Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer-related mortality worldwide (1,2).
Remedial therapies for HCC, such as surgery, radiofrequency
ablation, liver transplantation and chemotherapy, remain limited
(3). Nevertheless, surgery may
have adverse effects and is not suitable for patients with advanced
disease (4,5). Advanced tumors are not usually
resistant to single-sequence therapy; however, combination
chemotherapy can prolong the 5-year survival rate and reduce
recurrence rates.
Tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL) is a type II transmembrane
protein and a member of the TNF superfamily. TRAIL selectively
initiates the apoptosis of cancer cells without apparent toxic
side-effects on normal cells, thus making it a promising
chemotherapeutic agent for the treatment of various types of cancer
(6-9). However, recent findings have revealed
that some cancer cells, including HCC cells, can develop resistance
to TRAIL-mediated apoptosis (10).
TRAIL crosslinks to its receptors, namely death receptor (DR)4 and
DR5, and triggers their trimerization and intracellular adaptor
death domain clustering to generate a death-inducing signaling
complex (11,12). This complex then recruits the
Fas-associated death domain accessory molecule and subsequently
incites caspase-8. In turn, caspase-8 incites downstream ‘effector’
caspases, such as caspase-3 (13).
Luteolin (chemical name,
3′,4′,5,7-tetrahydroxyflavone) is a widely used flavonoid compound
produced in numerous types of plants, such as vegetables, fruits
and medicinal herbs. Previous studies have revealed that luteolin
exhibits diverse biological properties, such as antioxidant
(14), anti-inflammatory (15) and anti-proliferative effects
(16). The anticancer effects of
luteolin have been demonstrated in numerous cancer cells, such as
prostate, pancreatic, colorectal, breast and ovarian cancer cells
(17-20). It has been demonstrated that
luteolin, as a flavonoid, can cross the blood brain barrier
(21,22). However, the regulatory mechanisms
underlying the effects of luteolin in HCC cells remain unclear
(23).
Autophagy is a catabolic degradation mechanism that
results in the autophagosomal-lysosomal degeneration of cytosolic
proteins and other cellular components (24). It is induced under numerous
cellular stresses, such as starvation, infection, protein
aggregation and organelle damage (25,26).
Autophagy has complex functions in various stages of cancer and
functions as a tumor suppressor that inhibits tumor initiation
(27-29). In addition, autophagy reduces tumor
growth and development by attenuating cellular metabolic stress,
and thus acts as a survival pathway (23). Autophagosome construction is
mediated by the autophagy-related 12 (ATG12)-ATG5-ATG16 system and
LC3-I-phospholipid conjugate LC3-II, which is widely employed as an
autophagy marker (30,31).
Albeit the well-established anticancer effects of
luteolin (17-19), its synergistic effects with TRAIL
and the related molecular pathways are currently unclear.
Therefore, in this study, we aimed to elucidate the molecular
mechanisms underlying the outcome of luteolin and its synergistic
effects when administered in combination with TRAIL in Huh7 liver
cancer cells.
Materials and methods
Cells and cell culture
Human liver cancer cells (Huh7 and Hep3B) were
obtained from the American Type Culture Collection (Global
Bioresource Center, Manassas, VA, USA) and maintained in Dulbecco’s
modified Eagle’s medium (Gibco BRL, Grand Island, NY, USA)
containing 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO,
USA). For experimentation, the medium of the cells was changed to
DMEM containing 1% FBS. Cells were cultured at 37°C with 5%
CO2 in humidified incubator.
Reagents
Luteolin (solvent, DMSO) was acquired from Cayman
Chemical Co. (Ann Arbor, MI 48108, USA), TRAIL [solvent,
phosphate-buffered saline (PBS)] was acquired from AbFrontier Co.,
Ltd. (Seoul, Korea). Chloroquine (CQ) diphosphate salt was
purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and
CQ was dissolved in water to produce a 10 mM stock solution.
SP600125 (10 µM) was acquired from the Beyotime Institute of
Biotechnology (Shanghai, China). SP600125 and CQ added to the cells
1 h prior to treatment luteolin or TRAIL.
Cell viability assay
The liver cancer cells were plated in 12-well plates
and treated with luteolin (0, 5, 10 and 20 µM) for 18 h and
exposed to TRAIL (200 ng/ml) for a further 2 h. Cell morphology was
assessed under a microscope (Nikon, Tokyo, Japan, magnification,
×100), and cell viability was evaluated by the crystal violet
staining solution kit from Sigma-Aldrich (Merck KGaA, Darmstadt,
Germany) according to the manufacturer’s instructions.
Trypan blue exclusion assay
Cell viability was evaluated by trypan blue
exclusion assay (Sigma-Aldrich, St. Louis, MO, USA) using a
hemocytometer (#02-671-10; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Following each treatment, cells were stained with 0.4%
trypan blue for 5 min at room temperature. Unstained cells were
regarded as viable, and stained cells were regarded as dead. The
total cell number and the number of trypan blue-positive cells were
counted using a light microscope in a blinded manner. The
percentage of surviving cells was calculated using the formula:
Number of stained cells/number of total cells ×100. Each
experiments was performed in triplicate.
Transmission electron microscopy (TEM)
analysis
Following fixation of the cells in 2% glutaraldehyde
(Electron Microscopy Sciences, Hatfield, PA, USA) and 2%
paraformaldehyde (Electron Microscopy Sciences) in 0.05 M sodium
cacodylate (pH 7.2; Electron Microscopy Sciences) for 2 h at 4°C
specimens were fixed in 1% osmium tetroxide (Electron Microscopy
Sciences) for 1 h at 4°C, dehydrated with increasing ethanol (25,
50, 70, 90 and 100%) for 5 min each and embedded in epoxy resin
(Embed 812; Electron Microscopy Sciences) for 48 h at 60°C
according to the manufacturers’ instructions. Ultrathin sections
(60 nm) were prepared using an LKB-III ultratome (Leica
Microsystems GmbH, Wetzlar, Germany) and were stained with 0.5%
uranyl acetate (Electron Microscopy Sciences) for 20 min and 0.1%
lead citrate (Electron Microscopy Sciences) for 7 min at room
temperature. Images were recorded on a Hitachi H7650 electron
microscope (Hitachi, Ltd., Tokyo, Japan; magnification, ×10,000)
installed at the Center for University-Wide Research Facilities
(CURF) at Chonbuk National University.
Immunofluorescence staining
The Huh7 cells were cultured on poly-L-lysine-coated
coverslips. Following differentiation and specific treatment, the
cells were adjusted with 4% paraformaldehyde and permeablized with
0.1% Triton X-100. The cells were then incubated in a blocking
solution followed by overnight incubation at 4°C with anti-p62
(cat. no. 5114; 1:1,000) and p-c-Jun N-terminal kinase (JNK, cat.
no. 9255s) antibodies were from Cell Signaling Technology, Inc.
(Danvers, MA, USA). After washing with PBS, the cells were
incubated with secondary antibodies (Alexa Fluor®
488-conjugate; donkey polyclonal anti-rabbit, 1:500; Thermo Fisher
Scientific, Inc.; cat. no. A-21206) and Texas-Red-X-conjugate (goat
poly-clonal anti-mouse, 1:500; Thermo Fisher Scientific, Inc.; cat.
no. T-6390) for 2 h in the dark. In addition,
4′,6′-diamidino-2-phenyl-indole (DAPI; D9564; Sigma-Aldrich, St.
Louis, MO, USA) was used to non-specifically stain the nuclei.
Finally, immunostaining was visualized under a fluorescence
microscope (#451203, Nikon ECLIPSE 80i; Nikon Corporation, Tokyo,
Japan; magnification, ×400).
Western blot analysis
Western blot analysis was carried out as previously
described (32). Briefly, RIPA
lysis buffer was used to extract total proteins. The supernatant
was collected by centrifuging at 13,282 × g at 4°C for 10 min.
Protein concentration was tested using the Pierce BCA Protein Assay
kit (Thermo Fisher Scientific, Inc.). The samples (30 µg)
were separated on SDS-PAGE (10%) and then blotted onto a
polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Bedford,
MA, USA). The membrane was blocked with 5% non-fat dry milk at 25°C
for 1 h, and then incubated with primary antibodies overnight for 1
h at 4°C. The β-actin antibody was from Sigma-Aldrich (cat. no.
A2228; 1:2,000; Merck KGaA, Darmstadt, Germany), The antibodies
against DR4 (cat. no. ab8414; 1:1,000) and DR5 (cat. no. ab181846;
1:1,000) were from Abcam (Cambridge, MA, USA); against LC3A/B (cat.
no. 3868; 1:1,000), cleaved caspase-3 (cat. no. 9661; 1:500), p-JNK
(cat. no. 9255s, 1:1,000), p62 (cat. no. 5114; 1:1,000), ATG5 (cat.
no. 2630; 1:1,000) were from Cell Signaling Technology (Danvers,
MA, USA) and the antibody against cleaved caspase-8 (cat. no.
551242; 1:1,000) was from BD Pharmingen/BD Biosciences (San Jose,
CA, USA). The membrane was incubated with the corresponding
horseradish peroxidase-conjugated secondary antibody (cat. no.
4410; Cell Signaling Technology; 1:2,000) at 25°C for 1 h. The
immunoreactive protein bands were visualized using an enhanced
chemiluminescence detection system (GE Healthcare Life Sciences,
Chalfont, UK).
Small interfering ATG5 RNA
transfection
The Huh7 cells were seeded into 6-well plates at a
density of 1×104 cells/well. Following 48 h of
incubation the cells reached 80% confluence and were transiently
transfected with ATG5 siRNA using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The sequences for
ATG5 siRNA (siRNA; oligo ID HSS114103; Invitrogen/Thermo Fisher
Scientific) were as follows: Sense, 5′-GGCCUUUCAUUCAGAAGCUTT-3′ and
antisense, 5′-AGCUUCUGAAUGAAAGGCCTT-3′; the sequences for negative
control (NC) were sense, 5′-UCUCCGAACGUGUCACGUTT-3′ and anti-sense,
5′-ACGUGACACGUUCGGAGAATT-3′. The oligodeoxynucleotides for the NC
were obtained following scrambling of the siRNA
oligodeoxynucleotide for ATG5, and were determined to not be
associated with any mRNA sequence by BLAST. Cell transfection was
performed, according to the protocol of the transfection kit
manufacturer. Briefly, each sequence of MIF siRNA and 10 µl
Lipofectamine 2000 was diluted in serum-free medium (250 µl)
at room temperature for 5 min, mixed together, and incubated for 30
min at room temperature. The mixture was subsequently administered
to the Tca8113, SCC25 and HN5 cells, and after 5 h of incubation,
the medium was replaced with complete medium.
Statistical analysis
Statistical analyses were carried out using GraphPad
Prism software (version 5.03; GraphPad Software, Inc., La Jolla,
CA, USA). All experiments were performed 3 times, and the data are
expressed as the means ± standard error. Significant differences
between the control and treated samples were analyzed using one-way
factorial analysis of variance (ANOVA), followed by Duncan’s
post-hoc test. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Luteolin sensitizes HCC cells to
TRAIL-induced apoptosis
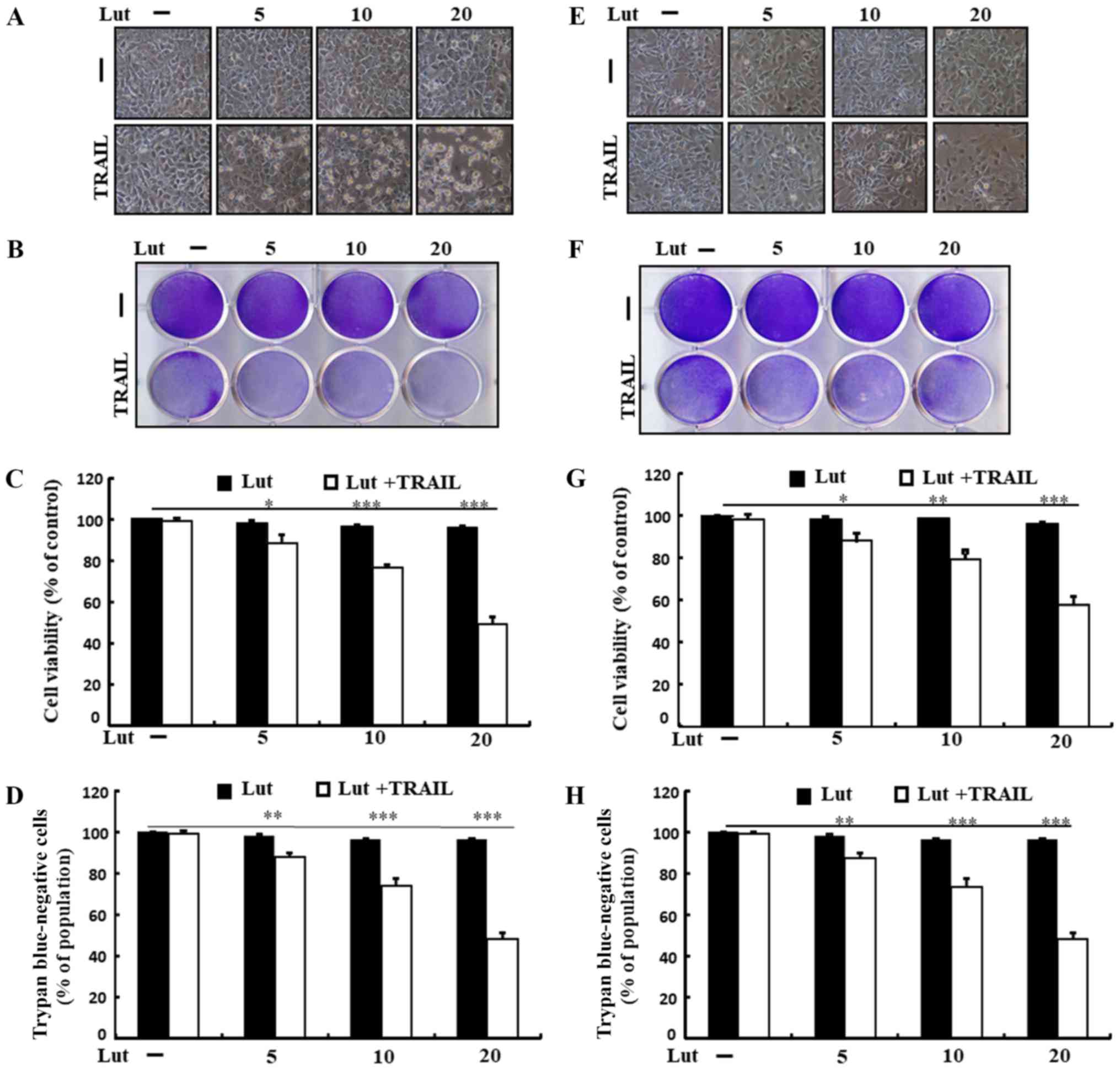
Treatment with TRAIL or luteolin alone induced
minimal or no cell death (Fig. 1).
In addition, the treated cells exhibited no morphological
differences from those of the control cells, revealing that the HCC
cells were immensely resistant to TRAIL-mediated apoptosis.
Co-treatment with TRAIL and increasing concentrations of luteolin
markedly attenuated cell viability, increasing apoptotic bodies and
decreasing attached cells compared to treatment with luteolin or
TRAIL alone (Fig. 1A and E).
Luteolin treatment sensitized both cells to TRAIL, but the
sensitivity was higher in Huh7 cells than in Hep3B cells, and we
focused on the Huh7 cells in this study. These findings suggested
that luteolin effectively enhanced the sensitivity of human HCC
cells to TRAIL-induced apoptosis.
Luteolin induces autophagy and sensitizes
cells to TRAIL-induced apoptosis
As shown in Fig. 2A and
B, luteolin treatment significantly upregulated LC3-II and
downregulated p62 expression, which is consistent with DR5
upregulation. The results of TEM analysis revealed that numerous
autophagic and vacant vacuoles were secreted in the
luteolin-treated cells (Fig. 2C).
The cells co-treated with luteolin and TRAIL also exhibited a
higher expression of cleaved caspase-3 and cleaved caspase-8
(Fig. 2D). These outcomes
suggested that luteolin can initiate autophagy in Huh7 cells.
Luteolin-mediates the augmentation of
TRAIL-initiated apoptosis which is suppressed by the attenuation of
autophagy
Co-treatment with luteolin, chloroquine and TRAIL
suppressed cell death. Cell morphological analysis revealed that
the apoptotic bodies were markedly reduced and the attaching cells
were increased in the luteolin-, chloroquine- and TRAIL-treated
cells. These results also confirmed that treatment with chloroquine
attenuated the cell death induced by combined treatment with
luteolin and TRAIL (Fig. 3A).
Co-treatment with luteolin, TRAIL and chloroquine markedly
increased the viability of the human HCC Huh7 cells (Fig. 3B-D). These outcomes suggested that
chloroquine blocked TRAIL-induced and luteolin-mediated liver
cancer cell death.
The inhibition of autophagy attenuates
the enhancing effects of luteolin on the sensitivity of liver
cancer cells to TRAIL-induced apoptosis mediated by the promotion
of the autophagic flux
The origination of autophagy was further confirmed
based on the suppression of the autophagic flux following
chloroquine treatment, which resulted in an increase in the
accumulation of membrane-bound LC3-II, an increase in p62 levels,
and the partial suppression of DR5 upregulation (Fig. 4A). The results of
immunofluorescence staining also confirmed the increased p62 levels
following treatment with chloroquine (Fig. 4B). Chloroquine also partially
inhibited the upregulation DR5 which had been induced by luteolin
and TRAIL (Fig. 4C). In addition,
co-treatment with luteolin, TRAIL and chloroquine inhibited the
increase in cleaved caspase-8 and cleaved caspase-3 induced by
luteolin and TRAIL (Fig. 4D).
These outcomes suggested that the luteolin-induced augmentation of
TRAIL-initiated apoptosis and the promotion of the autophagic flux
were suppressed by chloroquine.
The enhancing effects of luteolin on the
sensitivity of the cells to TRAIL-initiated apoptosis are
suppressed by the genetic attenuation of autophagy
Co-treatment with luteolin, ATG5 siRNA and TRAIL
prevented cell death. Cell morphological analysis revealed that the
apoptotic bodies were markedly reduced and the attaching cells were
increased in the luteolin-, ATG5 siRNA- and TRAIL-treated cells.
The results confirmed that autophagy inhibition suppressed the cell
death induced by treatment with luteolin and TRAIL, and also
compared to transfection with the negative control (NC) (Fig. 5A). Co-treatment with luteolin and
TRAIL, and transfection with ATG5 siRNA markedly increased the
viability of the Huh7 HCC cells and significantly inhibited cell
death (Fig. 5B-D). These outcomes
suggested that autophagy inhibition blocked TRAIL- and
luteolin-induced liver cancer cell death.
The genetic attenuation of autophagy
suppresses luteolin-induced TRAIL-initiated apoptosis mediated by
the activation of the autophagic flux
ATG5 knockdown decreased the luteolin-induced
expression of LC3-II and DR5 and markedly increased the p62 protein
levels (Fig. 6A). The p62 protein
levels determined based on immunofluorescence staining were
consistent with the results of western blot analysis (Fig. 6B). However, co-treatment with
luteolin, ATG5 siRNA and TRAIL attenuated the increase in Ac-cas3
and Ac-cas8 expression (Fig. 6C).
These findings suggested that the luteolin-induced augmentation of
TRAIL-initiated apoptosis mediated by the activation of the
autophagic flux was suppressed by the genetic attenuation of
autophagy.
Luteolin induces DR expression via JNK
activation
Cell morphological analysis revealed that the
apoptotic bodies were markedly reduced and the attaching cells were
increased in JNK inhibitor (SP600125)-, luteolin- and TRAIL-treated
cells (Fig. 7A). Co-treatment with
luteolin, SP600125 and TRAIL reduced the cell apoptosis induced by
luteolin and TRAIL (Fig. 7A and
B). The results revelaed that luteolin increased JNK and
decreased Akt phosphorylation (Fig.
7C). Pre-treatment with the JNK inhibitor (SP600125)
significantly attenuated JNK phosphorylation and inhibited DR5
upregulation (Fig. 7D). The
results of immunofluorescence staining also confirmed that SP600125
treatment significantly reduced JNK phosphorylation (Fig. 7E).
Discussion
TRAIL has been established as a significant
potential cancer treatment due to its tumor specificity and safety
(33). Untagged recombinant human
TRAIL has been demonstrated to not induce any toxicity to normal
human hepatocytes and is currently used as a clinical drug in the
treatment of liver diseases (34).
DRs, DR5 and DR4, are pro-apoptotic receptors of TRAIL and can
interact with TRAIL to activate the extrinsic apoptotic signaling
pathway (11). The antitumor
effects of luteolin have been demonstrated in various tumor cells
(18,35-37).
Luteolin can initiate cell cycle arrest and apoptosis, attenuate
cell proliferation and metastatic advancement, and can inhibit
doxorubicin-initiated cyto-toxicity (19,38,39).
Autophagy is a lysosomal degradation system in which damaged
proteins sustain cellular homeostasis (25,40,41).
Autophagy not only functions as a cell survival pathway, but also
causes autophagic cell death when cells are subjected to stress
(42-45).
Recent studies have demonstrated that HCC cells can
develop resistance to TRIAL-initiated apoptosis (46,47).
In the current study, we demonstrated that treatment with TRAIL or
luteolin alone initiated minimal or no apoptosis (Fig. 1). However, co-treatment with
luteolin and TRAIL markedly initiates the death of Huh7 HCC cells,
which are highly resistant to treatment with luteolin or TRAIL
alone. This indication may lead to the development of a more
effective and established mouse xenograft model and follow-up
assessments using luteolin and perhaps, this may also treatment
strategy may also be used in the future in the treatment of
patients.
A recent study revealed that luteolin attenuated
cancer cell proliferation and autophagy initiation (23). Furthermore, TRAIL-resistant tumor
cells can be sensitized by chemotherapeutic agents that induce DR5
upregulation. Therefore, sensitization to TRAIL-induced apoptosis
via DR5 upregulation is a promising approach for the treatment of
TRAIL-resistant hepatoma cells (48-51).
In the present study, the results from western blot analysis and
immunocytochemistry indicated that luteolin treatment significantly
increased the LC3-II levels and decreased the p62 levels in
parallel with DR5 upregulation (Fig.
2). It has recently been demonstrated that autophagy is
involved in drug-induced DR upregulation (52). The results of this study also
suggested that chloroquine promoted cell survival and partially
inhibited DR5 upregulation in Huh7 liver cancer cells (Figs. 3 and 4). However, for a reliable conclusion
regarding DR5, additional studies are warranted with DR5
modulation, including DR5 knockout or knockdown experiments. In
addition, the genetic attenuation of autophagy blocked the
TRAIL-initiated and luteolin-mediated apoptosis of Huh7 cells
(Figs. 5 and 6). Recent reports have also revealed that
Akt, ERK and JNK can mediate DR upregulation (53-55).
The findings of this study suggested that luteolin induced JNK
activation via DR upregulation (Fig.
7).
In conclusion, the findings of this study
demonstrate that JNK-mediated DR5 upregulation by luteolin
sensitizes Huh7 liver cancer cells to TRAIL-induced cell death by
promoting an autophagic flux. Combined treatment with luteolin and
TRAIL may thus prove to be a useful and effective therapeutic
regimen for some TRAIL-resistant cancers, including HCC.
Funding
This study was supported by a grant from the
National Research Foundation of Korea (NRF), funded by the Korean
Government (2016R1A2B2009293).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
UMN and SYP designed the study. UMN performed the
experiments. UMN and SYP analyzed the data and wrote the
manuscript. Both authors reviewed the results and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sherman M: Epidemiology of hepatocellular
carcinoma. Oncology. 78(Suppl 1): 7–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balogh J, Victor D III, Asham EH,
Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour
HP Jr: Hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
3:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arii S: Molecularly targeted therapy for
hepatocellular carcinoma from the basic and clinical aspects. Int J
Clin Oncol. 15:2342010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Di Bisceglie AM, Bruix J,
Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M,
Talwalkar J, et al Panel of Experts in HCC-Design Clinical Trials:
Design and endpoints of clinical trials in hepatocellular
carcinoma. J Natl Cancer Inst. 100:698–711. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S: The promise of cancer therapeutics
targeting the TNF-related apoptosis-inducing ligand and TRAIL
receptor pathway. Oncogene. 27:6207–6215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahmood Z and Shukla Y: Death receptors:
Targets for cancer therapy. Exp Cell Res. 316:887–899. 2010.
View Article : Google Scholar
|
|
8
|
Allen JE and El-Deiry WS: Regulation of
the human TRAIL gene. Cancer Biol Ther. 13:1143–1151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Micheau O, Shirley S and Dufour F: Death
receptors as targets in cancer. Br J Pharmacol. 169:1723–1744.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen CY, Yiin SJ, Hsu JL, Wang WC, Lin SC
and Chern CL: Isoobtusilactone A sensitizes human hepatoma Hep G2
cells to TRAIL-induced apoptosis via ROS and CHOP-mediated
up-regulation of DR5. J Agric Food Chem. 60:3533–3539. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R,
Ebner R, Ni J and Dixit VM: The receptor for the cytotoxic ligand
TRAIL. Science. 276:111–113. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walczak H, Degli-Esposti MA, Johnson RS,
Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA,
Smith CA, et al: TRAIL-R2: A novel apoptosis-mediating receptor for
TRAIL. EMBO J. 16:5386–5397. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashokkumar P and Sudhandiran G: Protective
role of luteolin on the status of lipid peroxidation and
antioxidant defense against azoxymethane-induced experimental colon
carcinogenesis. Biomed Pharmacother. 62:590–597. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishitani Y, Yamamoto K, Yoshida M, Azuma
T, Kanazawa K, Hashimoto T and Mizuno M: Intestinal
anti-inflammatory activity of luteolin: Role of the aglycone in
NF-κB inactivation in macrophages co-cultured with intestinal
epithelial cells. Biofactors. 39:522–533. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashokkumar P and Sudhandiran G: Luteolin
inhibits cell proliferation during Azoxymethane-induced
experimental colon carcinogenesis via Wnt/β-catenin pathway. Invest
New Drugs. 29:273–284. 2011. View Article : Google Scholar
|
|
17
|
Huang X, Dai S, Dai J, Xiao Y, Bai Y, Chen
B and Zhou M: Luteolin decreases invasiveness, deactivates STAT3
signaling, and reverses interleukin-6 induced
epithelial-mesenchymal transition and matrix metalloproteinase
secretion of pancreatic cancer cells. OncoTargets Ther.
8:2989–3001. 2015. View Article : Google Scholar
|
|
18
|
Han K, Meng W, Zhang JJ, Zhou Y, Wang YL,
Su Y, Lin SC, Gan ZH, Sun YN and Min DL: Luteolin inhibited
proliferation and induced apoptosis of prostate cancer cells
through miR-301. OncoTargets Ther. 9:3085–3094. 2016. View Article : Google Scholar
|
|
19
|
Naso LG, Badiola I, Marquez Clavijo J,
Valcarcel M, Salado C, Ferrer EG and Williams PAM: Inhibition of
the metastatic progression of breast and colorectal cancer in vitro
and in vivo in murine model by the oxidovanadium(IV) complex with
luteolin. Bioorg Med Chem. 24:6004–6011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dia VP and Pangloli P:
Epithelial-to-Mesenchymal Transition in Paclitaxel-Resistant
Ovarian Cancer Cells Is Downregulated by Luteolin. J Cell Physiol.
232:391–401. 2017. View Article : Google Scholar
|
|
21
|
Youdim KA, Qaiser MZ, Begley DJ,
Rice-Evans CA and Abbott NJ: Flavonoid permeability across an in
situ model of the blood-brain barrier. Free Radic Biol Med.
36:592–604. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jang S, Kelley KW and Johnson RW: Luteolin
reduces IL-6 production in microglia by inhibiting JNK
phosphorylation and activation of AP-1. Proc Natl Acad Sci USA.
105:7534–7539. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen
Y, Chen H, Chu F and Zhang Y: Luteolin promotes cell apoptosis by
inducing autophagy in hepatocellular carcinoma. Cell Physiol
Biochem. 43:1803–1812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
El-Khattouti A, Selimovic D, Haikel Y and
Hassan M: Crosstalk between apoptosis and autophagy: Molecular
mechanisms and therapeutic strategies in cancer. J Cell Death.
6:37–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang C: Negative regulation of autophagy.
Cell Death Differ. 17:1807–1815. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giampietri C, Petrungaro S, Padula F,
D’Alessio A, Marini ES, Facchiano A, Filippini A and Ziparo E:
Autophagy modulators sensitize prostate epithelial cancer cell
lines to TNF-alpha-dependent apoptosis. Apoptosis. 17:1210–1222.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lorenzi PL, Claerhout S, Mills GB and
Weinstein JN: A curated census of autophagy-modulating proteins and
small molecules: Candidate targets for cancer therapy. Autophagy.
10:1316–1326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yao D, Wang P, Zhang J, Fu L, Ouyang L and
Wang J: Deconvoluting the relationships between autophagy and
metastasis for potential cancer therapy. Apoptosis. 21:683–698.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autopha-gosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tanida I, Minematsu-Ikeguchi N, Ueno T and
Kominami E: Lysosomal turnover, but not a cellular level, of
endogenous LC3 is a marker for autophagy. Autophagy. 1:84–91. 2005.
View Article : Google Scholar
|
|
32
|
Nazim UM, Jeong JK and Park SY:
Ophiopogonin B sensitizes TRAIL-induced apoptosis through
activation of autophagy flux and downregulates cellular FLICE-like
inhibitory protein. Oncotarget. 9:4161–4172. 2017.
|
|
33
|
Kelley SK and Ashkenazi A: Targeting death
receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol.
4:333–339. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Volkmann X, Fischer U, Bahr MJ, Ott M,
Lehner F, Macfarlane M, Cohen GM, Manns MP, Schulze-Osthoff K and
Bantel H: Increased hepatotoxicity of tumor necrosis factor-related
apoptosis-inducing ligand in diseased human liver. Hepatology.
46:1498–1508. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yee SB, Choi HJ, Chung SW, Park DH, Sung
B, Chung HY and Kim ND: Growth inhibition of luteolin on HepG2
cells is induced via p53 and Fas/Fas-ligand besides the TGF-β
pathway. Int J Oncol. 47:747–754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park SH, Ham S, Kwon TH, Kim MS, Lee DH,
Kang JW, Oh SR and Yoon DY: Luteolin induces cell cycle arrest and
apoptosis through extrinsic and intrinsic signaling pathways in
MCF-7 breast cancer cells. J Environ Pathol Toxicol Oncol.
33:219–231. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun DW, Zhang HD, Mao L, Mao CF, Chen W,
Cui M, Ma R, Cao HX, Jing CW and Wang Z: Luteolin inhibits breast
cancer development and progression in vitro and in vivo by
suppressing notch signaling and regulating miRNAs. Cell Physiol
Biochem. 37:1693–1711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sato Y, Sasaki N, Saito M, Endo N, Kugawa
F and Ueno A: Luteolin attenuates doxorubicin-induced cytotoxicity
to MCF-7 human breast cancer cells. Biol Pharm Bull. 38:703–709.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sui JQ, Xie KP and Xie MJ: Inhibitory
effect of luteolin on the proliferation of human breast cancer cell
lines induced by epidermal growth factor. Sheng Li Xue Bao.
68:27–34. 2016.PubMed/NCBI
|
|
40
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Quan W, Jung HS and Lee MS: Role of
autophagy in the progression from obesity to diabetes and in the
control of energy balance. Arch Pharm Res. 36:223–229. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Szczesny B, Brunyánszki A, Ahmad A, Oláh
G, Porter C, Toliver-Kinsky T, Sidossis L, Herndon DN and Szabo C:
Time-dependent and organ-specific changes in mitochondrial
function, mitochondrial DNA integrity, oxidative stress and
mononuclear cell infiltration in a mouse model of burn injury. PLoS
One. 10:e01437302015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Xu HL, Liu YX, An N, Zhao S and Bao
JK: Autophagy modulation as a target for anticancer drug discovery.
Acta Pharmacol Sin. 34:612–624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lamy L, Ngo VN, Emre NC, Shaffer AL III,
Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landgren O, et al:
Control of autophagic cell death by caspase-10 in multiple myeloma.
Cancer Cell. 23:435–449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo YH, Wu SB, Wei YH, Chen YC, Tsai MH,
Ho CC, Lin SY, Yang CS and Lin P: Cadmium-based quantum dot induced
autophagy formation for cell survival via oxidative stress. Chem
Res Toxicol. 26:662–673. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamanaka T, Shiraki K, Sugimoto K, Ito T,
Fujikawa K, Ito M, Takase K, Moriyama M, Nakano T and Suzuki A:
Chemotherapeutic agents augment TRAIL-induced apoptosis in human
hepatocellular carcinoma cell lines. Hepatology. 32:482–490. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shankar S and Srivastava RK: Enhancement
of therapeutic potential of TRAIL by cancer chemotherapy and
irradiation: mechanisms and clinical implications. Drug Resist
Updat. 7:139–156. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen JJ, Chou CW, Chang YF and Chen CC:
Proteasome inhibitors enhance TRAIL-induced apoptosis through the
intronic regulation of DR5: involvement of NF-kappa B and reactive
oxygen species-mediated p53 activation. J Immunol. 180:8030–8039.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim H, Kim EH, Eom YW, Kim WH, Kwon TK,
Lee SJ and Choi KS: Sulforaphane sensitizes tumor necrosis
factor-related apoptosis-inducing ligand (TRAIL)-resistant hepatoma
cells to TRAIL-induced apoptosis through reactive oxygen
species-mediated up-regulation of DR5. Cancer Res. 66:1740–1750.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jung EM, Lim JH, Lee TJ, Park JW, Choi KS
and Kwon TK: Curcumin sensitizes tumor necrosis factor-related
apop-tosis-inducing ligand (TRAIL)-induced apoptosis through
reactive oxygen species-mediated upregulation of death receptor 5
(DR5). Carcinogenesis. 26:1905–1913. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Horinaka M, Yoshida T, Shiraishi T, Nakata
S, Wakada M and Sakai T: The dietary flavonoid apigenin sensitizes
malignant tumor cells to tumor necrosis factor-related
apoptosis-inducing ligand. Mol Cancer Ther. 5:945–951. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen L, Meng Y, Guo X, Sheng X, Tai G,
Zhang F, Cheng H and Zhou Y: Gefitinib enhances human colon cancer
cells to TRAIL-induced apoptosis of via autophagy- and JNK-mediated
death receptors upregulation. Apoptosis. 21:1291–1301. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cheng H, Hong B, Zhou L, Allen JE, Tai G,
Humphreys R, Dicker DT, Liu YY and El-Deiry WS: Mitomycin C
poten-tiates TRAIL-induced apoptosis through p53-independent
upregulation of death receptors: Evidence for the role of c-Jun
N-terminal kinase activation. Cell Cycle. 11:3312–3323. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Thamkachy R, Kumar R, Rajasekharan KN and
Sengupta S: ERK mediated upregulation of death receptor 5 overcomes
the lack of p53 functionality in the diaminothiazole DAT1 induced
apoptosis in colon cancer models: Efficiency of DAT1 in Ras-Raf
mutated cells. Mol Cancer. 15:222016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shoeb M, Ramana KV and Srivastava SK:
Aldose reductase inhibition enhances TRAIL-induced human colon
cancer cell apoptosis through AKT/FOXO3a-dependent upregulation of
death receptors. Free Radic Biol Med. 63:280–290. 2013. View Article : Google Scholar : PubMed/NCBI
|