Introduction
Lung cancer is one of the most common cancers and
the major cause of cancer deaths worldwide, with 1.6 million new
lung cancer cases and 1.4 million lung cancer deaths each year
(1). Although surgery is used to
treat the early phase of lung cancer, the diagnosis of most
patients is confirmed at an advanced stage when surgery is
impossible (2). Chemotherapy and
radiotherapy are used in clinical practice for lung cancer
treatment, but the prognosis is poor, and the survival rate of
patients with lung cancer is <15% after 5 years (3). Therefore, it is critical to find
novel strategies and effective methods for the treatment of lung
cancer.
Histones are basic nuclear proteins that are
involved in chromatin condensation in eukaryotic cells. There are
five types of histones: H1/H5, H2A, H2B, H3 and H4 with no introns
in their encoding genes, and no poly A tail in their transcripts
(4–9). Increasing evidence has demonstrated
that histones regulate chromatin states via covalent modification
of amino acids, which further regulates gene expression. For
instance, the less compacted chromatin domains show high
transcriptional activity (10–12).
The properties of histones in cancer cells are known to differ from
those of noncancerous cells (13).
A recent study demonstrated that histone mutations result in
pediatric glioblastoma (14).
Prostate cancer is associated with histone H2A.Z deregulation
(15). HIST1H3D gene,
located on chromosome 6, encodes histone H3.1, a member of the H3
class of histones in humans (16).
H3.1 dimethylation of H3 lysine 9 is involved in gene-silencing and
heterochromatin formation (17).
Recently, it was shown that HIST1H3D expression is upregulated in
primary gastric cancer tissue (18).
In this study, we investigated HIST1H3D expression
in lung cancer tissues and human lung cancer cell lines, and
explored the effects of HIST1H3D expression on lung cancer and
signaling in proliferation and apoptosis.
Materials and methods
Patients and tissue samples
Samples of lung cancer tissue and adjacent
non-cancerous lung tissue were obtained from 74 patients
hospitalized in the Department of Thoracic Surgery at the First
Affiliated Hospital of Bengbu Medical College (Anhui, China) from
July 2004 to September 2007. All subjects provided written informed
consent. The experimental protocols were approved by the Ethics
Committee of Bengbu Medical College. Clinicopathological features
such as tumor size, pathological stage, lymph node metastasis and
survival were recorded. Overall survival was calculated from the
first surgical date to the date of death or the date of last
follow-up (July 2012).
Cell culture and MTT cell proliferation
assay
The H1299, H1688, H1975 and A549 human lung cancer
cells were cultured in RPMI-1640 medium (Sigma, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum.
H1299 cells were plated in 96-well plates at a
density of 2,000 cells/well. The cell viability was assessed on
days 1, 2, 3, 4 and 5. MTT (20 µl, 5 mg/ml) was added into
each well. DMSO (150 µl) was added to dissolve the crystals
after 4 h of incubation. The optical density (OD) of each well was
measured at 570 nm using an ELISA reader (Biotek®
Elx800; Bio-Tek Instruments, Inc., Winooski, VT, USA).
Immunohistochemistry evaluation
HIST1H3D immunoreactivity in the nucleus was
quantified based on a system in which scores of 0, 1, 2, 3 and 4
were assigned according to the percentage of positive cells at 0,
1–25, 26–50, 51–75 and 76–100%, respectively (19). The staining intensity scores were
defined as 0 (negative), 1 (+), 2 (++) and 3 (+++). The product of
the positive cells and intensity score was calculated. High and low
expression of HIST1H3D in the nucleus was defined as >1.5 and
≤1.5, respectively.
Lentiviral construction of
small-interfering RNA (siRNA) treatment
HIST1H3D-specific siRNA (5′-GCT GAT TCG CAA ACT GCC
ATT-3′) and the negative control (NC) sequence (5′-TTC TCC GAA CGT
GTC ACG T-3′) were cloned into the AgeI and EcoRI
sites of the pGCSIL-GFP lentiviral vector (GeneChem, Shanghai,
China) to generate the pGCSIL-GFP-HIST1H3D-siRNA plasmid for
specific inhibition of HIST1H3D expression and the
pGCSIL-GFP-NC plasmid as the NC.
Inhibition of HIST1H3D expression in H1299
cells with HIST1H3D-specific siRNA was performed. H1299 cells were
seeded into 6-well plates (5×104 cells/well). NC siRNA
(5′-TTC TCC GAA CGT GTC ACG T-3′) and HIST1H3D-specific siRNA
(5′-GCT GAT TCG CAA ACT GCC ATT-3′) were transfected into the cells
using a viral system at a multiplicity of infection (MOI) according
to the manufacturer's protocol.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from H1299 cells using
TRIzol reagent and cDNA was generated from a sample of total RNA (2
µg) by reverse transcription using the
PrimeScript® RT kit. Total RNA was used for reverse
transcription. RT-PCR was performed with the SYBR®
PrimeScript™ RT-PCR kit (Takara, Kyoto, Japan) according to the
manufacturer's instructions. The specific sequences of primers were
as follows: HIST1H3D (forward, 5'-TTC GCA AAC TGC CAT TCC-3′ and
reverse, 5′-GAG CCT TTG GGT TTT GGT T-3′), and GAPDH (forward,
5′-TGA CTT CAA CAG CGA CAC CCA-3′ and 5′-CAC CCT GTT GCT GTA GCC
AAA-3′). GAPDH was used as an internal control. Primers were
designed by Beacon Designer 2 software. The 2−ΔΔCT
method was used to analyze the relative changes in gene
expression.
Western blot analysis
Lung cancer tissue and adjacent non-cancerous
tissues or H1299 cells were harvested and homogenized in a
pre-chilled lysis buffer (Sangon Biotech). Homogenates were
centrifuged at 12,000 × g for 20 min at 4°C. Equal amounts of
denatured proteins were separated by SDS-PAGE and then transferred
to PVDF membranes. To block non-specific binding, membranes were
incubated in TBST containing 5% fat-free milk for 2 h. The
membranes were then incubated with primary antibodies (polyclonal
anti-HIST1H3D, anti-CCNE2, anti-CDK6, anti-CDKN1A, anti-THBS1,
anti-TP53I3 and anti-GAPDH, 1:1,000 dilution) overnight at 4°C.
After washing with TBST, membranes were incubated with an
HRP-conjugated secondary antibody (1:1,000 dilution; Sangon
Biotech) for 2 h at room temperature. After being rinsed with
washing buffer, immunoreactive bands were visualized using EasyBlot
ECL kit (Sangon Biotech).
Cell growth assay
Cell growth was determined using a Cellomics
ArrayScan (GeneChem). Infected cells (800 cells/well) were seeded
into 96-well plates and cell growths was assayed at 1, 2, 3, 4 and
5 days using a Cellomics ArrayScan (Cellomics Inc., Pittsburgh, PA,
USA). Cell growth curves were constructed for each cell type.
Flow cytometric analyses
For cell cycle analysis, cells (2,000 cells/well)
were cultured in 6-well plates. At 3 days post-infection with
HIST1H3D-siRNA lentivirus or control lentivirus, cells were then
collected, washed twice with ice-cold PBS and fixed with 70%
ethanol at 4°C. The cells were then stained with PBS containing 50
µg/ml propidium iodide (PI, 2 mg/ml; Sigma-Aldrich) in the
presence of 100 µg/ml of RNase (10 mg/ml; Sangon Biotech).
For analysis of cell apoptosis, the cells were stained with 100
µl binding buffer containing 5 µl Annexin V-APC (BD
Bioscience, San Diego, CA, USA) at room temperature in the dark for
10–15 min. Cells were analyzed by flow cytometry using FACSCalibur
(BD Biosciences).
Colony formation assay
H1299 cells were plated in 6-well plates (500
cells/well) and cultured at 37°C in a humidified atmosphere
containing 5% CO2 for 14 days to allow colony formation.
At the indicated time-points, cells were washed twice with PBS,
treated with Giemsa stain for 20 min, and then washed several times
with ddH2O. The number of colonies was counted under a
fluorescence microscope (MicroPublisher 3.3RTV; Olympus, Tokyo,
Japan).
Gene expression profiling
After transfection, total RNA was extracted from
H1299 cells using TRIzol reagent. RNA quality was assessed by
NanoDrop 2000 and Agilent Bioanalyzer 2100. Gene expression
profiling was performed using human gene arrays on the Affymetrix
GeneChip PrimeView platform according to the manufacturer's
protocol. Differentially expressed genes were identified on the
basis of a 1.5-fold change in expression with P-values <0.05.
KEGG and BioCarta pathway analyses were performed to investigate
the functional and pathway annotation of all the selected
genes.
Statistical analyses
Data were expressed as mean ± standard deviation
(SD). The difference between two groups was determined by Student's
t-test or Wilcoxon's rank test. Correlations between HIST1H3D
expression and clinicopathological characteristics were determined
using Pearson's χ2 test. Survival analysis was performed
using the log-rank test. All statistical analyses were performed
using SPSS 12.0 software. A value of P<0.05 was considered to
indicate statistical significance.
Results
HIST1H3D expression in lung cancer cells
and tissues
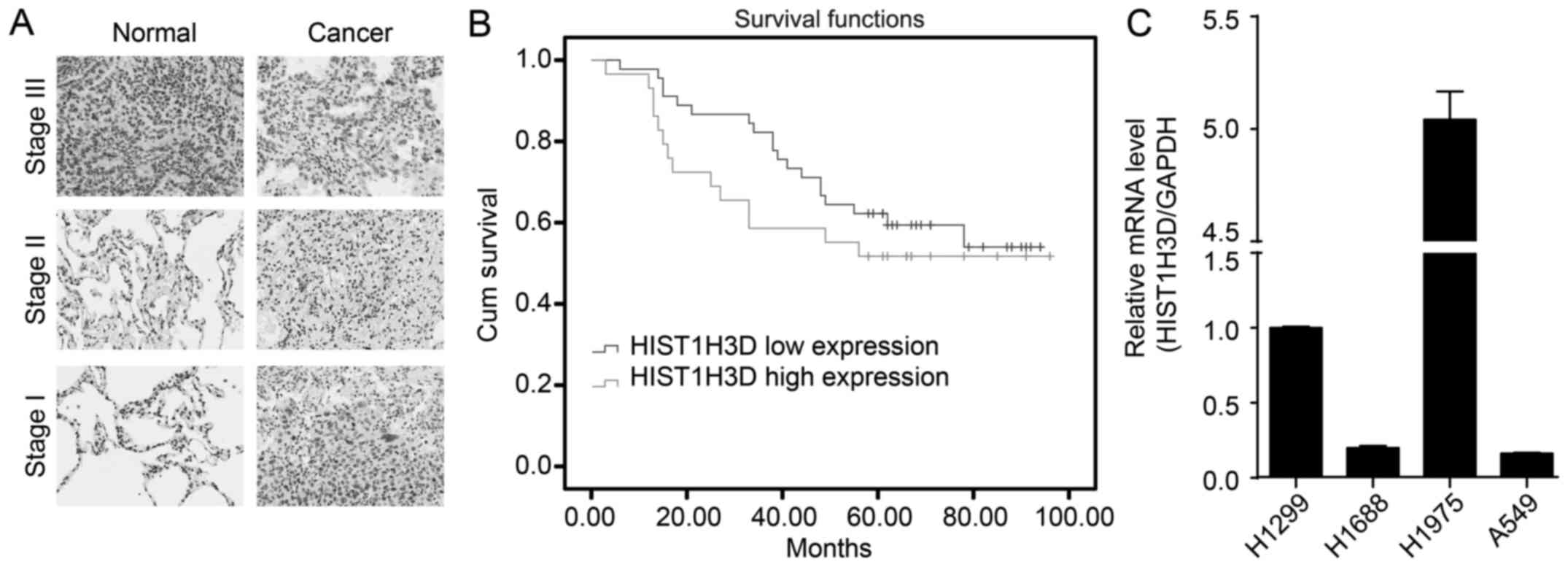
Our investigation of the expression of HIST1H3D mRNA
in lung cancer and the adjacent non-cancerous tissues showed that
the level of HIST1H3D protein in lung cancer tissues was
significantly increased, compared with the adjacent non-cancerous
tissues (P<0.01) (Fig. 1A).
Kaplan-Meier survival analysis suggested that HIST1H3D expression
in lung cancer tissues was not associated with that in the adjacent
non-cancerous tissues (P-value for log-rank, 0.35) (Fig. 1B). Furthermore, we determined that
HIST1H3D mRNA was highly expressed in the lung cancer cell lines
H1299 and H1975 (Fig. 1C).
However, HIST1H3D mRNA was expressed at low levels in the lung
cancer cell lines A459 and H1688. The discrepancy of HIST1H3D
expression in different lung cancer cell lines may be due to tumor
heterogeneity.
In addition, higher levels of HIST1H3D were observed
in lung cancer tissues at stage III compared with the levels
observed at stage I/II (P<0.05) (Table I), with higher expression in the
cases with lymph node metastasis compared to those without
(P<0.05) (Table I). However,
there was no significant difference in HIST1H3D expression between
the genders, nor a significant association with age, tumor size,
and distant metastasis (P>0.05) (Table I).
 | Table IRelationship between HIST1H3D
expression and clinicopathological characteristics of patients with
lung cancer. |
Table I
Relationship between HIST1H3D
expression and clinicopathological characteristics of patients with
lung cancer.
| Variable | HIST1H3D expression
| P-value |
|---|
| Low | High |
|---|
| Gender |
| Male | 24 | 15 | 0.89 |
| Female | 21 | 14 | |
| Age (years) |
| ≤60 | 25 | 16 | 0.89 |
| >60 | 20 | 13 | |
| Tumor size
(cm) |
| ≤3.5 | 25 | 13 | 0.37 |
| >3.5 | 20 | 16 | |
| Stage |
| I | 11 | 0 | 0.02 |
| II | 22 | 19 | |
| III | 12 | 10 | |
| Lymph node
metastasis |
| Nx/N0 | 35 | 16 | 0.04a |
| N1/N2/N3 | 10 | 13 | |
| Distant
metastasis |
| M0 | 43 | 28 | 0.81 |
| M1 | 2 | 1 | |
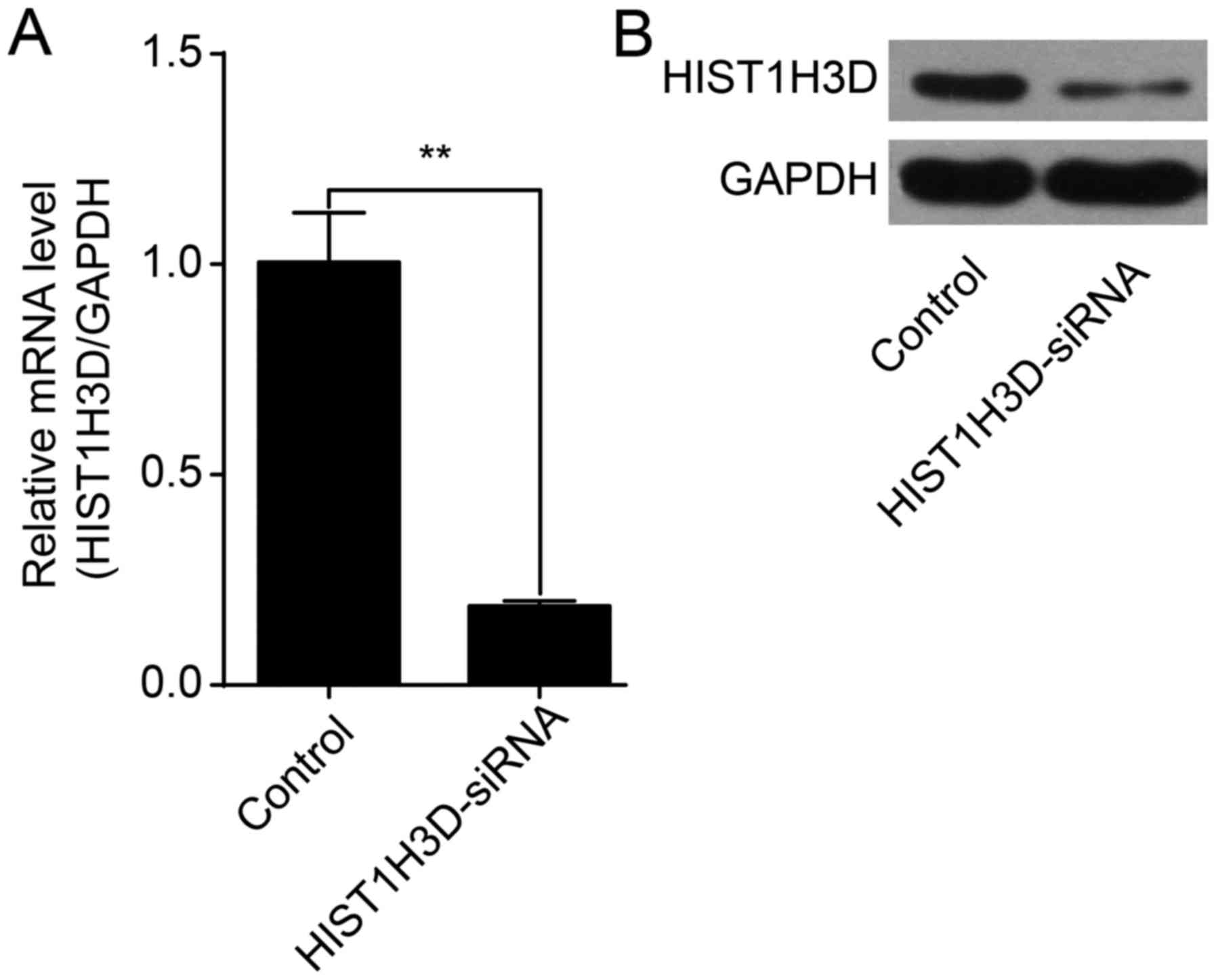
Lentiviral transfection with HIST1H3D
suppresses HIST1H3D expression
HIST1H3D siRNA or control lentivirus was transfected
into H1299 cells. HIST1H3D mRNA levels were significantly reduced
by 81% in cells transfected with HIST1H3D siRNA compared to cells
transfected with NC siRNA (P<0.01) (Fig. 2A). Western blot analysis showed
that HIST1H3D protein expression was decreased in cells transfected
with HIST1H3D siRNA (Fig. 2B).
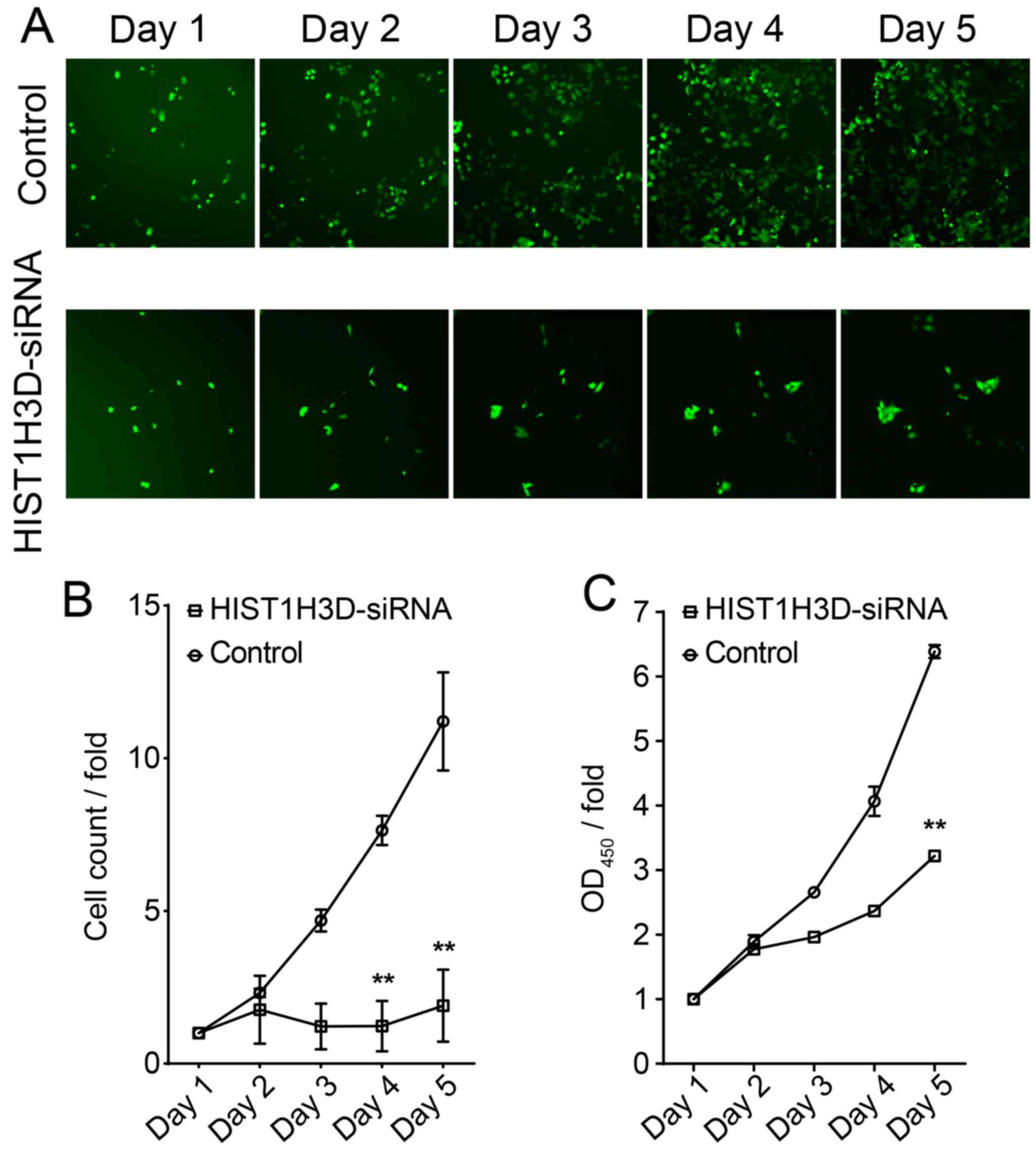
Effect of siRNA-mediated knockdown of
HIST1H3D on cell proliferation
To evaluate the effect of HIST1H3D on cell
proliferation, we determined proliferation of H1299 cells
transfected with HIST1H3D-specific siRNA. The number of cells in
the control group increased in a time-dependent manner, while only
a slight increase in cell numbers was observed in the
HIST1H3D-siRNA group (Fig. 3A and
B), with a significant difference in cell numbers between the
Control and HIST1H3D-siRNA groups observed at 5 days post-infection
(P<0.01) (Fig. 3B).
H1299 cell proliferation was also evaluated by MTT
assay. The results showed that HIST1H3D-siRNA group cell growth at
day 5 were significantly inhibited compared to control group
(P<0.01) (Fig. 3C).
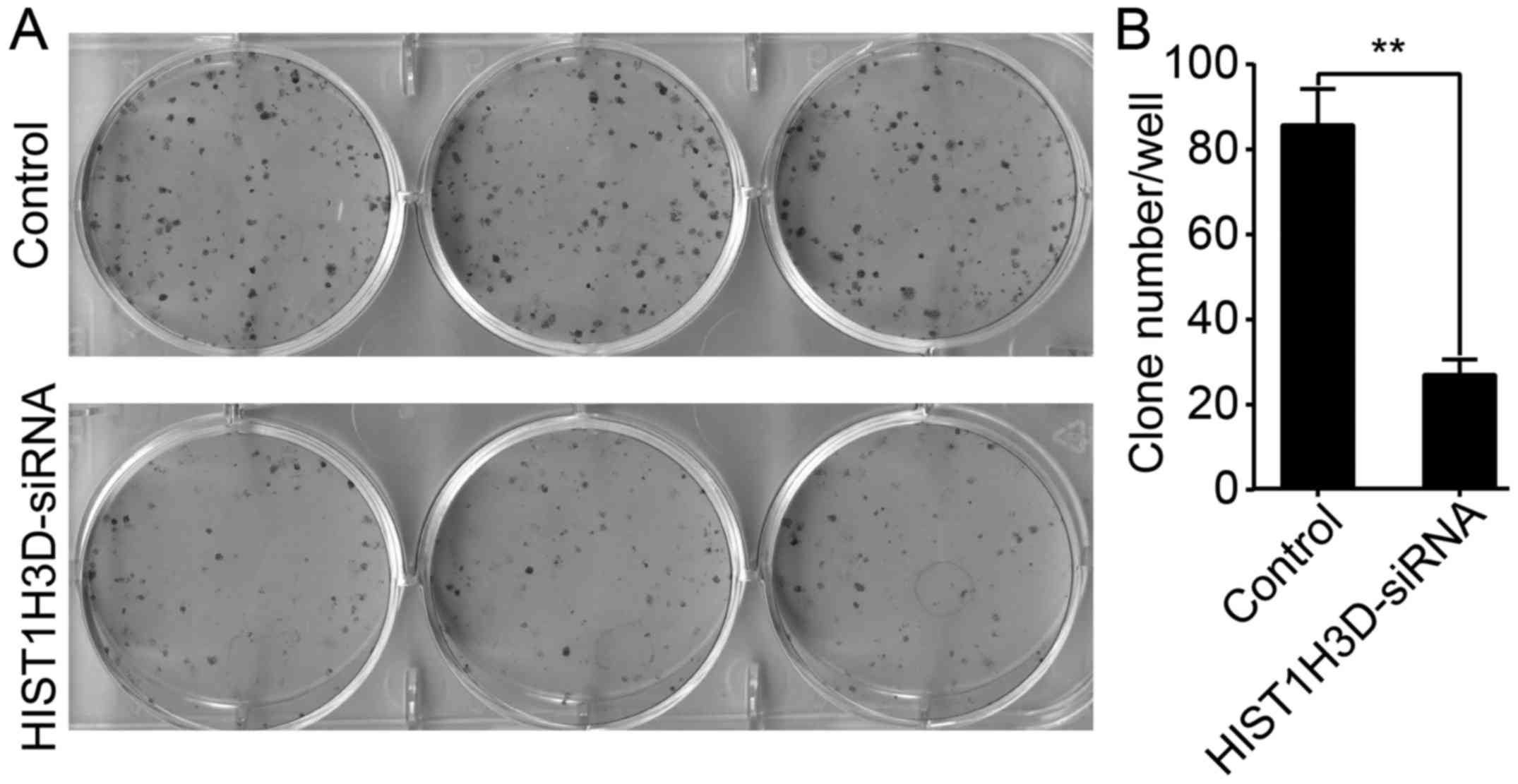
Effect of HIST1H3D siRNA on cell colony
formation
As shown in Fig. 4,
the number of colonies was decreased by 64.6% in H1299 cells
transfected with HIST1H3D siRNA compared with those transfected
with NC siRNA (P<0.01).
Effect of HIST1H3D siRNA on cell
apoptosis
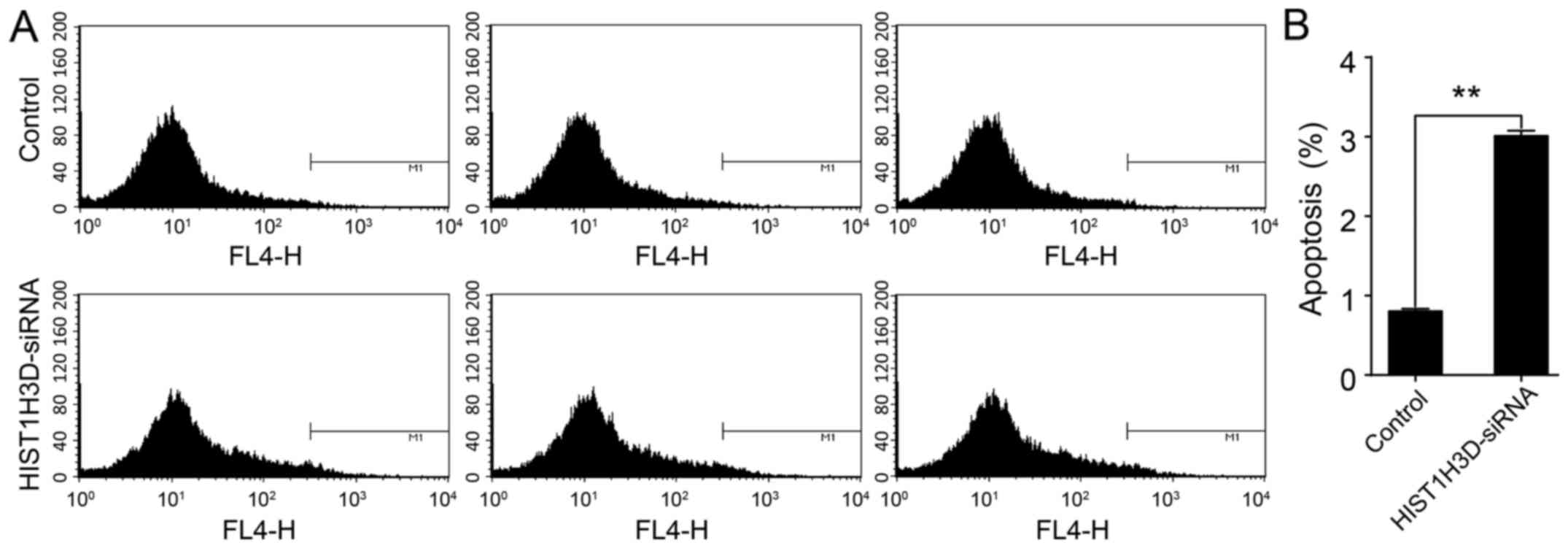
To determine the effect of HIST1H3D expression on
cell survival, cell apoptosis was analyzed by a flow cytometry. As
shown in Fig. 5, the number of
apoptotic cells transfected with HIST1H3D siRNA was significantly
greater than that when compared with the number of NC siRNA
transfected cells (P<0.01).
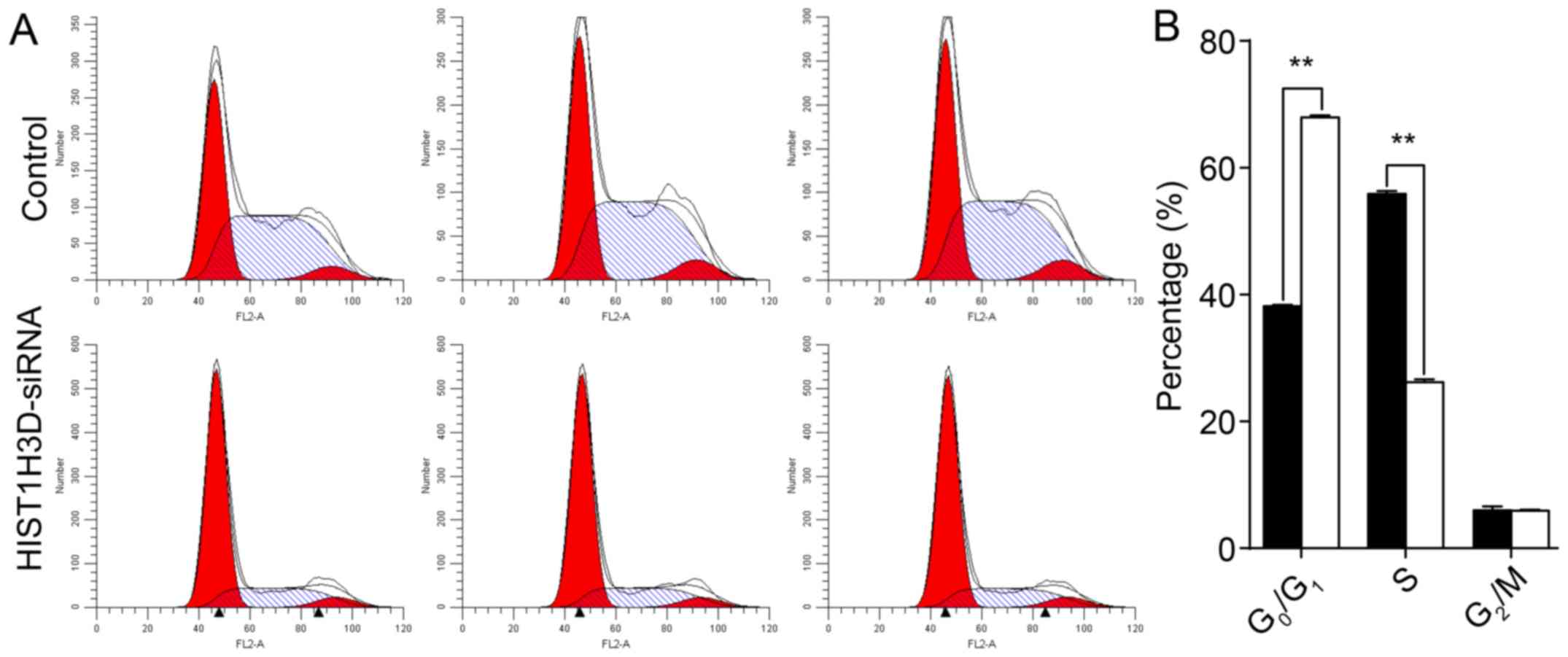
Effect of HIST1H3D siRNA on cell cycle
distribution
Cell cycle analysis showed that, following
transfection with HIST1H3D siRNA, 67.9, 26.2 and 5.9% of the cells
were in the G0/G1, S and G2/M
phases, respectively (Fig. 6).
Following transfection with NC siRNA, 38.2, 55.9 and 5.9% of the
cells were in the G0/G1, S and
G2/M phases, respectively. The result suggested that
inhibition of HIST1H3D-siRNA arrested cells in the
G0/G1 phase (P<0.01).
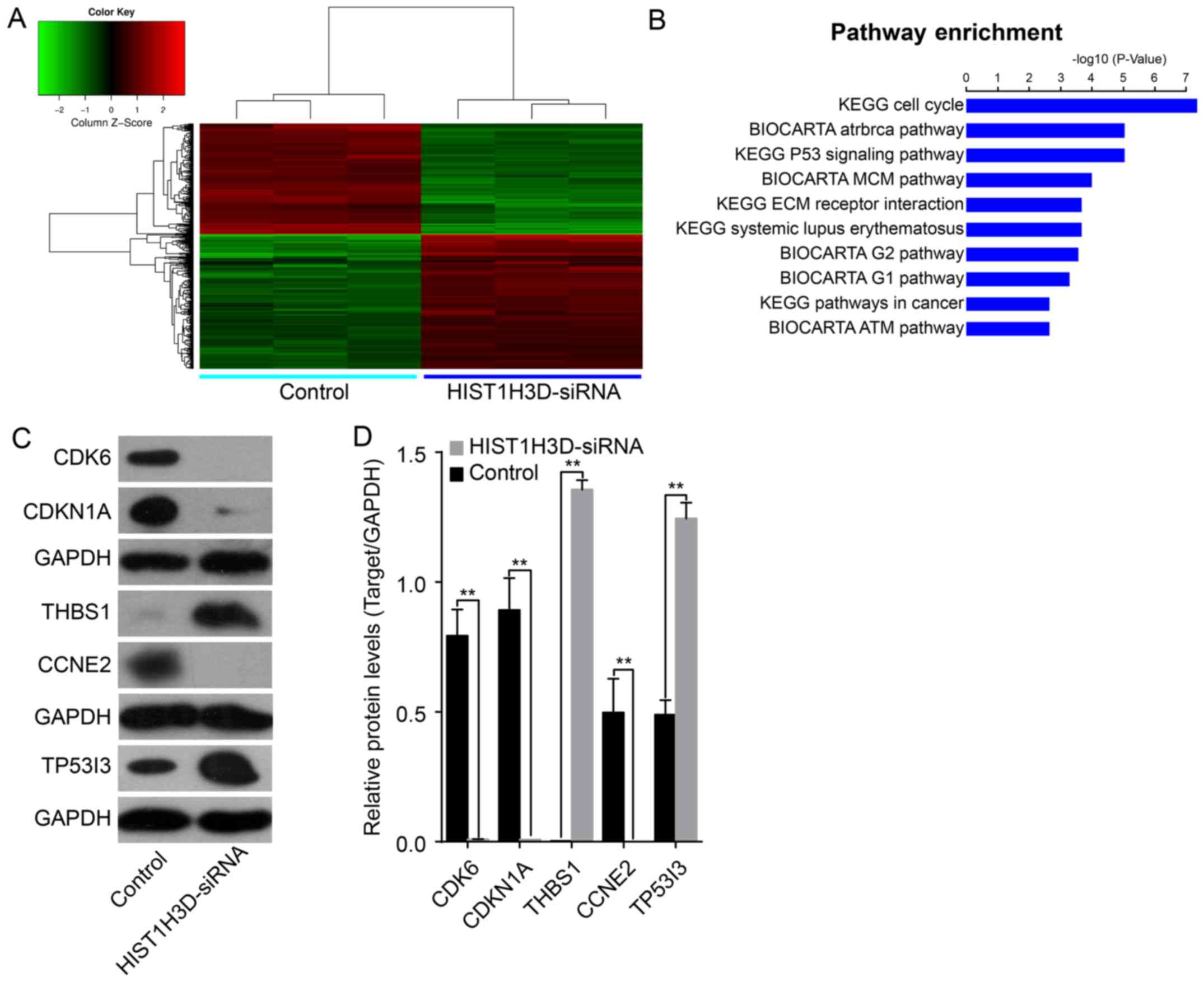
Profiling of expression of genes
regulated by HIST1H3D
To determine the effects of HIST1H3D on the
potential pathways involved in the development of lung cancer, we
performed microarray analysis to identify the differential gene
expression profiles between cells following siRNA-mediated
knockdown of HIST1H3D. A total of 522 differentially expressed
genes (284 upregulated and 238 downregulated) were identified
following HIST1H3D knockdown (Fig.
7A). KEGG and BioCarta pathway analysis showed that
HIST1H3D-regulated genes were involved mainly in the cell cycle
progression, p53 signaling, and MCM (Fig. 7B).
Most of the DNA microarray results were validated by
western blot assays (Fig. 7C). We
observed upregulated expression of the THBS1 and
TP53I3 genes, and downregulated expression of the
CDK6, CDKN1 and CCNE2 genes in si-HIST1H3D
group when compared to the control group (Fig. 7D).
Discussion
Histones are basic nuclear proteins that are
involved in regulation of chromatin structure in eukaryotic cells.
Histones undergo post-translational modifications of the their
N-terminal tail, such as methylation, acetylation, phosphorylation,
citrullination, SUMOylation, ubiquitination and ADP-ribosylation
(12). These modifications alter
chromatin structure, with a critical role in regulating gene
expression. Abnormal histone modifications can lead to changes in
the chromosome structure and gene transcription, and also plays a
critical role in cancer cell proliferation, differentiation and
apoptosis (20,21).
Accumulating evidence shows that histones contribute
to the pathogenesis of various malignancies (22,23).
Previous studies have shown that histone H3 mRNA is accumulated in
tumors, and upregulation of histone H3 mRNA expression increases
the proliferative activity of tumor cells (24,25).
Furthermore, recent studies suggest that histone cluster genes are
overexpressed in several types of malignancies. Sadikovic et
al performed integrative whole-genome analysis of DNA copy
number, promoter methylation and gene expression in 10
osteosarcomas, and identified overexpression of histone cluster 2
genes at chromosome 1q21.1–q21.3 (26). In microarray-based expression
profiling of meningiomas, and Pérez-Magán et al identified
overexpression of 16 histone cluster 1 genes located on chromosome
6p in recurrent meningiomas (27).
In the present study, we first determined the
HIST1H3D expression in lung cancer tissues and adjacent
non-cancerous tissues. Results indicated that HIST1H3D expression
was increased in lung cancer tissues compared with that in adjacent
non-cancerous tissues. We observed higher expression of HIST1H3D in
patients with advanced disease (TNM II/III) and lymph node
metastasis (N1/N2/N3), while there were no significant differences
in HIST1H3D expression between the genders. Furthermore, there were
no significant associations of HIST1H3D expression with age, tumor
size or distant metastasis. Furthermore, we demonstrated increased
HIST1H3D expression in two lung cancer cell lines H1299 and H1975
and decreased HIST1H3D expression in A459 and H1688 cells. These
findings suggest that HIST1H3D expression may be associated with
tumor heterogeneity in lung cancer. Iwaya et al found that
HIST1H3D is associated with the development of high-risk lung
cancer (18). Thus, association
between HIST1H3D expression and the risk of lung cancer requires
further investigation.
To assess the effects of HIST1H3D expression on lung
cancer, we reduced the HIST1H3D expression by RNA interference
(RNAi) in H1299 human lung cancer cells. RNAi is a powerful genetic
tool for silencing gene expression through degradation of specific
mRNA (28–31). After confirming siRNA-mediated
knockdown of HIST1H3D in H1299 cells, we observed reduced
proliferation and colony formation as well as increased apoptosis.
These results suggest that upregulation of HIST1H3D expression
promotes tumor cell development. However, the mechanisms by which
HIST1H3D knockdown inhibits proliferation and increases apoptosis
remain to be clarified due to the discrepancy between suppression
of cell proliferation and increase of apoptosis ratio.
The cell cycle is the series of events that lead to
cell division. We performed flow cytometric cell cycle analysis to
elucidate the mechanism underlying the effects of HIST1H3D on the
control of cell growth. Interestingly, we found that HIST1H3D
knockdown induced G0/G1 cell cycle arrest,
indicating that HIST1H3D promotes human lung cancer cell
proliferation and colony formation by modulating cell cycle
progression. However, the mechanism responsible for cell cycle
arrest of HIST1H3D-silenced cells remains to be clarified in future
studies.
The tumor suppressor p53 plays key roles in
regulating the cell cycle and apoptosis by modulating the
expression of target genes (32,33).
In this study, cDNA microarray analysis indicated that the p53
signaling pathway is triggered following HIST1H3D-knockdown, these
results were validated by western blot analysis. Differential
expression of a number of genes in the p53 signaling pathway was
detected, with upregulation of genes such as THBS1 and
TP53I3 and downregulation of other genes, including
CDK6, CDKN1 and CCNE2. These results suggest
that the HIST1H3D is involved in the development of lung cancer
through regulating the expression of these genes. Although the
current data are too limited to support an in-depth hypothesis, our
results provide a direction for future research.
In conclusion, our results show that HIST1H3D
expression is upregulated in lung cancer tissues and cell lines.
Furthermore, this study demonstrates that siRNA-mediated
downregulation of HIST1H3D expression inhibits cell growth,
proliferation and colony formation, while promoting cell apoptosis,
and cycle arrest in the G0/G1 phase. These
findings indicate that HIST1H3D plays an important role in lung
cancer development and implicate HIST1H3D as a therapeutic target
in lung cancer.
Acknowledgments
This study was supported by the Personnel training
funds of Anhui Province Health and Family Planning Commission, the
Natural Science Fund of Education Department of Anhui Province
(KJ2015B070), and the Doctor's Scientific Research Foundation of
Bengbu Medical College.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Li Z, Bai L and Lin Y: NF-kappaB
in lung cancer, a carcinogenesis mediator and a prevention and
therapy target. Front Biosci (Landmark Ed). 16:1172–1185. 2011.
View Article : Google Scholar
|
|
3
|
Vischioni B, Oudejans JJ, Vos W, Rodriguez
JA and Giaccone G: Frequent overexpression of aurora B kinase, a
novel drug target, in non-small cell lung carcinoma patients. Mol
Cancer Ther. 5:2905–2913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birnstiel ML, Busslinger M and Strub K:
Transcription termination and 3′ processing: The end is in site!
Cell. 41:349–359. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu TJ, Levine BJ, Skoultchi AI and
Marzluff WF: The efficiency of 3′-end formation contributes to the
relative levels of different histone mRNAs. Mol Cell Biol.
9:3499–3508. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Osley MA: The regulation of histone
synthesis in the cell cycle. Annu Rev Biochem. 60:827–861. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zanier K, Luyten I, Crombie C, Muller B,
Schümperli D, Linge JP, Nilges M and Sattler M: Structure of the
histone mRNA hairpin required for cell cycle regulation of histone
gene expression. RNA. 8:29–46. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meshi T, Taoka KI and Iwabuchi M:
Regulation of histone gene expression during the cell cycle. Plant
Mol Biol. 43:643–657. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schümperli D: Cell-cycle regulation of
histone gene expression. Cell. 45:471–472. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Turner BM: Cellular memory and the histone
code. Cell. 111:285–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berger SL: Histone modifications in
transcriptional regulation. Curr Opin Genet Dev. 12:142–148. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jenuwein T and Allis CD: Translating the
histone code. Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cruft HJ, Mauritzen CM and Stedman E:
Abnormal properties of histones from malignant cells. Nature.
174:580–585. 1954. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rheinbay E, Louis DN, Bernstein BE and
Suvà ML: A tell-tail sign of chromatin: Histone mutations drive
pediatric glioblastoma. Cancer Cell. 21:329–331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dryhurst D and Ausió J: Histone H2A.Z
deregulation in prostate cancer. Cause or effect? Cancer Metastasis
Rev. 33:429–439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Albig W, Kioschis P, Poustka A, Meergans K
and Doenecke D: Human histone gene organization: Nonregular
arrangement within a large cluster. Genomics. 40:314–322. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peters AH and Schübeler D: Methylation of
histones: Playing memory with DNA. Curr Opin Cell Biol. 17:230–238.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iwaya T, Fukagawa T, Suzuki Y, Takahashi
Y, Sawada G, Ishibashi M, Kurashige J, Sudo T, Tanaka F, Shibata K,
et al: Contrasting expression patterns of histone mRNA and microRNA
760 in patients with gastric cancer. Clin Cancer Res. 19:6438–6449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Tang J, Liao D, Wang G, Zhang M,
Sang Y, Cao J, Wu Y, Zhang R, Li S, et al: Chromobox homolog 4 is
correlated with prognosis and tumor cell growth in hepatocellular
carcinoma. Ann Surg Oncol. 20(Suppl 3): S684–S692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berdasco M and Esteller M: Aberrant
epigenetic landscape in cancer: How cellular identity goes awry.
Dev Cell. 19:698–711. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turner G and Hancock RL: Histone methylase
activity of adult, embryonic and neoplastic liver tissues. Life Sci
II. 9:917–922. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ballestar E and Esteller M: The epigenetic
breakdown of cancer cells: From DNA methylation to histone
modifications. Prog Mol Subcell Biol. 38:169–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sakamoto R, Nitta T, Kamikawa Y, Sugihara
K, Hasui K, Tsuyama S and Murata F: The assessment of cell
proliferation during 9,10-dimethyl-1,2-benzanthracene-induced
hamster tongue carcinogenesis by means of histone H3 mRNA in situ
hybridization. Med Electron Microsc. 37:52–61. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Piscopo M, Campisi G, Colella G,
Bilancione M, Caccamo S, Di Liberto C, Tartaro GP, Giovannelli L,
Pulcrano G and Fucci L: H3 and H3.3 histone mRNA amounts and ratio
in oral squamous cell carcinoma and leukoplakia. Oral Dis.
12:130–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sadikovic B, Yoshimoto M, Chilton-MacNeill
S, Thorner P, Squire JA and Zielenska M: Identification of
interactive networks of gene expression associated with
osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol
Genet. 18:1962–1975. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pérez-Magán E, Rodríguez de Lope A,
Ribalta T, Ruano Y, Campos-Martín Y, Pérez-Bautista G, García JF,
García-Claver A, Fiaño C, Hernández-Moneo JL, et al: Differential
expression profiling analyses identifies downregulation of 1p, 6q,
and 14q genes and overexpression of 6p histone cluster 1 genes as
markers of recurrence in meningiomas. Neuro Oncol. 12:1278–1290.
2010.PubMed/NCBI
|
|
28
|
Sen GL and Blau HM: A brief history of
RNAi: The silence of the genes. FASEB J. 20:1293–1299. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong Q: RNAi: A novel strategy for the
treatment of prion diseases. J Clin Invest. 116:3101–3103. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Z, Li G, Wu L, Weng D, Li X and Yao K:
Cripto-1 overexpression is involved in the tumorigenesis of
nasopharyngeal carcinoma. BMC Cancer. 9:3152009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sumimoto H, Hirata K, Yamagata S, Miyoshi
H, Miyagishi M, Taira K and Kawakami Y: Effective inhibition of
cell growth and invasion of melanoma by combined suppression of
BRAF (V599E) and Skp2 with lentiviral RNAi. Int J Cancer.
118:472–476. 2006. View Article : Google Scholar
|
|
32
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levine AJ and Oren M: The first 30 years
of p53: Growing ever more complex. Nat Rev Cancer. 9:749–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|