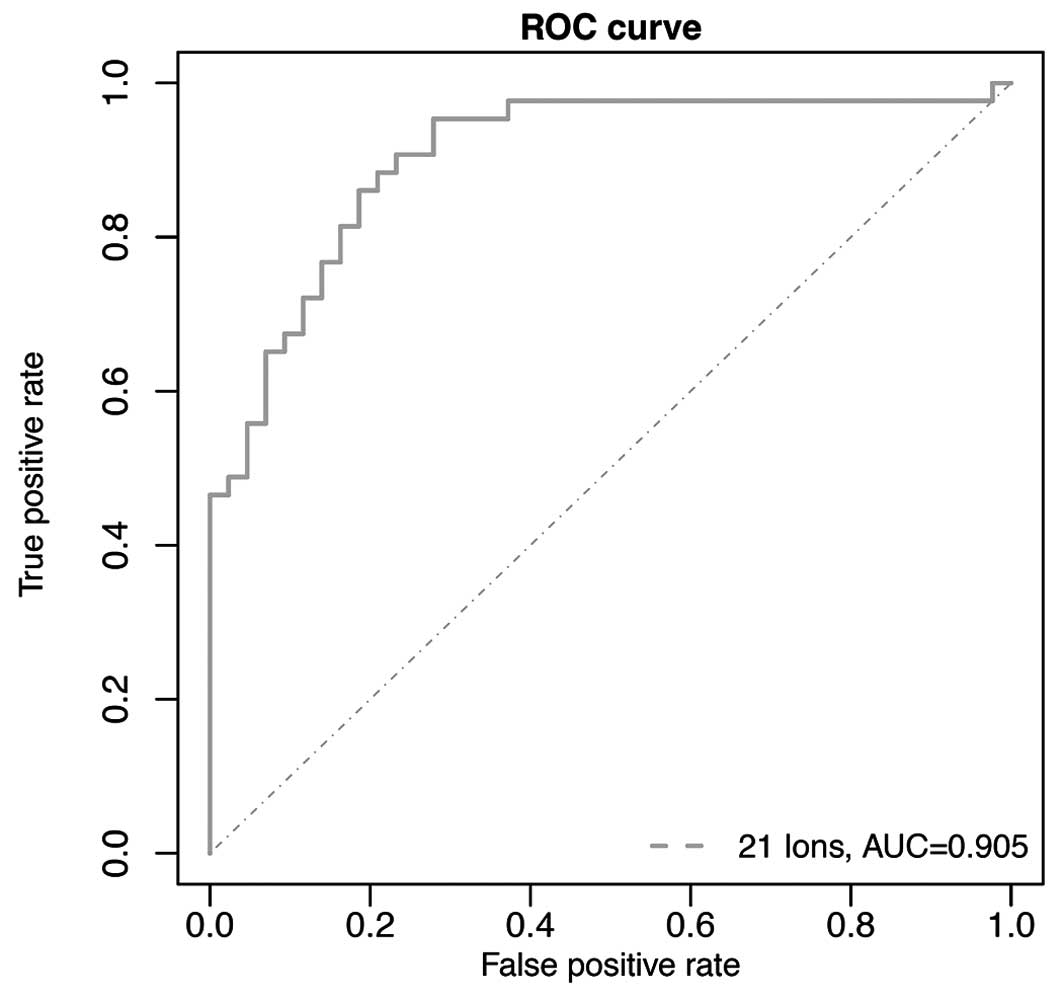
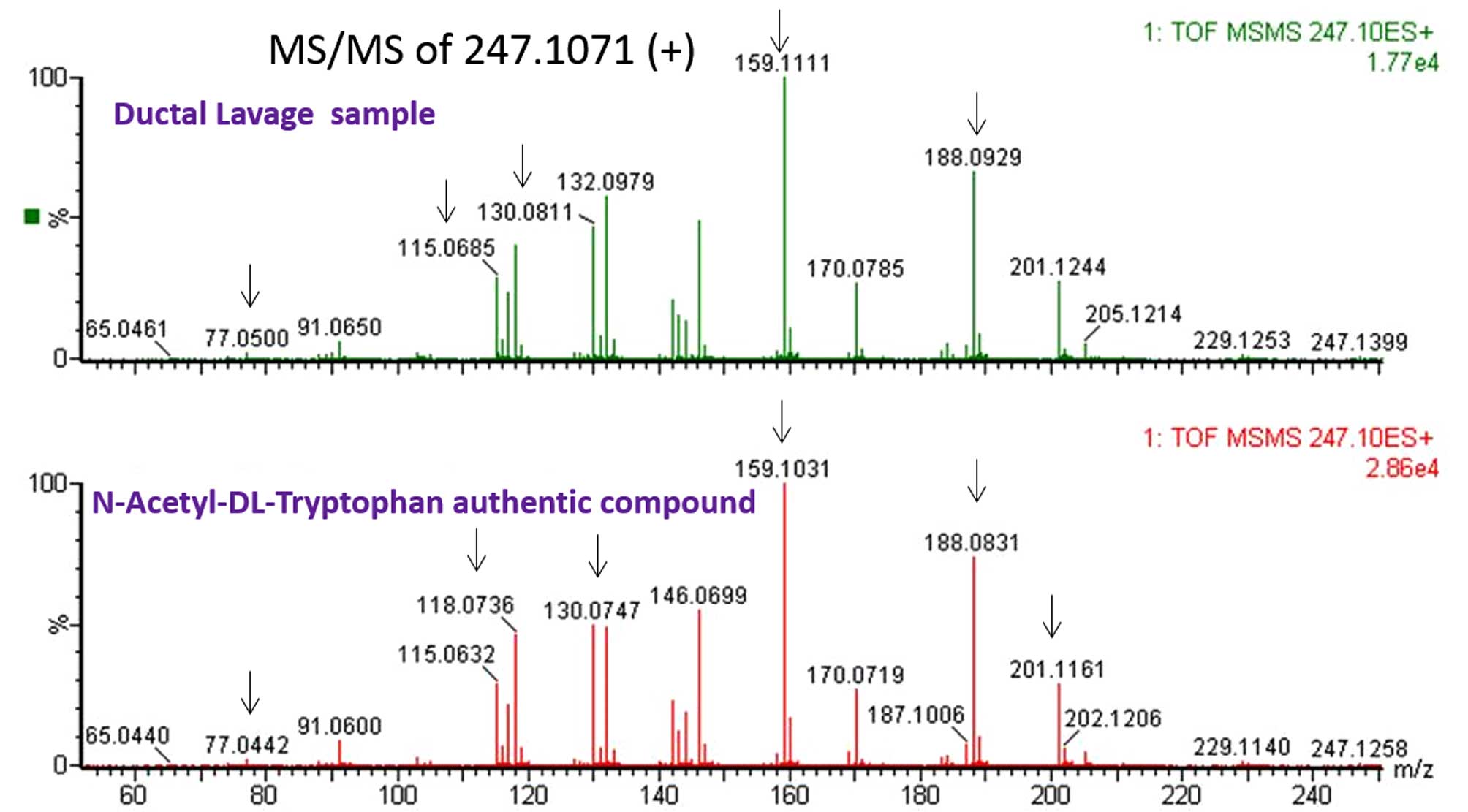
|
1
|
American Cancer Society. Global Cancer
Facts and Figures. 3rd edition. American Cancer Society; Atlanta,
GA: 2015
|
|
2
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Denkert C, Bucher E, Hilvo M, Salek R,
Orešič M, Griffin J, Brockmöller S, Klauschen F, Loibl S, Barupal
DK, et al: Metabolomics of human breast cancer: New approaches for
tumor typing and biomarker discovery. Genome Med.
4:372012.PubMed/NCBI
|
|
4
|
Sengupta D and Pratx G: Imaging metabolic
heterogeneity in cancer. Mol Cancer. 15:42016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Long JP, Li XN and Zhang F: Targeting
metabolism in breast cancer: How far we can go? World J Clin Oncol.
7:122–130. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mannello F and Ligi D: Resolving breast
cancer heterogeneity by searching reliable protein cancer
biomarkers in the breast fluid secretome. BMC Cancer. 13:3442013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shajahan-Haq AN, Cheema MS and Clarke R:
Application of metabolomics in drug resistant breast cancer
research. Metabolites. 5:100–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Claudino WM, Quattrone A, Biganzoli L,
Pestrin M, Bertini I and Di Leo A: Metabolomics: Available results,
current research projects in breast cancer, and future
applications. J Clin Oncol. 25:2840–2846. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Zuo Y, Man YG, Avital I,
Stojadinovic A, Liu M, Yang X, Varghese RS, Tadesse MG and Ressom
HW: Pathway and network approaches for identification of cancer
signature markers from omics data. J Cancer. 6:54–65. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sitter B, Lundgren S, Bathen TF, Halgunset
J, Fjosne HE and Gribbestad IS: Comparison of HR MAS MR
spectroscopic profiles of breast cancer tissue with clinical
parameters. NMR Biomed. 19:30–40. 2006. View Article : Google Scholar
|
|
11
|
Mountford CE, Somorjai RL, Malycha P,
Gluch L, Lean C, Russell P, Barraclough B, Gillett D, Himmelreich
U, Dolenko B, et al: Diagnosis and prognosis of breast cancer by
magnetic resonance spectroscopy of fine-needle aspirates analysed
using a statistical classification strategy. Br J Surg.
88:1234–1240. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Budczies J, Denkert C, Müller BM,
Brockmöller SF, Klauschen F, Györffy B, Dietel M,
Richter-Ehrenstein C, Marten U, Salek RM, et al: Remodeling of
central metabolism in invasive breast cancer compared to normal
breast tissue - a GC-TOFMS based metabolomics study. BMC Genomics.
13:3342012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Budczies J, Brockmöller SF, Müller BM,
Barupal DK, Richter-Ehrenstein C, Kleine-Tebbe A, Griffin JL,
Orešič M, Dietel M, Denkert C, et al: Comparative metabolomics of
estrogen receptor positive and estrogen receptor negative breast
cancer: Alterations in glutamine and beta-alanine metabolism. J
Proteomics. 94:279–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jobard E, Pontoizeau C, Blaise BJ,
Bachelot T, Elena-Herrmann B and Trédan O: A serum nuclear magnetic
resonance-based metabolomic signature of advanced metastatic human
breast cancer. Cancer Lett. 343:33–41. 2014. View Article : Google Scholar
|
|
15
|
Asiago VM, Alvarado LZ, Shanaiah N, Gowda
GA, Owusu-Sarfo K, Ballas RA and Raftery D: Early detection of
recurrent breast cancer using metabolite profiling. Cancer Res.
70:8309–8318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Budhu A, Terunuma A, Zhang G, Hussain SP,
Ambs S and Wang XW: Metabolic profiles are principally different
between cancers of the liver, pancreas and breast. Int J Biol Sci.
10:966–972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Armitage EG and Barbas C: Metabolomics in
cancer biomarker discovery: Current trends and future perspectives.
J Pharm Biomed Anal. 87:1–11. 2014. View Article : Google Scholar
|
|
18
|
Tredwell GD, Miller JA, Chow HH, Thompson
PA and Keun HC: Metabolomic characterization of nipple aspirate
fluid by 1H NMR spectroscopy and GC-MS. J Proteome Res.
13:883–889. 2014. View Article : Google Scholar
|
|
19
|
Do Canto LM, Marian C, Willey S, Sidawy M,
Da Cunha PA, Rone JD, Li X, Gusev Y and Haddad BR: MicroRNA
analysis of breast ductal fluid in breast cancer patients. Int J
Oncol. 48:2071–2078. 2016.PubMed/NCBI
|
|
20
|
Isaacs C, Cavalli LR, Cohen Y, Pennanen M,
Shankar LK, Freedman M, Singh B, Liu M, Gallagher A, Rone JD, et
al: Detection of LOH and mitochondrial DNA alterations in ductal
lavage and nipple aspirate fluids from high-risk patients. Breast
Cancer Res Treat. 84:99–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masood S: Development of a novel approach
for breast cancer prediction and early detection using minimally
invasive procedures and molecular analysis: How cytomorphology
became a breast cancer risk predictor. Breast J. 21:82–96. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sheikh KD, Khanna S, Byers SW, Fornace A
Jr and Cheema AK: Small molecule metabolite extraction strategy for
improving LC/MS detection of cancer cell metabolome. J Biomol Tech.
22:1–4. 2011.PubMed/NCBI
|
|
23
|
Kaur P, Rizk N, Ibrahim S, Luo Y, Younes
N, Perry B, Dennis K, Zirie M, Luta G and Cheema AK: Quantitative
metabolomic and lipidomic profiling reveals aberrant amino acid
metabolism in type 2 diabetes. Mol Biosyst. 9:307–317. 2013.
View Article : Google Scholar
|
|
24
|
Patterson AD, Li H, Eichler GS, Krausz KW,
Weinstein JN, Fornace AJ Jr, Gonzalez FJ and Idle JR:
UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics
inspector self-organizing metabolomic maps as tools for
understanding the cellular response to ionizing radiation. Anal
Chem. 80:665–674. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuhl C, Tautenhahn R, Böttcher C, Larson
TR and Neumann S: CAMERA: An integrated strategy for compound
spectra extraction and annotation of liquid chromatography/mass
spectrometry data sets. Anal Chem. 84:283–289. 2012. View Article : Google Scholar
|
|
26
|
Storey J: False discovery rates.
International Encyclopedia of Statistical Science. 1st edition.
Lovric M: Springer; pp. 16732011
|
|
27
|
Westerhuis JA, van Velzen EJ, Hoefsloot HC
and Smilde AK: Multivariate paired data analysis: Multilevel PLSDA
versus OPLSDA. Metabolomics. 6:119–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reid S and Tibshirani R: Regularization
paths for conditional logistic regression: The clogitL1 package. J
Stat Softw. 58:122014. View Article : Google Scholar
|
|
30
|
Zhou B, Wang J and Ressom HW:
MetaboSearch: Tool for mass-based metabolite identification using
multiple databases. PLoS One. 7:e400962012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wishart DS, Knox C, Guo AC, Eisner R,
Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al:
HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res.
37(Database): D603–D610. 2009. View Article : Google Scholar :
|
|
32
|
Smith CA, O'Maille G, Want EJ, Qin C,
Trauger SA, Brandon TR, Custodio DE, Abagyan R and Siuzdak G:
METLIN: A metabolite mass spectral database. Ther Drug Monit.
27:747–751. 2005. View Article : Google Scholar
|
|
33
|
Cui Q, Lewis IA, Hegeman AD, Anderson ME,
Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR and Markley
JL: Metabolite identification via the Madison Metabolomics
Consortium Database. Nat Biotechnol. 26:162–164. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sud M, Fahy E, Cotter D, Brown A, Dennis
EA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CR, Russell DW, et
al: LMSD: LIPID MAPS structure database. Nucleic Acids Res.
35(Database): D527–D532. 2007. View Article : Google Scholar
|
|
35
|
Fan Y, Zhou X, Xia TS, Chen Z, Li J, Liu
Q, Alolga RN, Chen Y, Lai MD, Li P, et al: Human plasma
metabolomics for identifying differential metabolites and
predicting molecular subtypes of breast cancer. Oncotarget.
7:9925–9938. 2016.PubMed/NCBI
|
|
36
|
Mishra P and Ambs S: Metabolic signatures
of human breast cancer. Mol Cell Oncol. 2:22015.
|
|
37
|
Qiu Y, Zhou B, Su M, Baxter S, Zheng X,
Zhao X, Yen Y and Jia W: Mass spectrometry-based quantitative
metabolomics revealed a distinct lipid profile in breast cancer
patients. Int J Mol Sci. 14:8047–8061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen J, Yan L, Liu S, Ambrosone CB and
Zhao H: Plasma metabolomic profiles in breast cancer patients and
healthy controls: By race and tumor receptor subtypes. Transl
Oncol. 6:757–765. 2013. View Article : Google Scholar
|
|
39
|
Willmann L, Schlimpert M, Halbach S, Erbes
T, Stickeler E and Kammerer B: Metabolic profiling of breast
cancer: Differences in central metabolism between subtypes of
breast cancer cell lines. J Chromatogr B Analyt Technol Biomed Life
Sci. 1000:95–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamashita J, Ogawa M and Sakai K:
Prognostic significance of three novel biologic factors in a
clinical trial of adjuvant therapy for node-negative breast cancer.
Surgery. 117:601–608. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamashita S, Yamashita J, Sakamoto K,
Inada K, Nakashima Y, Murata K, Saishoji T, Nomura K and Ogawa M:
Increased expression of membrane-associated phospholipase A2 shows
malignant potential of human breast cancer cells. Cancer.
71:3058–3064. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Takayama T, Tsutsui H, Shimizu I, Toyama
T, Yoshimoto N, Endo Y, Inoue K, Todoroki K, Min JZ, Mizuno H, et
al: Diagnostic approach to breast cancer patients based on target
metabolomics in saliva by liquid chromatography with tandem mass
spectrometry. Clin Chim Acta. 452:18–26. 2016. View Article : Google Scholar
|
|
43
|
Hadi NI and Jamal Q: ‘OMIC’ tumor markers
for breast cancer: A review. Pak J Med Sci. 31:1256–1262. 2015.
View Article : Google Scholar : PubMed/NCBI
|