Introduction
Hepatocellular carcinoma (HCC) is the most common
primary malignancy of the liver in adults. It is the 5th most
frequent cancer worldwide and the 3rd leading cause of cancer
mortality (1). Several approaches
have been attempted for the treatment of HCC, such as surgical
resection, transarterial chemoembolization, radiofrequency ablation
and orthotopic liver transplantation. More recently, an oral
multikinase inhibitor, sorafenib, has become a key drug for
non-resectable HCC (2). Sorafenib
inhibits the serine/threonine kinase activity of Raf-1 and B-Raf,
the receptor tyrosine kinase activity of vascular endothelial
growth factor receptors (VEGFRs) 1, 2 and 3, and platelet-derived
growth factor receptor β (PDGFR-β), the cellular signalings of
which are implicated in the molecular pathogenesis of HCC.
Despite the variety of therapeutic options, and the
many points of action of sorafenib, the prognosis of HCC is still
very poor. Several factors account for the limited efficacy of
these treatments. First, most patients have underlying liver
disease [e.g., liver cirrhosis due to chronic hepatitis C virus
(HCV) or hepatitis B virus (HBV) infection]. Second, HCC has a high
rate of recurrence that is caused by intrahepatic metastasis or
multicentric occurrence. Another reason for its poor outcome is
that HCC is a complex and heterogeneous tumor, and many key signal
transduction pathways and molecules other than those targeted by
sorafenib are possibly implicated in the pathogenesis of HCC. In
addition, many genetic and epigenetic alterations such as copy
number alteration (CNA), DNA methylation, or DNA mutation have been
suggested as being related to the development of HCC (3,4).
Therefore, it is important to have a clear landscape of the genomic
aberrations in order to understand the multistep process of HCC
progression.
Among the genetic alterations, CNAs can be found in
almost all human malignancies. Several attempts have been made to
identify CNAs by searching for new genes that are causative for HCC
carcinogenesis. In fact, frequent copy number gains at chromosomes
1q, 8q and 20q, and frequent copy number losses at 1p, 4q, 8p, 16q
and 17q have been identified in HCC using array-comparative genomic
hybridization (CGH) (5–7). However, the roles of these CNAs in
the pathogenesis of HCC have yet to be elucidated.
Given these facts, the purpose of the current study
was to identify new HCC-related genes by investigating CNAs in the
whole genome using array-CGH, and by focusing on specific genes
included in the CNA region. Furthermore, by performing functional
assays of the identified genes, we aimed to elucidate the role of
the genes in the pathogenesis and progression of HCC, and to
clarify whether the genes have the potential to be new therapeutic
targets.
Materials and methods
Patients
Paired tumor and surrounding non-tumor liver tissues
(liver cirrhosis) were collected from patients who underwent
radical surgery for HCC at Mie University Hospital (Tsu, Japan).
Normal liver tissues were obtained from trimmed scrap portions of
donor livers used for living donor liver transplantation. In total,
liver tissues were obtained from 15 patients and 15 normal
subjects. DNA copy number alterations (CNAs) were analyzed in HCC
tissues, and gene expression was compared between HCC and normal
tissues. Table I shows the patient
characteristics. This study was approved by the Institutional
Review Board of Mie University Hospital (authorization no. 285).
Written informed consent was obtained from each patient included in
the study. The study protocol conforms to the ethical guidelines of
the 1975 Declaration of Helsinki as reflected in a priori
approval by the institution’s human research committee.
 | Table ICharacteristics of HCC patients
studied. |
Table I
Characteristics of HCC patients
studied.
| Gender | Male | 11 |
| Female | 4 |
| Age | | 62±8.7a |
| HBV serology | Positive | 7 |
| Negative | 8 |
| HCV serology | Positive | 8 |
| Negative | 7 |
| AFP (ng/ml) | | 885±3,107a |
| DCP (mAU/ml) | | 2,168±6,243a |
Copy number analysis
GeneChip 50K single nucleotide polymorphism (SNP)
mapping array analysis was performed according to the standard
Single Primer GeneChip Mapping Assay protocol using a Human Mapping
50K Array Hind III (Affymetrix, Santa Clara, CA, USA). Individual
SNP copy numbers and chromosomal regions with gains or deletions
were evaluated with CNAG 2.0 (8).
Expression profiling
Oligonucleotide microarray experiments were carried
out using Human Genome U133 Plus 2.0 arrays according to the
manufacturer’s instructions (Affymetrix). Data were analyzed with
GeneSpring GX 7.3.1 (Silicon Genetics, Redwood City, CA, USA).
HCC cell lines
The human HCC cell lines HepG2 (RCB1648) and Huh7
(RCB1942) were purchased from the Riken Cell Bank (Tsukuba, Japan),
Hep3B (ATCC HTB-52) and SK-Hep1 (ATCC HB-8064) were purchased from
the American Type Culture Collection (Manassas, VA), and HLE
(JCRB0404) and PLC/PRF/5 (JCRB0406) were purchased from the Health
Science Research Resources Bank (Osaka, Japan). All cell lines were
cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life
Technologies, Tokyo, Japan) supplemented with 1%
penicillin/streptomycin (Life Technologies) and 10% fetal calf
serum (FCS) (Life Technologies) in a humidified atmosphere
containing 5% CO2 at 37°C.
Qualitative reverse transcription
polymerase chain reaction (PCR)
The expression of CTHRC1 mRNA in the HCC cell lines
was determined by reverse transcription PCR of total RNA. Total RNA
was extracted from approximately 107 cells of each cell
line with the RNeasy mini kit (Qiagen, Tokyo, Japan), and cDNA was
synthesized by extension of oligo dT primers with PrimeScript
reverse transcriptase (Takara Bio, Inc., Otsu, Japan). PCR of the
cDNA was performed with Ex Taq (Takara Bio). The sense primer used
for amplification of CTHRC1 was 5′-AGGGAGGTGGTGGACCTGTAT-3′ and
antisense primer was 5′-GCCAACCCAGATAGCAACAT-3′.
Quantitative real-time PCR
The cDNA of HCC tissues, non-tumorous tissues and
HCC cell lines was synthesized from 1 μg of total RNA and
quantitative real-time PCR (qRT-PCR) was performed using the ABI
prism 7300 Real-time PCR system (Applied Biosystems, Foster City,
CA, USA) with EagleTaq master mix kits (Roche Molecular Systems,
Branchburg, NJ, USA). The expression levels of target genes from
triplicate reactions were determined by normalization to β-actin
according to the manufacturer’s instructions. Primer sets are as
follows: CTHRC1 forward, 5′-CCAAGGGGAAGCAAAAGG-3′; reverse,
5′-CCCTTGTAAGCACATTCCATTA-3′. Human integrin β-2 forward,
5′-CAGCAATGTGGTCCAACTCA-3′; reverse, 5′-GAGGGCGTTGTGATCCAG-3′.
Human integrin β-3 forward, 5′-CGCTAAATTTGAGGAAGAACG-3′; reverse,
5′-GAAGGTAGACGTGGCCTCTTT-3′.
Western blot analysis
Polyclonal antibody for CTHRC1 was generated by
immunization of rabbits. HepG2 cells were fractionated using the
ProteoExtract Subcellular Proteome Extraction Kit (Merck Millipore,
Darmstadt, Germany) according to the manufacturer’s instructions,
and localization of CTHRC1 in HCC cells was determined by western
blot analysis. Protein lysates of each fraction were separated by
SDS-polyacrylamide gel electrophoresis (12.5%) and transferred to
polyvinylidene difluoride membranes. Blots were blocked with 5%
milk in Tris-HCl (pH 7.5) with 0.1% Tween-20 for 2 h and proved
with primary antibody at 4°C overnight. The immunoblots were then
probed with horseradish peroxidase-conjugated anti-rabbit secondary
antibody (GE Healthcare, Amersham Place, UK) and visualized using
ECL plus (GE Healthcare, Munich, Germany).
Knockdown of CTHRC1 mRNA
Three types of short hairpin RNA (ShRNA) against
CTHRC1 and control ShRNA were constructed using the piGENE vector
(Igene, Tokyo, Japan). Their target sequences are listed as
follows: Sh1, GAAATGA ATTCAACAATTA; Sh2, AAGGAAGCCCTGAAATGAA; Sh3,
AGGGAAAGCTTTGAGGAGT; and control (T7STOP), CACCTTTTTTTT. These
ShRNAs and control plasmid were transfected into HepG2 cells and
Huh7 cells with FuGENE HD (Roche, Mannheim, Germany), followed by
the addition of 1 μg/ml of puromycin after 24 h for selecting
transfected cells. Cells were harvested 72 h later for analysis of
gene expression, cell proliferation, migration and invasion.
Cell proliferation assay
Cell proliferation was assessed with the xCELLigence
system (Roche Inc., Basel, Switzerland) according to the
manufacturer’s instructions. Briefly, each well of a 16-well
microtiter plate (E-Plate 16) was filled with 100 μl of DMEM to
equilibrate the well membrane, and each plate was incubated for 30
min at 37°C in 5% CO2. HCC cells transfected with ShRNA
against CTHRC1 or control plasmid were suspended in 100 μl of DMEM
and seeded at a density of 1×104 cells per well. Cells
were cultured for 48 h with the Real-Time Cell Analyzer (RTCA)
single plate (SP) instrument placed in a standard incubator at 37°C
in 5% CO2. Then, cell proliferation was monitored by
recording cell index (CI) values at 15-min intervals for 48 h.
Cell migration and invasion assays
Cell migratory and invasive abilities were also
assessed by the xCELLigence system. Briefly, cell migration was
assessed by seeding HCC cells at 2×104/well on a
fibronectin-coated CIM-16 plate, and cell invasion was assessed by
seeding HCC cells at 3×104/well on a CIM-16 plate coated
with Matrigel (Becton-Dickinson, Tokyo, Japan). Both cell invasion
and migration were monitored by recording the CI at 15-min
intervals for 48 h.
Real-time PCR array
The changes in gene expression related to cell
migration and invasion by CTHRC1 knockdown in HCC cells were
analyzed with the Human Extracellular Matrix and Adhesion Molecules
RT2 Profiler PCR Array (SABiosciences, Frederick, MD, USA)
according to the manufacturer’s instructions. These changes were
confirmed also by qRT-PCR.
Immunohistochemistry
Immunohistochemical staining for CTHRC1 was
performed on surgically resected HCC tissues using the Vectastain
ABC kit (Vector Laboratories, Burlingame, CA, USA). Deparaffinized
sections were heated for 5 min in citrate buffer at 100°C with a
pressure cooker to reactivate the antigen, and treated with 0.3%
H2O2 in methanol for 30 min to abolish
endogenous peroxidase activity. Sections were blocked with 1% goat
serum in phosphate-buffered saline, covered with primary antibody
at 4°C overnight, and then covered with second-step biotinylated
antibody for 30 min, and incubated with peroxidase-labeled
streptavidin for 30 min. After washing, sections were incubated
with 0.05% diaminobenzidine/0.15% H2O2 and
counterstained with 10% hematoxylin (Wako Pure Chemical Industries,
Ltd., Osaka, Japan).
Statistical analysis
Comparisons of gene expression, cell proliferation,
migration and invasion were performed using the two-tailed
Student’s t-test. For differences between rates, Fisher’s exact
test was used. p<0.05 was considered statistically
significant.
Results
CTHRC1 was identified as a new
HCC-related gene
The chromosomes which had frequent CNAs (≥20%) in 15
HCC tissues are listed in Table
II. Among them, we focused on 8q, in which CNAs were detected
in 53% of HCC tissues, because CNAs in 8q have been also reported
very frequently by several groups (9–11),
but have not been studied in detail. Through gene expression
profiling of 8q, we identified CTHRC1 located at 8q22.3 as a
new HCC-related gene, of which the expression level was higher in
HCC compared with normal tissues by 13.5-fold (Fig. 1A). Moreover, re-validation with
qRT-PCR revealed that expression levels of CTHRC1 mRNA increased
from normal tissues to liver cirrhosis and HCC in a stepwise manner
(Fig. 1B). We investigated the
expression of CTHRC1 mRNA in HCC cell lines by RT-PCR. As shown in
Fig. 1C, CTHRC1 transcripts were
detected in all 6 HCC cell lines. Localization of the CTHRC1
protein in HCC cells was also investigated using HepG2. HepG2 cells
were fractionated and each fraction was analyzed for CTHRC1
expression by western blot analysis. Fig. 1D shows that the 26-kDa CTHRC1
protein was expressed in membrane fractions of the HepG2 cells.
 | Table IIChromosomes which had frequent copy
number alterations in HCC. |
Table II
Chromosomes which had frequent copy
number alterations in HCC.
| Gain | Loss |
|---|
|
|
|---|
| Chromosome | Frequency (%) | Chromosome | Frequency (%) |
|---|
| 1q | 60 | 17p | 47 |
| 8q | 53 | 8p | 27 |
| 13q | 27 | 16q | 27 |
| 20q | 27 | 1p | 20 |
| 6p | 27 | 2q | 20 |
| 17q | 20 | 13q | 20 |
CTHRC1 promotes proliferation of HCC
cells
As it has been reported that CTHRC1 is involved in
tissue remodeling in rheumatoid arthritis and injured arteries by
promoting the migration of fibroblasts (12), we hypothesized that CTHRC1 might
have some role in the proliferation and motility of HCC cells.
First, we investigated the effect of CTHRC1 on the proliferation of
HCC cells. Three types of ShRNA were used to suppress the
expression of CTHRC1 in HepG2 and Huh7 cells. The knockdown
efficiency was confirmed by qRT-PCR (Fig. 2A and B). As shown in Fig. 2C and D, cell proliferation after 24
and 48 h assessed with xCELLignece system was significantly
decreased in both the HepG2 and Huh7 cells with CTHRC1 knockdown
compared with the control cells.
CTHRC1 promotes migration and invasion of
HCC cells
We next examined the influence of CTHRC1 knockdown
on HCC cell migratory activity. As shown in Fig. 2E, CTHRC1 knockdown caused a
significant reduction in cell migration after 24 h in the Huh7
cells compared with the control cells. In addition, the results of
the cell invasion assay using the Matrigel-coated plate showed that
the cell invasions of both the HepG2 and Huh7 cells after 24 h were
also significantly reduced in the CTHRC1-depleted cells compared
with the control cells (Fig. 2F and
G).
CTHRC1 knockdown reduces integrin β mRNA
in HCC cells
To elucidate the mechanism of the suppression of
cell migratory and invasive activity in HCC cells by CTHRC1
knockdown, we compared the mRNA expression, which related to cell
migration and invasion between the CTHRC1-depleted HepG2 cells and
the control cells, using real-time PCR array. The results suggested
that the expression of integrin β was affected by suppression of
CTHRC1 mRNA (data not shown). To confirm these results, we next
performed qRT-PCR to examine the changes in the expression levels
of integrin β mRNA in HCC cells after knockdown of CTHRC1. As shown
in Fig. 3, mRNA expressions of
integrin β-2 and integrin β-3 were significantly reduced with
CTHRC1 depletion in both the HepG2 and Huh7 cells. On the other
hand, no cell proliferation, migration/invasion, or amount of
integrin β mRNA in HCC changed with overexpression of CTHRC1 (data
not shown).
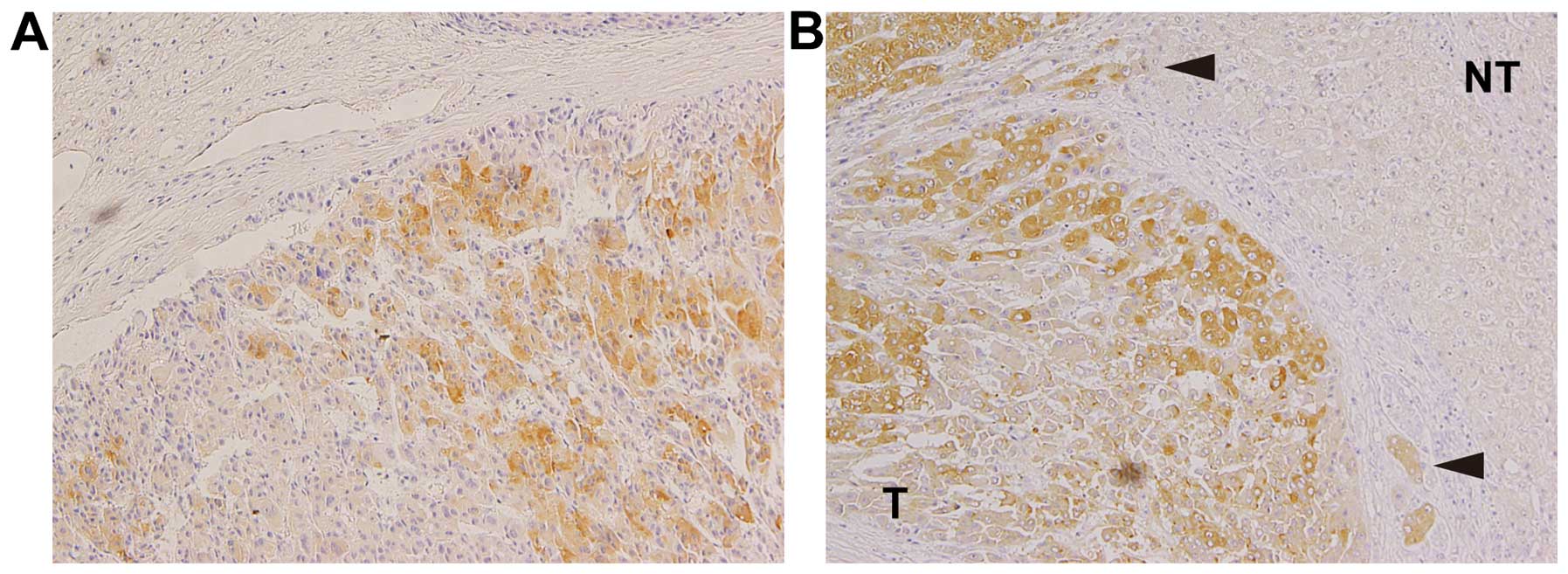
CTHRC1 is expressed in the invasive area
of HCC tissues
Forty-one surgically resected HCC specimens and
adjacent non-tumor tissues were examined for CTHRC1 expression by
immunohistochemistry. The HCC tissues were divided into three
groups according to their degree of differentiation:
well-differentiated HCC, moderately-differentiated HCC and
poorly-differentiated HCC. The levels of CTHRC1 expression were
analyzed in each group and the percentages of positive staining
area were divided into three categories. The results are summarized
in Table III. The rate for the
positive area for CTHRC1 greater than 66% was significantly higher
in poorly-differentiated HCC than that of well-differentiated HCC
(71.4 vs. 18.2%, p<0.05). The differences of these rates in both
between well-differentiated HCC and moderately-differentiated HCC,
and between moderately-differentiated HCC and poorly-differentiated
HCC were not significant, probably due to the small sample number.
None of the patients had positive CTHRC1 staining in the adjacent
non-tumorous areas. Interestingly, some patients showed strong
positive staining for CTHRC1 in the HCC cells close to fibrous
boundaries (Fig. 4) and in
invasive areas (Fig. 4B,
arrowheads), suggesting high migratory and invasive activity of
these cells.
 | Table IIISummary of immunohistochemical
staining. |
Table III
Summary of immunohistochemical
staining.
| | Extent of staining
area |
|---|
| |
|
|---|
| Case | 0–33% | 34–66% | 67–100% |
|---|
| Total HCC | 41 | 36.6 | 24.4 | 39.0 |
| Well diff | 11 | 63.6 | 18.2 | 18.2 |
| Mod diff | 23 | 26.1 | 34.8 | 39.1 |
| Poorly diff | 7 | 28.6 | 0 | 71.4a |
Discussion
Despite the availability of several therapeutic
options, it is still difficult to control the progression of HCC
completely. One reason is the complexity of signal transduction in
HCC; in other words, a large number of molecules are involved in
the pathogenesis of HCC. Therefore, it is important to search for
the novel molecules related to HCC progression in order to
understand the mechanism of its pathogenesis and to achieve better
prognosis. In this study, we identified CTHRC1 located at
chromosome 8q22.3 as a novel HCC-related gene. The results of
functional assay showed that CTHRC1-deletion caused reduced cell
proliferation and motility in HCC cells. In addition, integrin β
mRNA expression was decreased by knockdown of CTHRC1 in HCC cells.
Moreover, the CTHRC1 protein was overexpressed in HCC tissues,
especially in poorly-differentiated HCCs.
We first analyzed the whole genome for CNA using
array-CGH and found copy number gain of 8q in 53% of the HCC
tissues. We next performed gene expression analysis of 8q and
identified CTHRC1 as a novel HCC-related gene. Our results
showed that CTHRC1 was overexpressed in HCC tissues compared with
the surrounding non-tumor tissues. Furthermore, CTHRC1 mRNA was
upregulated in all HCC cell lines examined, and the CTHRC1 protein
was located in the cell membranes of the HCC cells. The mammalian
CTHRC1 gene was first identified in balloon-injured arteries
by Pyagay et al (12). This
gene is highly conserved among vertebrates. The CTHRC1 protein
contains an NH2-terminal peptide for extracellular secretion, a
short collagen triple helix repeat of 36 amino acids, and a
COOH-terminal globular domain. Although CTHRC1 expression was not
detectable in normal arteries, on injury, it was transiently
expressed by fibroblasts of the remodeling adventitia and by smooth
muscle cells of the neointima. Durmus et al (13) reported that CTHRC1 was highly
expressed in developing bones and cartilage, the bone matrix and
periosteum of adult bones, and in the epithelium-mesenchymal
interface including epidermis, airway epithelium, and esophageal
epithelium. In addition, these sites of CTHRC1 expression were
found to overlap considerably with those reported for the
transforming growth factor β (TGF-β) family members and
interstitial collagens (13).
Kimura et al also reported that CTHRC1 increased bone mass
as a positive regulator of postnatal bone formation (14). These investigators found that
CTHRC1-null mice had a decreased number of osteoblasts and
low bone mass, while CTHRC1 transgenic mice had an increased
number of osteoblasts and displayed high bone mass. Furthermore,
expression of CTHRC1 has been reported in stromal cells of breast
cancer (15). Using cDNA arrays,
Tang et al suggested that the CTHRC1 gene was aberrantly
expressed in several types of human solid cancer cells, especially
in cancers of the gastrointestinal tract, lung, breast, thyroid,
ovarian, cervix, liver and pancreas (16). The results of the present study
were congruent with the report by Tang et al (16). Although it is still unclear whether
CTHRC1 has any function in carcinogenesis, several reports have
suggested that inflammation and tissue repair are tightly linked
with the development of cancer (17,18).
Therefore, it is possible that CTHRC1 plays a certain role in the
early stage of carcinogenesis of several cancers, including
HCC.
To further elucidate the potential roles of CTHRC1
in HCC progression, we next analyzed the effect of CTHRC1 deletion
on the proliferation, migration and invasion of HCC cells. Our
results showed that HCC cell proliferation was reduced by the
deletion of CTHRC1. Moreover, knockdown of CTHRC1 also reduced the
migratory and invasive ability of HCC cells. Several previous
studies have suggested that CTHRC1 is related to the motility and
invasion of both non-tumor and tumor cells. Using a microarray,
Turashvili et al found that the CTHRC1 gene was
upregulated in invasive lobular breast carcinoma, suggesting the
relation of CTHRC1 to carcinogenesis and cancer progression
(19). Using the scratch wound
healing assay, Pyagay et al showed that both PAC1 cells and
embryonic fibroblasts overexpressing CTHRC1 increased migratory
activities compared with control cells (12). Tang et al reported that
CTHRC1 expression was significantly higher in invasive melanoma
than in non-invasive melanoma (16). In addition, the experimental
results of Tang et al with the Boyden chamber also showed
that when CTHRC1 expression was inhibited with small interfering
RNA (siRNA), migration of melanoma cells was significantly reduced
(16). Our experimental results
with the real-time cell analyzer indicated that CTHRC1 plays
important roles, not only in proliferation or migration, but also
in invasion of HCC cells, supporting the findings of previous
reports. Therefore, there is a possibility that CTHRC1 is a
new target for therapy of HCC especially for preventing
metastasis.
Next, to elucidate the mechanism of how
CTHRC1 affects the motility of HCC cells, we analyzed the
changes in mRNA expression. The results of the mRNA array suggested
that expression of integrin β-2 and β-3 mRNA was reduced by CTHRC1
deletion; this was confirmed by qRT-PCR. Integrins are a
superfamily of transmembrane receptors that form heterodimers of α-
and β-subunits. By binding to extracellular matrix (ECM)
components, integrins mediate cell adhesion and direct a number of
cellular processes such as proliferation, migration and
differentiation (20–22). Integrins are also important in
promoting cell survival by preventing anoikis, which is apoptosis
induced by anchorage-dependent cells detaching from the surrounding
ECM (23,24). There has been accumulating evidence
showing that integrins β-2 and β-3 have a relationship with cancer
progression and migration, including HCC (25–31).
For example, Li et al reported that the downregulation of
integrin β-3 in small cell lung cancer cells reduced cell
migration/invasion and induced apoptosis (30). Our results suggest that CTHRC1
promotes the migration/invasion of HCC cells, and also increases
cell survival of HCC by inhibiting anoikis via integrin β
expression. However, no cell proliferation, migration/invasion, or
amount of integrin β mRNA in HCC changed with overexpression of
CTHRC1 (data not shown). Presumably, the expression level of CTHRC1
in HCC cells is already sufficient; therefore, increasing its
expression does not affect these cell processes to any greater
degree. In addition, the detailed mechanism on how CTHRC1 regulates
the expression of integrin β remains to be elucidated.
Another possible mechanism by which CTHRC1 mediates
cell motility is activation of Wnt signaling. Yamamoto et al
found that CTHRC1 is a Wnt cofactor protein that selectively
activates the planar cell polarity (PCP) pathway by stabilizing
ligand-receptor interaction (32).
The PCP pathway of Wnt signaling is mediated by the activation of
small GTP-binding proteins, including Rac, Rho, Jun N-terminal
kinase and Rho-associated kinase, and regulates actin
polymerization and cell migration (33,34).
Therefore, it is possible that CTHRC1 promotes cell migration
through activation of the PCP pathway of Wnt signaling.
Finally, our results of immunohistochemical staining
for CTHRC1 showed that poorly-differentiated HCC had a higher
expression level of CTHRC1 compared with well-differentiated HCC,
supporting previous findings in invasive breast cancer (19). Moreover, strongly positive staining
was observed in tumor cells around the cancer borderlines or
invasive areas in several cases, suggesting that those cells had a
higher activity level of proliferation and migration. These results
indicate that CTHRC1 can be a new biomarker for aggressive
HCC.
During preparation of this manuscript, Park et
al reported the roles of CTHRC1 in pancreas cancer (35) and very recently, Chen et al
also reported CTHRC1 expression in HCC (36). They found CTHRC1 increased motility
and adhesiveness of pancreas cancer cells and HCC cells. Moreover,
Chen et al also reported HCC with higher CTHRC1 mRNA
expression had worse prognosis (36). Our results are clearly supporting
these findings. In addition, our data add the information that
CTHRC1 protein is actually expressed in human HCC tissues, more
prominently in cancer cells which are supposed to be
aggressive.
Taken together, the results of the present study
obtained from cultured cell lines and human HCC tissues indicated
that CTHRC1 is upregulated in HCC cells and promotes cell
proliferation, migration and invasion. CTHRC1 has the
potential to be a new biomarker for types of HCC with poor
prognosis, and to be a new therapeutic target for HCC.
Acknowledgements
We thank Mina Tenpaku and Hiromi Nishii for their
technical assistance.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
CTHRC1
|
collagen triple helix repeat
containing 1
|
|
HCV
|
hepatitis C virus
|
|
HBV
|
hepatitis B virus
|
|
CNA
|
copy number alteration
|
|
CGH
|
comparative genomic hybridization
|
|
SNP
|
single nucleotide polymorphism
|
|
qRT-PCR
|
quantitative real-time polymerase
chain reaction
|
|
ShRNA
|
short hairpin RNA
|
References
|
1
|
Bosch FX, Ribes J, Diaz M and Cleries R:
Primary liver cancer: worldwide incidence and trends.
Gastroenterology. 127:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Ricci S, Mazzaferro V, et al:
Sorafenib in advanced hepatocellular carcinoma. N Engl J Med.
359:378–390. 2008. View Article : Google Scholar
|
|
3
|
Aleksic K, Lackner C, Geigl JB, et al:
Evolution of genomic instability in diethylnitrosamine-induced
hepatocarcinogenesis in mice. Hepatology. 53:895–904. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neumann O, Kesselmeier M, Geffers R, et
al: Methylome analysis and integrative profiling of human HCCs
identify novel protumorigenic factors. Hepatology. 56:1817–1827.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nishida N, Nishimura T, Ito T, Komeda T,
Fukuda Y and Nakao K: Chromosomal instability and human
hepatocarcinogenesis. Histol Histopathol. 18:897–909. 2003.
|
|
6
|
Patil MA, Gutgemann I, Zhang J, et al:
Array-based comparative genomic hybridization reveals recurrent
chromosomal aberrations and Jab1 as a potential target for 8q gain
in hepatocellular carcinoma. Carcinogenesis. 26:2050–2057. 2005.
View Article : Google Scholar
|
|
7
|
Katoh H, Ojima H, Kokubu A, et al:
Genetically distinct and clinically relevant classification of
hepatocellular carcinoma: putative therapeutic targets.
Gastroenterology. 133:1475–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nannya Y, Sanada M, Nakazaki K, et al: A
robust algorithm for copy number detection using high-density
oligonucleotide single nucleotide polymorphism genotyping arrays.
Cancer Res. 65:6071–6079. 2005. View Article : Google Scholar
|
|
9
|
Collonge-Rame MA, Bresson-Hadni S, Koch S,
et al: Pattern of chromosomal imbalances in non-B virus related
hepatocellular carcinoma detected by comparative genomic
hybridization. Cancer Genet Cytogenet. 127:49–52. 2001. View Article : Google Scholar
|
|
10
|
Chochi Y, Kawauchi S, Nakao M, et al: A
copy number gain of the 6p arm is linked with advanced
hepatocellular carcinoma: an array-based comparative genomic
hybridization study. J Pathol. 217:677–684. 2009. View Article : Google Scholar
|
|
11
|
Midorikawa Y, Yamamoto S, Ishikawa S, et
al: Molecular karyotyping of human hepatocellular carcinoma using
single-nucleotide polymorphism arrays. Oncogene. 25:5581–5590.
2006. View Article : Google Scholar
|
|
12
|
Pyagay P, Heroult M, Wang Q, et al:
Collagen triple helix repeat containing 1, a novel secreted protein
in injured and diseased arteries, inhibits collagen expression and
promotes cell migration. Circ Res. 96:261–268. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Durmus T, LeClair RJ, Park KS, Terzic A,
Yoon JK and Lindner V: Expression analysis of the novel gene
collagen triple helix repeat containing-1 (Cthrc1). Gene Expr
Patterns. 6:935–940. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kimura H, Kwan KM, Zhang Z, et al: Cthrc1
is a positive regulator of osteoblastic bone formation. PLoS One.
3:e31742008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
West RB, Nuyten DS, Subramanian S, et al:
Determination of stromal signatures in breast carcinoma. PLoS Biol.
3:e1872005. View Article : Google Scholar
|
|
16
|
Tang L, Dai DL, Su M, Martinka M, Li G and
Zhou Y: Aberrant expression of collagen triple helix repeat
containing 1 in human solid cancers. Clin Cancer Res. 12:3716–3722.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beachy PA, Karhadkar SS and Berman DM:
Tissue repair and stem cell renewal in carcinogenesis. Nature.
432:324–331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Turashvili G, Bouchal J, Baumforth K, et
al: Novel markers for differentiation of lobular and ductal
invasive breast carcinomas by laser microdissection and microarray
analysis. BMC Cancer. 7:552007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oktay M, Wary KK, Dans M, Birge RB and
Giancotti FG: Integrin-mediated activation of focal adhesion kinase
is required for signaling to Jun NH2-terminal kinase and
progression through the G1 phase of the cell cycle. J Cell Biol.
145:1461–1469. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Clark EA and Brugge JS: Integrins and
signal transduction pathways: the road taken. Science. 268:233–239.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giancotti FG and Ruoslahti E: Integrin
signaling. Science. 285:1028–1032. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maubant S, Saint-Dizier D, Boutillon M, et
al: Blockade of alpha v beta3 and alpha v beta5 integrins by RGD
mimetics induces anoikis and not integrin-mediated death in human
endothelial cells. Blood. 108:3035–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benoit YD, Larrivee JF, Groulx JF,
Stankova J, Vachon PH and Beaulieu JF: Integrin alpha8beta1 confers
anoikis susceptibility to human intestinal epithelial crypt cells.
Biochem Biophys Res Commun. 399:434–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takeichi T, Mocevicius P, Deduchovas O, et
al: Alpha v beta2 integrin is indispensable for
CD8+T-cell recruitment in experimental pancreatic and
hepatocellular cancer. Int J Cancer. 130:2067–2076. 2012.PubMed/NCBI
|
|
26
|
Oberyszyn TM, Conti CJ, Ross MS, et al:
Beta2 integrin/ICAM-1 adhesion molecule interactions in cutaneous
inflammation and tumor promotion. Carcinogenesis. 19:445–455. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, Zuo J, Ji G, et al: Proapoptotic
function of integrin beta(3) in human hepatocellular carcinoma
cells. Clin Cancer Res. 15:60–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeanes AI, Wang P, Moreno-Layseca P, et
al: Specific beta-containing integrins exert differential control
on proliferation and two-dimensional collective cell migration in
mammary epithelial cells. J Biol Chem. 287:24103–24112. 2012.
View Article : Google Scholar
|
|
29
|
Jung CW, Song TJ, Lee KO, et al:
Characterization of hepatocellular carcinoma cell lines based on
cell adhesion molecules. Int J Mol Med. 29:1158–1164. 2012.
|
|
30
|
Li N, Zhang JP, Guo S, et al:
Down-regulation of beta3-integrin inhibits bone metastasis of small
cell lung cancer. Mol Biol Rep. 39:3029–3035. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Papas MG, Karatzas PS, Papanikolaou IS, et
al: LFA-1 expression in a series of colorectal adenocarcinomas. J
Gastrointest Cancer. 43:462–466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamamoto S, Nishimura O, Misaki K, et al:
Cthrc1 selectively activates the planar cell polarity pathway of
Wnt signaling by stabilizing the Wnt-receptor complex. Dev Cell.
15:23–36. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Veeman MT, Axelrod JD and Moon RT: A
second canon. Functions and mechanisms of beta-catenin-independent
Wnt signaling. Dev Cell. 5:367–377. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kikuchi A and Yamamoto H: Tumor formation
due to abnormalities in the beta-catenin-independent pathway of Wnt
signaling. Cancer Sci. 99:202–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park EH, Kim S, Jo JY, et al: Collagen
triple helix repeat containing-1 promotes pancreatic cancer
progression by regulating migration and adhesion of tumor cells.
Carcinogenesis. 34:694–702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen YL, Wang TH, Hsu HC, Yuan RH and Jeng
YM: Overexpression of CTHRC1 in hepatocellular carcinoma promotes
tumor invasion and predicts poor prognosis. PLoS One. 8:e703242013.
View Article : Google Scholar : PubMed/NCBI
|