Introduction
Esophageal squamous cell carcinoma (ESCC) is the
most common type of esophageal cancer in Japan. Despite recent
advances in the use of combinations of chemo- and radiotherapy,
ESCC remains one of the most lethal malignancies worldwide, and has
a five-year survival rate of only 20–40% even after curative
surgery (1,2). Nucleic acid-based therapy is a
promising new approach, and the development of such a therapy might
improve the morbidity and mortality of ESCC.
Epigenetic changes, such as DNA methylation, histone
modifications and miRNA biogenesis, regulate gene expression
(3). Histone deacetyltransferases,
which deacetylate the lysine residues of histone proteins,
facilitate the access of many transcriptional factors to DNA and
consequently suppress the expression of the target genes (4,5).
Histone deacetylase inhibitors (HDACIs) are anticancer agents with
a potent HDAC-inhibiting activity (4,5).
Numerous HDACIs have been identified and some of which have
recently been used in clinical trials of cancer treatment (6). The microRNAs (miRs) are non-coding
RNAs that are 21–25 nucleotides in length that silence gene
expression, usually by targeting mRNAs for cleavage or
translational repression (7). They
are known to be involved in gene functions in a broad range of
biological processes, including development, cell differentiation,
proliferation, apoptosis, metabolism, carcinogenesis and growth
control (8–11).
Studies have analyzed the function of microRNA-375
(miR-375) as a tumor suppressor in head and neck squamous cell
carcinoma, maxillary sinus squamous cell carcinoma, hepatocellular
cancer, pancreatic cancer and other cancers (12–15).
Some of them reported that miR-375 plays a tumor-suppressive role
in various cancers (12–15). On the other hand, miR-375 was
upregulated in the tumor tissue from prostate cancer and papillary
thyroid carcinoma, which revealed that the upregulation of miR-375
can serve as a novel marker of these cancers (16,17).
We have reported that the expression of miR-375 in
HDACI-treated cells was upregulated >400-fold in ESCC cell lines
(18). This result indicates the
possibility that the transcription of miRs is regulated by histone
modification, similar to that observed in other forms of gene
transcription. The miR-375 expression in ESCC was lower than that
in the normal epithelium. miR-375 inhibits the proliferation,
migration and invasion of ESCC cells. Moreover, we showed that
LDHB and AEG-1/MTDH were miR-375-targeted genes.
Clinical specimens of ESCC exhibited a high level of LDHB
expression at both the mRNA and protein levels. Knockdown of
LDHB by RNA interference (RNAi) showed that it had a
tumor-suppressive function in ESCC cells. LDHB, which is
regulated by the tumor-suppressive miR-375, may therefore act as an
oncogene in ESCC (18).
However, the correlation between miR-375 expression
and the clinicopathological features of ESCC have been unclear. In
addition, the relationship between miR-375 expression and
LDHB and AEG-1/MTDH mRNA expression in ESCC specimens
has not yet been examined. Nor has the tumor-suppressive effect of
miR-375 on ESCC been examined in in vivo assays.
The aim of this study was to clarify the
correlations between the expression of miR-375 and the
clinicopathological features of ESCC, the LDHB and
AEG-1/MTDH mRNA expression in ESCC and to elucidate the
tumor-suppressive effect of miR-375 in vivo, in assays using
a non-viral delivery system.
Materials and methods
Clinical ESCC specimens
ESCC specimens and normal epithelial tissue
specimens were collected from 85 patients who underwent surgical
treatment for histologically proven ESCC in the Department of
Frontier Surgery, Chiba University Graduate School of Medicine
(Chiba, Japan), from 2001 to 2012. All patients gave their informed
consent for tissue donation. Surgical treatments were performed
without any preoperative radio- or chemotherapy. Normal esophageal
epithelial tissue specimens were obtained far from the cancer.
After excision, the tissue specimens were immediately frozen in
liquid nitrogen until the subsequent analysis. The clinical stage
of ESCC was assessed on the basis of the tumor-node-metastasis
(TNM) classification system recommended by the International Union
Against Cancer (7th edition, 2009).
RNA extraction
The tissue specimens were treated with the TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer’s instructions, for total RNA
extraction. The purity and concentration of all RNA samples were
evaluated by their absorbance ratio at 260/280 nm, which was
determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop
Technologies, Inc., Rockland, DE, USA).
Real-time quantitative RT-PCR for
measuring the miR-375 expression
The expression of miR-375 was determined by
quantitative RT-PCR using TaqMan® MicroRNA Assay kits
(Applied Biosystems, Foster City, CA, USA). First-strand cDNA was
synthesized from 10 ng of total RNA for each sample. The individual
assays used a 15 μl reaction mixture containing 5 μl of RNA
extract, 0.15 μl of 100 mM dNTPs, 1 μl of MultiScribe Reverse
Transcriptase (50 U/μl), 1.5 μl of 10X reverse transcription
buffer, 0.19 μl of RNase inhibitor (20 U/μl), 1 μl of gene-specific
TaqMan primer and 4.16 μl of nuclease-free water according to the
manufacturer’s instructions. The reaction mixture was incubated at
16°C for 30 min, 42°C for 30 min and 85°C for 5 min and 4°C until
use. Subsequently, 1.33 μl of the DNA template was amplified using
10 μl of TaqMan Universal PCR Master Mix, 7.67 μl of nuclease-free
water and 1 μl of gene-specific TaqMan primers/probe mix in a final
volume of 20 μl. The amplification, detection and data analysis
were performed with the iCycler IQ Real-Time Detection System
(Bio-Rad, Hercules, CA, USA). The reaction mixture was incubated at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 60 sec. The RT-qPCR experiments were performed in duplicate.
The expression of RNU6B was used as internal control.
Real-time quantitative RT-PCR to assess
the mRNA expression of LDHB and AEG-1/MTDH
The mRNA expression levels of LDHB and
AEG-1/MTDH were examined by real-time quantitative PCR. The
cDNA templates for real-time PCR were synthesized from 1 μg of
total RNA using a High Capacity RNA-to-cDNA kit (Applied
Biosystems). The Actin α1 (ACTA1) gene served as an
internal control. The PCR reaction consisted of SsoFast EvaGreen
Supermix (containing dNTPs, Sso7d fusion polymerase,
MgCl2, EvaGreen dye and stabilizers; Bio-Rad), 1 μM each
primer and cDNA. All reactions were run in duplicate on the MyiQ2
Two-Color Real-Time PCR Detection System (Bio-Rad). The PCR process
was as follows: initial denaturation at 95°C for 30 sec, followed
by 40 cycles of denaturation at 95°C for 5 sec and annealing at
55°C for 10 sec. The following primer sequences were used: LDHB,
5′-TGG CGTGTGCTATCAGCATT-3′ and 5′-GCTTATCTTCCA AAACATCCACAAG-3′;
AEG-1/MTDH, 5′-TCCGAGAAG CCCAAACCAAAT-3′ and 5′-CTTCACCCTCAGCCACTT
CAA-3′; ACTA1, 5′-CCTTCATCGGTATGGAGTC-3′ and
5′-GTTGGCATACAGGTCCTT-3′. The comparative cycle threshold
(CT) method was applied to quantify the expression
levels of the mRNAs. The relative amount of LDHB and
AEG-1/MTDH to ACTA1 mRNA was calculated using the
2−ΔCT equation, where ΔCT = (CT LDHB or
AEG-1/MTDH − CT ACTA1).
Statistical analysis
The correlation(s) between the expression of miR-375
and the clinicopathological factors were assessed by the
χ2 test. The overall survival was calculated as the time
from the surgical treatment until death or the last follow-up date.
The correlation between the overall survival and the expression of
miR-375 was calculated using the log-rank test, and the results are
presented as curves determined using the Kaplan-Meier method. A
linear regression analysis was performed to determine whether there
was a correlation between the expression of miR-375 and the mRNA
expression of LDHB and MTDH.
Validation of the prolonged accumulation
of miRNA in xenografts by atelocollagen
To examine the antitumor effects of miR-375 in an
in vivo assay, we selected atelocollagen
(Atelogene®; Koken Co., Ltd., Tokyo, Japan) as a
non-viral delivery system. To explore the effects of atelocollagen
on the accumulation of the miRNA, we prepared two types of
tumor-bearing mice, those bearing a fluorescently-labeled miRNA (10
μM)/atelocollagen complex and those bearing a fluorescently-labeled
miRNA (10 μM)/PBS complex. Alexa Fluor 555, premiR-Neg and Pre-miR
miRNA Precursor (all from Life Technologies, Grand Island, NY, USA)
was used as the fluorescently-labeled miRNA. The
fluorescently-labeled miRNA was adjusted to a concentration of 10
μM. Equal volumes of atelocollagen (0.1% in PBS at pH 7.4) and
fluorescently-labeled miRNA solution were combined and mixed by
rotating for 20 min at 4°C. The labeled miR and PBS complex was
prepared in an identical manner. We injected 200 μl of
fluorescently-labeled miRNA (10 μM)/atelocollagen complex into one
tumor-bearing mouse and the fluorescently-labeled miRNA (10 μM)/PBS
complex into another mouse via subcutaneous (s.c.) injection.
Fifteen minutes and 24 h after the injection, we observed the
fluorescence signal using an in vivo imaging system
(IVIS)®.
In vivo assay of the effects of the
non-viral delivery of miR-375 on ESCC tumors
T.Tn and TE2 (5.0×105 cells each) human
ESCC cells were injected into the backs of BALB/c nu/nu mice. When
the estimated tumor volume reached ~300 mm3, mice were
randomly divided into four groups containing five animals each.
The miR-375 and negative control miRNA were obtained
from Bonac Corp. (Fukuoka, Japan). Both the miR-375 and negative
control miRNA levels were adjusted to a concentration of 10 μM. To
prepare the miR-375 (10 μM)/atelocollagen and control-miR (10
μM)/atelocollagen complex, equal volumes of atelocollagen (0.1% in
PBS at pH 7.4) and miR solution were combined and mixed by rotating
for 20 min at 4°C. Five weeks after tumor injection, we injected
the mice with 200 μl of the miR-375/atelocollagen or
control-miRNA/atelocollagen complex by s.c. injection. The
treatment was performed twice a week for a total of six injections.
The tumor size was measured in two dimensions using calipers, and
the tumor volume (mm3) was calculated as a2 ×
b/2 mm3 (a, minor axis; b, major axis). The tumor size
was measured for 22 days.
The Animal Research Committee of the Chiba
University, Japan, approved all studies. All animal procedures were
performed according to the guidelines for the Animal Research
Committee of the Chiba University, Japan. Either was used for mouse
euthanasia and anesthesia.
Determination of the LDHB and AEG-1/MTDH
mRNA expression in tumors treated with the miR-375/atelocollagen
and control-miR/atelocollagen complex
We prepared TE2 and T.Tn tumor-bearing mice, and
divided them into four groups containing three animals each. As
stated above, we injected the miR-375/atelocollagen or
control-miRNA/atelocollagen complex into the xenografts by s.c.
injection. The treatment was conducted one time, and the tumors
were excised 24 h after the treatment. We extracted total RNA from
the tumors and determined the LDHB and AEG-1/MTDH
mRNA expression level. The mRNA expression level of these genes was
compared between the miR-375 and control-miRNA groups.
Results
The correlation between the expression of
miR-375 in ESCC specimens and the clinicopathological factors
We performed RNA extraction and RT-PCR on ESCC
specimens and normal epithelial tissue specimens from 85 patients
(Table I). We examined the
correlations between the miR375 expression and various
clinicopathological factors, including the patient age, gender, T
and N factor, lymphatic vessel and vessel invasion, stage and
differentiation. High expression of miR-375 was significantly
correlated with lymphatic vessel invasion (P=0.0352). None of the
other factors were found to be significantly related to the miR-375
expression (Table II).
 | Table IStatistical features of ESCC
patients. |
Table I
Statistical features of ESCC
patients.
|
Characteristics | No. |
|---|
| Total | 85 |
| Age |
| ≤65 | 37 |
| >65 | 48 |
| Gender |
| Male | 70 |
| Female | 15 |
| Location |
| Ph | 1 |
| Ce | 3 |
| Ut | 8 |
| Mt | 32 |
| Lt | 36 |
| Ae | 4 |
| Unknown | 1 |
| pT factor |
| T1 | 27 |
| T2 | 8 |
| T3 | 38 |
| T4 | 12 |
| pN factor |
| N0 | 35 |
| N1 | 27 |
| N2 | 14 |
| N3 | 7 |
| N4 | 2 |
| Lymphatic vessel
invasion |
| 0 | 39 |
| 1 | 26 |
| 2 | 12 |
| 3 | 8 |
| Vessel
invasion |
| 0 | 26 |
| 1 | 21 |
| 2 | 27 |
| 3 | 11 |
| Pathology |
| Wel | 22 |
| Mod | 46 |
| Por | 13 |
| Other | 4 |
| pUICC stage |
| I | 22 |
| II | 18 |
| III | 44 |
| IV | 1 |
 | Table IIThe correlation between the
expression of miR-375 in ESCC specimens and clinicopathological
factors. |
Table II
The correlation between the
expression of miR-375 in ESCC specimens and clinicopathological
factors.
| Clinicopathological
feature | Low | High | P-value |
|---|
| Age |
| ≤65 | 7 | 30 | 0.8108 |
| >65 | 7 | 41 | |
| Gender |
| Male | 11 | 59 | 0.982 |
| Female | 3 | 12 | |
| pT factor |
| T1 | 3 | 24 | 0.5327 |
| T2, 3, 4 | 11 | 47 | |
| pN factor |
| N0 | 4 | 31 | 0.5327 |
| N1, 2, 3 | 10 | 40 | |
| Lymphatic vessel
invasion |
| 0 | 3 | 36 | 0.0352 |
| 1, 2, 3 | 11 | 35 | |
| Vessel
invasion |
| 0 | 4 | 22 | 0.8901 |
| 1, 2, 3 | 10 | 49 | |
| Pathology |
| Wel | 2 | 20 | 0.8803 |
| Mod | 9 | 37 | |
| Por | 3 | 10 | |
| Other | 0 | 4 | |
| pUICC stage |
| I | 3 | 19 | 1 |
| II, III, IV | 11 | 52 | |
Low expression of miR-375 was
significantly correlated with a poor prognosis
We also performed a statistical analysis of the
correlation between the expression of miR-375 and the prognosis of
the 85 patients. The low expression of miR-375 was significantly
correlated with a poor prognosis (P=0.007). In the group of
patients with high miR-375 expression, the five-year survival rate
was 58.2%, while that in the group with low miR-375 expression was
23.1% (Fig. 1).
A significant inverse correlation exists
between the miR-375 expression and LDHB mRNA expression in ESCC
specimens
We next examined the correlation between the
expression of miR-375 and the mRNA expression levels of LDHB
and AEG-1/MTDH using a linear regression analysis. We found
that there was a significant inverse correlation between the
expression of miR-375 and that of LDHB (P=0.039). No
significant correlation was observed with the AEG-1/MTDH
expression levels, although there was a tendency for an inverse
correlation with the miR-375 expression (P=0.149) (Fig. 2).
Atelocollagen prolongs the accumulation
of miRNA in xenograft tumors
We next examined whether atelocollagen could prolong
the accumulation of miRNA in the tumor using a
fluorescently-labeled miRNA and the IVIS imaging system. Fifteen
minutes after injection of the atelocollagen or PBS-miRNA mixture,
both xenografts exhibited the accumulation of fluorescence. After
24 h, we observed the accumulation of fluorescence in the tumors
injected with the fluorescently-labeled miRNA (10 μM)/atelocollagen
complex, but the fluorescence was not observed in another mouse
that was injected with fluorescently-labeled miRNA (10 μM)/PBS
(Fig. 3).
In vivo effects of miR-375 on ESCC
tumors
As shown in Fig. 4,
22 days after the first treatment, the growth of both TE2 and T.Tn
tumors in the miR-375/atelocollagen complex groups were
significantly suppressed compared with the miR-control groups. The
average volume of TE2 xenografts in the control-miR/atelocollagen
complex group was 1,900.8 mm3, while that of the TE2
xenografts in the miR-375/atelocollagen complex group was 668.0
mm3. The miR-375/atelocollagen complex therefore
suppressed the growth of the TE2 xenografts by 64.8%. For the T.Tn
xenografts, the average volume in the control-miRNA/atelocollagen
complex group was 1,938.5 mm3, while that in the
miR-375/atelocollagen complex group was 680.6 mm3. The
growth of the T.Tn xenografts was therefore reduced by 64.9%
(Fig. 4A–C).
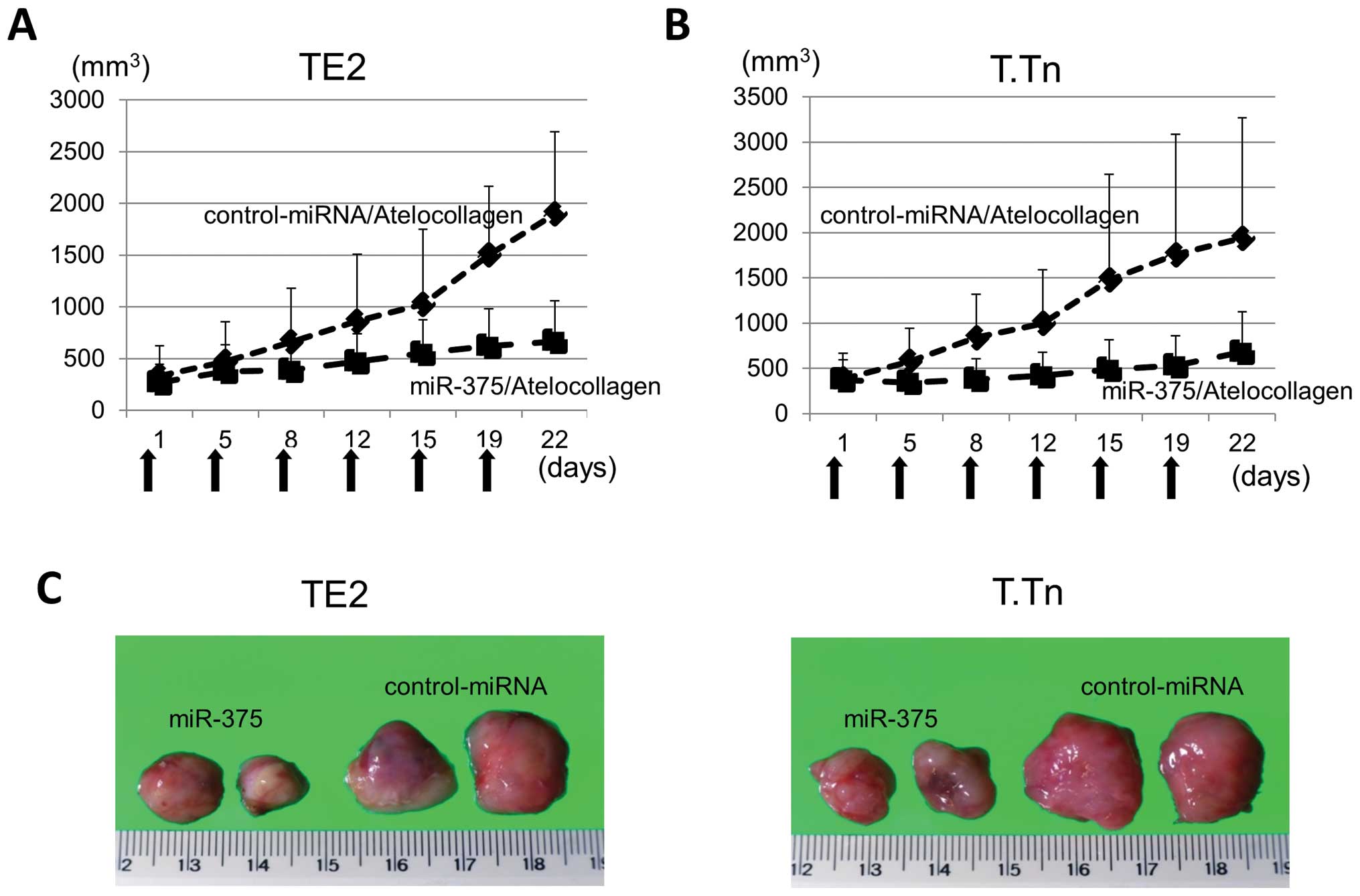 | Figure 4(A) The results of the in vivo
assay using the non-viral delivery system to examine the antitumor
effect of microRNA-375 (miR-375). The TE2 and T.Tn tumor-bearing
mice were injected with 200 μl of miR-375/atelocollagen and
control-miRNA/atelocollagen complex by subcutaneous (s.c.)
injection. The treatment was performed twice a week for a total of
six injections. The arrows indicate the days the mice were treated.
The average volume of TE2 xenografts in the control-microRNA
(miR)/atelocollagen complex group was 1,900.8 mm3 on day
22, while that of the miR-375/atelocollagen complex group was 668.0
mm3. The data points indicate the average of tumor
sizes. (B) The average volume of T.Tn xenografts in the
control-miRNA/atelocollagen complex group was 1,938.5
mm3 on day 22, while that of the T.Tn xenografts in the
miR-375/atelocollagen complex group was 680.6 mm3. (C)
The TE2 and T.Tn xenograft tumors removed on day 22 are shown. From
the upper left panels, the xenografts were TE2 treated with:
miR-375/atelocollagen, miR-375/atelocollagen,
control-miRNA/atelocollagen and control-miRNA/atelocollagen. From
the lower left panels, the xenografts were T.Tn treated with:
miR-375/atelocollagen, miR-375/atelocollagen,
control-miRNA/atelocollagen and control-miRNA/atelocollagen. |
The mRNA expression levels of LDHB and
AEG-1/MTDH were downregulated in xenografts treated with
miR-375
The LDHB mRNA expression of the TE2
xenografts treated with miR-375 was significantly downregulated
(P=0.0067). The level of the T.Tn xenografts treated with miR-375
group was not significantly suppressed. There were no significant
differences in the AEG-1/MTDH mRNA expression levels between
the miR-375 and control groups. However, there was a tendency for
the LDHB expression of T.Tn xenografts and the
AEG-1/MTDH expression of TE2 and T.Tn xenografts to be
downregulated compared with the control groups (P=0.4240, P=0.1977,
P=0.2549) (Fig. 5).
Discussion
Previously we reported that there was HDACI-induced
miR-375 overexpression in ESCC cell lines (18). In the present study, we found that
the miR-375 expression in ESCC was lower than that in the normal
epithelium. miR-375 has a tumor-suppressive function in ESCC cells,
and miR-375 down-regulated LDHB and MTDH in ESCC cell
lines. Kong et al previously revealed that the
downregulation of miR-375 was frequently detected in primary ESCC,
which was significantly correlated with advanced stage disease,
distant metastasis, a poor overall survival and a shorter
disease-free survival (19). In
addition, a recent study estimated the expression of miR-375 in
ESCC cell lines and tissues using a tissue microarray (TMA) that
included 300 cases and confirmed the findings by miRNA in
situ hybridization (MISH) (20). The MISH results also showed that
miR-375 was significantly associated with an advanced clinical
stage, tumor metastasis and a poor outcome of ESCC (19,20).
In this study, only lymphatic vessel invasion was
found to significantly correlate with the high expression of
miR-375. However, we found that a low expression of miR-375 was
significantly correlated with a poor prognosis in our 85 patients.
It might therefore be possible to use the level of miR-375
expression as a prognostic index for ESCC patients. Of interest,
miR-375 might be regulated by epigenetic events, including histone
acetylation. Supporting this possibility, in our previous study,
miR-375 expression was restored by a HDACI in ESCC cells.
LDHB and AEG-1/MTDH were both
identified as miR-375-targeted genes in our previous study.
AEG-1/MTDH has been indicated to play a key role in the
progression, invasion, metastasis and the resistance to
chemotherapies of various types of tumors (21). In addition, the overexpression of
AEG-1/MTDH is considered to be a valuable marker of ESCC
progression (22).
In contrast to AEG-1/MTDH, there have been
only a few reports on whether LDHB expression is related to
the progression of cancers (23,24).
Our previous report showed that ESCC clinical specimens exhibited a
higher level of LDHB expression at both the mRNA and protein levels
compared with the normal esophageal epithelium. Kaplan-Meier curves
and log-rank tests revealed that positive immunoreactivity for the
LDHB protein had a tendency to indicate a poor prognosis. Although
it was verified that LDHB and AEG-1/MTDH were
targeted by miR-375, and that these genes might act as oncogenes,
the correlation between the miR-375 expression and LDHB and
AEG-1/MTDH expression levels in ESCC clinical specimens was
unclear. Therefore, in the present study, we evaluated the
correlation between the miR-375 expression and either the
LDHB or AEG-1/MTDH mRNA expression in ESCC specimens.
We found that there was a significant inverse correlation between
the expression of miR-375 and LDHB. Regarding
AEG-1/MTDH mRNA expression, there was a tendency for an
inverse correlation with the miR-375 expression. These results
confirmed that miR-375 regulates LDHB and AEG-1/MTDH
in clinical specimens and showed there is a possibility that
LDHB and AEG-1/MTDH play a role as oncogenes in
ESCC.
Although the use of miR-based therapy appears to be
an effective strategy, there are unresolved issues, including the
lack of tissue specificity, absence of an optimal delivery system,
poor cellular uptake, and risk of systemic toxicity (3). Regarding miR-375, several studies
have indicated that this miRNA has anticancer effects against
various cancers. However, only one previous report exists so far on
evaluation of the antitumor effect of miR-375 in vivo
(14). It was revealed that
therapeutic administration of cholesterol-conjugated
2′-O-methyl-modified miR-375 mimics (Chol-miR-375) could
significantly suppress the growth of hepatoma xenografts in nude
mice (14). Since no previous
reports had examined the tumor-suppressive effect of miR-375 in
vivo for ESCC xenografts, we determined whether miR-375 was
effective for ESCC in vivo using an atelocollagen complex to
deliver the miR-375.
Atelocollagen, which is prepared from the bovine
dermis, increases the cellular uptake, nuclease resistance and the
prolonged release of nucleic acids in various disease models in
vivo (25–27). Previous studies have already shown
the clear therapeutic efficacy of atelocollagen-mediated in
vivo delivery of nucleic acids (25–27).
In this study, we validated that the miR-375/atelocollagen complex
could significantly suppress the growth of ESCC xenografts.
Although major barriers to the clinical application
of miR-based therapeutics still exist, the administration of
tumor-suppressive miRNA using a non-viral delivery system might
provide a powerful new strategy for cancer therapy (3).
It was also found that the low expression of miR-375
was significantly correlated with a poor prognosis in ESCC
patients. A significant inverse correlation between the expression
of miR-375 and LDHB was observed, suggesting that
LDHB may play a role as an oncogene in ESCC. Moreover, we
validated that the miR-375/atelocollagen complex significantly
suppressed the growth of ESCC xenografts. The administration of a
tumor-suppressive miRNA using a non-viral delivery system might be
a powerful treatment for cancer.
Acknowledgements
We would like to thank the members of the Pathology
Laboratory of the Department of Frontier Surgery of Chiba
University for critical reading of the manuscript.
References
|
1
|
Ruol A, Castoro C, Portale G, et al:
Trends in management and prognosis for esophageal cancer surgery:
twenty-five years of experience at a single institution. Arch Surg.
144:247–254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morita M, Yoshida R, Ikeda K, et al:
Advances in esophageal cancer surgery in Japan: an analysis of 1000
consecutive patients treated at a single institute. Surgery.
143:499–508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoshino I and Matsubara H: MicroRNAs in
cancer diagnosis and therapy: from bench to bedside. Surg Today.
43:467–478. 2013. View Article : Google Scholar
|
|
4
|
Hoshino I, Matsubara H, Akutsu Y, et al:
Gene expression profiling induced by histone deacetylase inhibitor,
FK228, in human esophageal squamous cancer cells. Oncol Rep.
18:585–592. 2007.PubMed/NCBI
|
|
5
|
Hoshino I, Matsubara H, Hanari N, et al:
Histone deacetylase inhibitor FK228 activates tumor suppressor
Prdx1 with apoptosis induction in esophageal cancer cells. Clin
Cancer Res. 11:7945–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoshino I and Matsubara H: Recent advances
in histone deacetylase targeted cancer therapy. Surg Today.
40:809–815. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hua Z, Lv Q, Ye W, et al: MiRNA-directed
regulation of VEGF and other angiogenic factors under hypoxia. PLoS
One. 1:e1162006. View Article : Google Scholar
|
|
9
|
Nissan T and Parker R: Computational
analysis of miRNA- mediated repression of translation: implications
for models of translation initiation inhibition. RNA. 14:1480–1491.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eulalio A, Huntzinger E, Nishihara T,
Rehwinkel J, Fauser M and Izaurralde E: Deadenylation is a
widespread effect of miRNA regulation. RNA. 15:21–32. 2009.
View Article : Google Scholar :
|
|
11
|
Xu X, Chen Z, Zhao X, et al: MicroRNA-25
promotes cell migration and invasion in esophageal squamous cell
carcinoma. Biochem Biophys Res Commun. 421:640–645. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kinoshita T, Nohata N, Yoshino H, et al:
Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in
maxillary sinus squamous cell carcinoma. Int J Oncol. 40:185–193.
2012.
|
|
13
|
Ward A, Balwierz A, Zhang JD, et al:
Re-expression of microRNA-375 reverses both tamoxifen resistance
and accompanying EMT-like properties in breast cancer. Oncogene.
32:1173–1182. 2013. View Article : Google Scholar
|
|
14
|
He XX, Chang Y, Meng FY, et al:
MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and
suppresses liver cancer cell growth in vitro and in vivo. Oncogene.
31:3357–3369. 2012. View Article : Google Scholar
|
|
15
|
Lee EJ, Gusev Y, Jiang J, et al:
Expression profiling identifies microRNA signature in pancreatic
cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar
|
|
16
|
Dettmer M, Perren A, Moch H, Komminoth P,
Nikiforov YE and Nikiforova MN: Comprehensive MicroRNA expression
profiling identifies novel markers in follicular variant of
papillary thyroid carcinoma. Thyroid. 23:1383–1389. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nguyen HC, Xie W, Yang M, et al:
Expression differences of circulating microRNAs in metastatic
castration resistant prostate cancer and low-risk, localized
prostate cancer. Prostate. 73:346–354. 2013. View Article : Google Scholar
|
|
18
|
Isozaki Y, Hoshino I, Nohata N, et al:
Identification of novel molecular targets regulated by tumor
suppressive miR-375 induced by histone acetylation in esophageal
squamous cell carcinoma. Int J Oncol. 41:985–994. 2012.PubMed/NCBI
|
|
19
|
Kong KL, Kwong DL, Chan TH, et al:
MicroRNA-375 inhibits tumour growth and metastasis in oesophageal
squamous cell carcinoma through repressing insulin-like growth
factor 1 receptor. Gut. 61:33–42. 2012. View Article : Google Scholar
|
|
20
|
Li J, Li X, Li Y, et al: Cell-specific
detection of miR-375 down-regulation for predicting the prognosis
of esophageal squamous cell carcinoma by miRNA in situ
hybridization. PLoS One. 8:e535822013. View Article : Google Scholar
|
|
21
|
Hu G, Wei Y and Kang Y: The multifaceted
role of MTDH/AEG-1 in cancer progression. Clin Cancer Res.
15:5615–5620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu C, Chen K, Zheng H, et al:
Overexpression of astrocyte elevated gene-1 (AEG-1) is associated
with esophageal squamous cell carcinoma (ESCC) progression and
pathogenesis. Carcinogenesis. 30:894–901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kinoshita T, Hanazawa T, Nohata N, Okamoto
Y and Seki N: The functional significance of microRNA-375 in human
squamous cell carcinoma: aberrant expression and effects on cancer
pathways. J Hum Genet. 57:556–563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Zhang H, Xu A, et al: Elevation of
serum l-lactate dehydrogenase B correlated with the clinical stage
of lung cancer. Lung Cancer. 54:95–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Inaba S, Nagahara S, Makita N, et al:
Atelocollagen-mediated systemic delivery prevents immunostimulatory
adverse effects of siRNA in mammals. Mol Ther. 20:356–366. 2012.
View Article : Google Scholar :
|
|
26
|
Banno H, Takei Y, Muramatsu T, Komori K
and Kadomatsu K: Controlled release of small interfering RNA
targeting midkine attenuates intimal hyperplasia in vein grafts. J
Vasc Surg. 44:633–641. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takei Y, Kadomatsu K, Goto T and Muramatsu
T: Combinational antitumor effect of siRNA against midkine and
paclitaxel on growth of human prostate cancer xenografts. Cancer.
107:864–873. 2006. View Article : Google Scholar : PubMed/NCBI
|



















