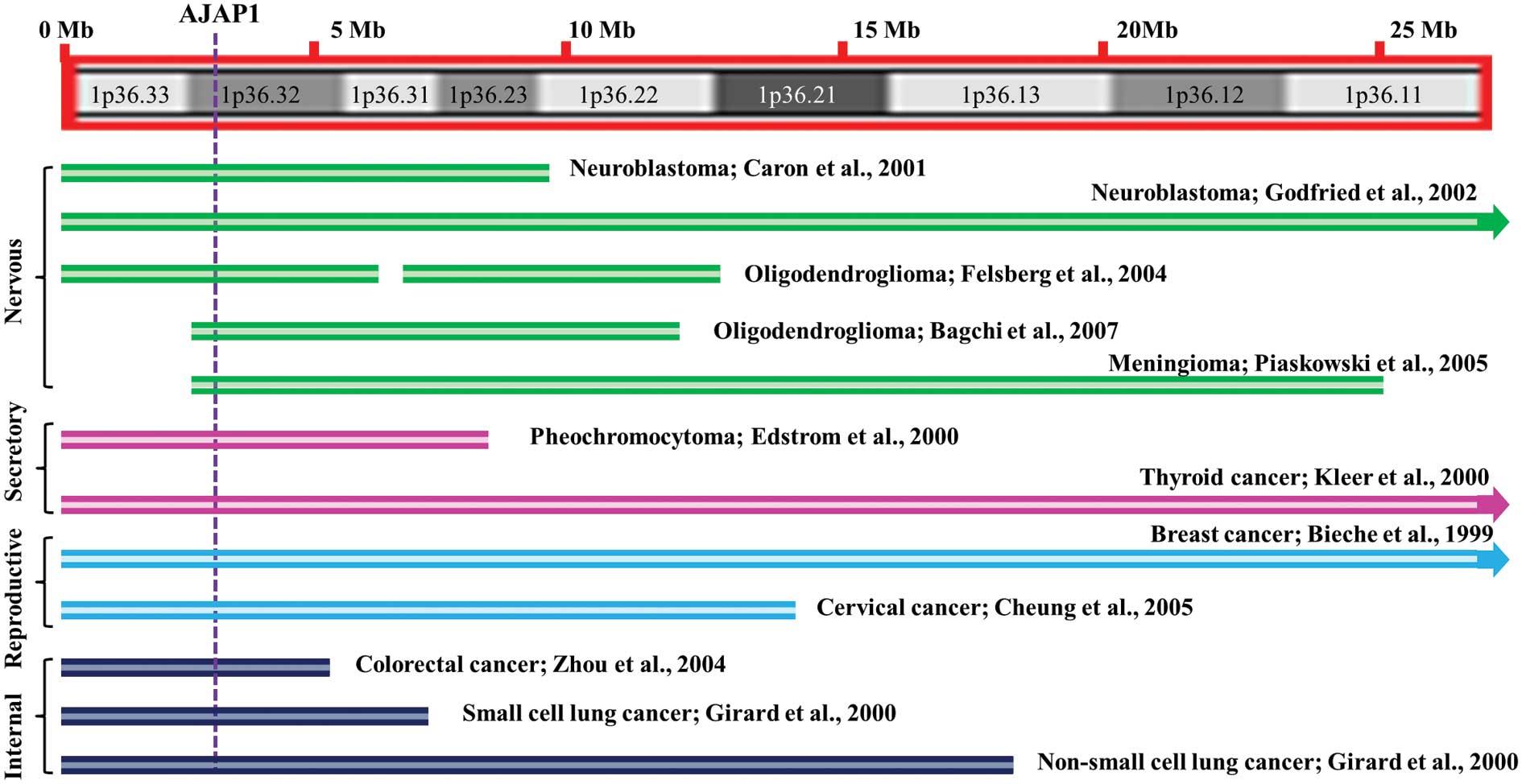
Caron H, Spieker N, Godfried M, et al:
Chromosome bands 1p35–36 contain two distinct neuroblastoma tumor
suppressor loci, one of which is imprinted. Genes Chromosomes
Cancer. 30:168–174. 2001.
|
|
2.
|
Godfried MB, Veenstra M, Valent A, et al:
Lack of interstitial chromosome 1p deletions in clinically-detected
neuroblastoma. Eur J Cancer. 38:1513–1519. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Felsberg J, Erkwoh A, Sabel MC, et al:
Oligodendroglial tumors: refinement of candidate regions on
chromosome arm 1p and correlation of 1p/19q status with survival.
Brain Pathol. 14:121–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Bagchi A, Papazoglu C, Wu Y, et al: CHD5
is a tumor suppressor at human 1p36. Cell. 128:459–475. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Piaskowski S, Rieske P, Szybka M, et al:
GADD45A and EPB41 as tumor suppressor genes in meningioma
pathogenesis. Cancer Genet Cytogenet. 162:63–67. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Edström Elder E, Nord B, Carling T, et al:
Loss of heterozygosity on the short arm of chromosome 1 in
pheochromocytoma and abdominal paraganglioma. World J Surg.
26:965–971. 2002.
|
|
7.
|
Bieche I, Khodja A and Lidereau R:
Deletion mapping of chromosomal region 1p32-pter in primary breast
cancer. Genes Chromosomes Cancer. 24:255–263. 1999. View Article : Google Scholar
|
|
8.
|
Cheung TH, Lo KW, Yim SF, et al:
Clinicopathologic significance of loss of heterozygosity on
chromosome 1 in cervical cancer. Gynecol Oncol. 96:510–515. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhou CZ, Qiu GQ, Zhang F, He L and Peng
ZH: Loss of heterozygosity on chromosome 1 in sporadic colorectal
carcinoma. World J Gastroenterol. 10:1431–1435. 2004.PubMed/NCBI
|
|
10.
|
Girard L, Zochbauer-Muller S, Virmani AK,
Gazdar AF and Minna JD: Genome-wide allelotyping of lung cancer
identifies new regions of allelic loss, differences between small
cell lung cancer and non-small cell lung cancer, and loci
clustering. Cancer Res. 60:4894–4906. 2000.PubMed/NCBI
|
|
11.
|
Kleer CG, Bryant BR, Giordano TJ, Sobel M
and Merino MJ: Genetic changes in chromosomes 1p and 17p in thyroid
cancer progression. Endocr Pathol. 11:137–143. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Schlisio S, Kenchappa RS, Vredeveld LC, et
al: The kinesin KIF1beta acts downstream from EglN3 to induce
apoptosis and is a potential 1p36 tumor suppressor. Genes Dev.
22:884–893. 2008. View Article : Google Scholar
|
|
13.
|
Okawa ER, Gotoh T, Manne J, et al:
Expression and sequence analysis of candidates for the 1p36.31
tumor suppressor gene deleted in neuroblastomas. Oncogene.
27:803–810. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Robbins CM, Tembe WA, Baker A, et al: Copy
number and targeted mutational analysis reveals novel somatic
events in metastatic prostate tumors. Genome Res. 21:47–55. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Sjoblom T, Jones S, Wood LD, et al: The
consensus coding sequences of human breast and colorectal cancers.
Science. 314:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ichimura K, Vogazianou AP, Liu L, et al:
1p36 is a preferential target of chromosome 1 deletions in
astrocytic tumours and homozygously deleted in a subset of
glioblastomas. Oncogene. 27:2097–2108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tanas MR, Sboner A, Oliveira AM, et al:
Identification of a disease-defining gene fusion in epithelioid
hemangioendothelioma. Sci Transl Med. 3:92–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Henrich KO, Fischer M, Mertens D, et al:
Reduced expression of CAMTA1 correlates with adverse outcome in
neuroblastoma patients. Clin Cancer Res. 12:131–138. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Zhang H, Zhai Y, Hu Z, et al: Genome-wide
association study identifies 1p36.22 as a new susceptibility locus
for hepatocellular carcinoma in chronic hepatitis B virus carriers.
Nat Genet. 42:755–758. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Caren H, Fransson S, Ejeskar K, Kogner P
and Martinsson T: Genetic and epigenetic changes in the common 1p36
deletion in neuroblastoma tumours. Br J Cancer. 97:1416–1424. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Liu Z, Yang X, Li Z, et al: CASZ1, a
candidate tumor-suppressor gene, suppresses neuroblastoma tumor
growth through reprogramming gene expression. Cell Death Differ.
18:1174–1183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar
|
|
23.
|
Guessous F, Zhang Y, Kofman A, et al:
microRNA-34a is tumor suppressive in brain tumors and glioma stem
cells. Cell Cycle. 9:1031–1036. 2010. View Article : Google Scholar
|
|
24.
|
Cogdell D, Chung W, Liu Y, et al:
Tumor-associated methylation of the putative tumor suppressor AJAP1
gene and association between decreased AJAP1 expression and shorter
survival in patients with glioma. Chin J Cancer. 30:247–253. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
White PS, Thompson PM, Gotoh T, et al:
Definition and characterization of a region of 1p36.3 consistently
deleted in neuroblastoma. Oncogene. 24:2684–2694. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Cheung NK and Dyer MA: Neuroblastoma:
developmental biology, cancer genomics and immunotherapy. Cancer.
13:397–411. 2013.PubMed/NCBI
|
|
27.
|
Fujita T, Igarashi J, Okawa ER, et al:
CHD5, a tumor suppressor gene deleted from 1p36.31 in
neuroblastomas. J Natl Cancer Inst. 100:940–949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Milde T, Pfister S, Korshunov A, et al:
Stepwise accumulation of distinct genomic aberrations in a patient
with progressively metastasizing ependymoma. Genes Chromosomes
Cancer. 48:229–238. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Nagasawa DT, Trang A, Choy W, et al:
Genetic expression profiles of adult and pediatric ependymomas:
molecular pathways, prognostic indicators, and therapeutic targets.
Clin Neurol Neurosurg. 115:388–399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Dong Z, Pang JS, Ng MH, Poon WS, Zhou L
and Ng HK: Identification of two contiguous minimally deleted
regions on chromosome 1p36.31-p36.32 in oligodendroglial tumours.
Br J Cancer. 91:1105–1111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Alentorn A, Sanson M and Idbaih A:
Oligodendrogliomas: new insights from the genetics and
perspectives. Curr Opin Oncol. 24:687–693. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
McDonald JM, Dunlap S, Cogdell D, et al:
The SHREW1 gene, frequently deleted in oligodendrogliomas,
functions to inhibit cell adhesion and migration. Cancer Biol Ther.
5:300–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Lin N, Di C, Bortoff K, et al: Deletion or
epigenetic silencing of AJAP1 on 1p36 in glioblastoma. Mol Cancer
Res. 10:208–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Ohgaki H and Kleihues P: The definition of
primary and secondary glioblastoma. Clin Cancer Res. 19:764–772.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Bharti S, Handrow-Metzmacher H,
Zickenheiner S, Zeitvogel A, Baumann R and Starzinski-Powitz A:
Novel membrane protein shrew-1 targets to cadherin-mediated
junctions in polarized epithelial cells. Mol Biol Cell. 15:397–406.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Le Bivic A, Sambuy Y, Mostov K and
Rodriguez-Boulan E: Vectorial targeting of an endogenous apical
membrane sialoglyco-protein and uvomorulin in MDCK cells. J Cell
Biol. 110:1533–1539. 1990.PubMed/NCBI
|
|
37.
|
Shore EM and Nelson WJ: Biosynthesis of
the cell adhesion molecule uvomorulin (E-cadherin) in Madin-Darby
canine kidney epithelial cells. J Biol Chem. 266:19672–19680.
1991.PubMed/NCBI
|
|
38.
|
Sako Y, Nagafuchi A, Tsukita S, Takeichi M
and Kusumi A: Cytoplasmic regulation of the movement of E-cadherin
on the free cell surface as studied by optical tweezers and single
particle tracking: corralling and tethering by the membrane
skeleton. J Cell Biol. 140:1227–1240. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Aberle H, Butz S, Stappert J, Weissig H,
Kemler R and Hoschuetzky H: Assembly of the cadherin-catenin
complex in vitro with recombinant proteins. J Cell Sci.
107:3655–3663. 1994.PubMed/NCBI
|
|
40.
|
Jakob V, Schreiner A, Tikkanen R and
Starzinski-Powitz A: Targeting of transmembrane protein shrew-1 to
adherens junctions is controlled by cytoplasmic sorting motifs. Mol
Biol Cell. 17:3397–3408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Schreiner A, Ruonala M, Jakob V, et al:
Junction protein shrew-1 influences cell invasion and interacts
with invasion-promoting protein CD147. Mol Biol Cell. 18:1272–1281.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Resch E, Quaiser S, Quaiser T, Schneider
G, Starzinski-Powitz A and Schreiner A: Synergism of shrew-1’s
signal peptide and transmembrane segment required for plasma
membrane localization. Traffic. 9:1344–1353. 2008.PubMed/NCBI
|
|
43.
|
Hiss JA, Resch E, Schreiner A, Meissner M,
Starzinski-Powitz A and Schneider G: Domain organization of long
signal peptides of single-pass integral membrane proteins reveals
multiple functional capacity. PloS One. 3:e27672008. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Resch E, Hiss JA, Schreiner A, Schneider G
and Starzinski-Powitz A: Long signal peptides of RGMa and DCBLD2
are dissectible into subdomains according to the NtraC model. Mol
Biosyst. 7:942–951. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Gottardi CJ and Peifer M: Terminal regions
of beta-catenin come into view. Structure. 16:336–338. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Lilien J and Balsamo J: The regulation of
cadherin-mediated adhesion by tyrosine
phosphorylation/dephosphorylation of beta-catenin. Curr Opin Cell
Biol. 17:459–465. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Castellone MD, Teramoto H, Williams BO,
Druey KM and Gutkind JS: Prostaglandin E2 promotes colon cancer
cell growth through a Gs-axin-beta-catenin signaling axis. Science.
310:1504–1510. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Liu X, Rubin JS and Kimmel AR: Rapid,
Wnt-induced changes in GSK3beta associations that regulate
beta-catenin stabilization are mediated by Galpha proteins. Curr
Biol. 15:1989–1997. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Zeng L, Kang C, Di C, et al: The adherens
junction-associated protein 1 is a negative transcriptional
regulator of MAGEA2, which potentiates temozolomide-induced
apoptosis in GBM. Int J Oncol. 44:1243–1251. 2014.
|
|
50.
|
Ernst A, Hofmann S, Ahmadi R, et al:
Genomic and expression profiling of glioblastoma stem cell-like
spheroid cultures identifies novel tumor-relevant genes associated
with survival. Clin Cancer Res. 15:6541–6550. 2009. View Article : Google Scholar
|
|
51.
|
Han L, Zhang KL, Zhang JX, et al: AJAP1 is
dysregulated at an early stage of gliomagenesis and suppresses
invasion through cytoskeleton re-organization. CNS Neurosci Ther.
20:429–437. 2014. View Article : Google Scholar : PubMed/NCBI
|