Introduction
Zinc finger proteins (ZNFs) regulate transcription
(1), replication (2), and other biologically significant
events, including cellular differentiation (3), and some have been suggested to be
responsible for human diseases (4,5).
However, the molecular mechanisms that regulate their gene
expression remain unclear. Surveillance of the human genomic DNA
database revealed that duplicated GGAA (TTCC) motifs, which are
frequently contained in the 5′ flanking regions of the DNA repair,
apoptosis, anti-viral response and mitochondrial function
associated factor encoding genes (6,7), are
present in the 5′ flanking regions of several ZNF family
genes (8). Interestingly, the
duplicated GGAA motif responds to interferon (IFN) β (9) and trans resveratrol (10, 12). In the ataxia telangiectasia and
Rad3 related, xeroderma pigmentosum type B, and retinoblastoma
(RB1) promoters, the motif notably responds to
12-O-tetra decanoylphorbol 13 acetate (TPA) in HL 60 cells
(13). These observations suggest
that ZNF family genes could be induced by the
cis-function of the GGAA motifs during the macrophage
like-differentiation of HL-60 cells.
The GGAA is a core sequence motif that E26
transformation-specific (ETS) family proteins recognize (14). Numerous transcription factors
(TFs), including GA-binding protein TF (GABP), nuclear factor
erythroid 2-related factor 2, nuclear factor κB/c Rel, STATs and
interferon regulatory factor proteins bind to the GGAA
motif-containing sequences and regulate gene transcription
(7). Notably, ETS family proteins
play essential roles in the regulation of cellular differentiation
(15,16). Duplicated GGAA motifs are contained
in the promoter region of the human CD41 (ITGA2B)
(17) and programmed death-1 genes
(18), suggesting that GGAA motif
binding proteins regulate megacaryocytic cell differentiation. ETS
family TF spleen focus forming virus proviral integration oncogene
(SPI1; PU.1) regulates the timing of the activation of the T
lineage developmental program (19). Other ETS family proteins, ETS
variant (ETV)4 and ETV5, regulate the differentiation of embryonic
stem cells (20). ETS1 regulates
neural crest development via the epigenetic control of bone
morphogenetic protein signaling (21). Moreover, ETS family proteins have
been suggested to affect tumorigenesis (22). The aberrant fusion protein EWS
friend leukemia integration 1 TF is known to induce osteosarcoma
development (23). In addition,
amplification of ETS1 is suggested to cause tumor invasion
(24). These lines of evidence
suggest that GGAA binding proteins are involved in the generation
and development of cancer.
In the present study, we isolated 5′upstream regions
of six human ZNF-encoding genes and found that the zinc finger
nfx-1-type containing 1 (ZNFX1) promoter is the most
prominently activated in response to TPA. Reverse transcription
quantitative polymerase chain reaction (RT-qPCR) and western
blotting revealed that amount of ZNFX1 transcripts and
degree of translation increase during the TPA-induced
differentiation of HL-60 cells. Deletion and point
mutation-introducing experiments demonstrated that the duplicated
GGAA motif, which is present 100-bp downstream of the transcription
start site (TSS), serves an essential role in the TPA response.
JASPAR analysis of the TPA responding duplicated GGAA motif
suggested that SPI1 (PU.1), ETS1, myeloid zinc finger 1 (MZF1) and
STAT1/2 are involved in the regulation of ZNFX1 gene
expression during the differentiation of HL-60 into macrophage like
cells. Taken together, it was suggested that investigations in the
transcription mechanism of the ZNFX1 gene may contribute to
the development of novel anti-cancer therapies.
Materials and methods
Materials
TPA was purchased from Sigma-Aldrich (Merck KGaA)
(13).
Cells and cell culture
The human promyelotic leukemia cell line HL-60
(Institute of Medical Science, Tokyo University) (25) was cultured in RPMI 1640 medium
(WAKO Pure Chemical), supplemented with 10% fetal calf serum (FCS)
(Biosera), containing 120 IU/ml penicillin G (Meiji Seika Pharma)
and 120 µg/ml streptomycin (Meiji Seika Pharma), at 37°C in
a humidified atmosphere with 5% CO2.
Construction of luciferase (Luc) reporter
plasmids
A 543-bp region of the human ZNFX1 gene and
other promoter regions of ZNF252, ZNF343,
ZNF555, ZNF782, zinc finger CCCH-type containing,
antiviral 1 (ZC3HAV1) genes were obtained by
PCR-amplification, which was carried out with Prime STAR (Takar
Bio, Inc.), appropriate primer pairs (Table I) and HeLa S3 genomic DNA as a
template. The reaction was as follows: 98°C for 15 sec, 55°C for 5
sec, and 72°C for 30 sec for 30 cycles. The PCR-amplified DNA
fragments were digested with restriction enzymes, then they were
ligated into the KpnI/HindIII or the
KpnI/XhoI site of pGL4.10[luc2] (Promega
Corporation), to make pGL4-ZNFX1, -ZNF252, -ZNF343, -ZNF555,
-ZNF782, -zinc finger CCCH-type containing, anti viral 1 (ZC3HAV1).
Similarly, the deletion of the 543-bp was carried out by PCR with
appropriate primer sets (Table I),
and pGL4-ZNFX1 as a template. The DNA fragments were ligated into
the MCS of pGL4.10[luc2] to make pGL4-X1-Δ1 to Δ7 plasmids.
Other plasmids,-Δ6mt1,-Δ6mt2,-Δ6mt12,-Δ7mt1,-Δ7mt2, and-Δ7mt12,
were constructed by a similar proce dure (13) with appropriate primer pairs
(Table I) and template plasmids,
pGL4-X1-Δ6 or pGL4-X1-Δ7, for the amplification of the DNA
fragments (Table I). Nucleotide
sequences were confirmed by a DNA sequencing service
(FASMAC-Greiner Japan, Inc.) with primers Rv
(5′-TAGCAAAATAGGCTGTCCCC-3′) and GL
(5′-CTTTATGTTTTTGGCGTCTTCC-3′).
 | Table IPrimer pairs used to amplify 5′
upstream regions of the human zinc finger motif containing
protein-encoding genes. |
Table I
Primer pairs used to amplify 5′
upstream regions of the human zinc finger motif containing
protein-encoding genes.
| Luciferase
plasmid | Primer | Sequence (5′ to
3′) |
|---|
| pGL4-ZNF252 | hZNF252-6191 |
TCGGTACCGCAATAGGTCTGAAACCTCTC |
| AhZNF252-5639 |
ATCTCGAGCGCGAACGCTAAATCCCGTGCC |
| pGL4-ZNF343 | hZNF343-0212 |
TCGGTACCGGGATCTTAGATAAGAGGCCAG |
| AhZNF343-9731 |
ATCTCGAGTGAAGTCTCTGTCCCCTTGGCC |
| pGL4-ZNF555 | hZNF555-1040 |
TCGGTACCTCCTGTCCGGACAAGGGGTCGC |
| AhZNF555-1536 |
ATCTCGAGCGCGACAGGAACCGGGACGCC |
| pGL4-ZNF782 | hZNF782-1294 |
TCGGTACCCGGGAAGCGGTTTGGGAAGCTC |
| AhZNF782-0834 |
ATCTCGAGAAACCTGACTCTCATCCACGTC |
| pGL4-ZC3HAV1 | hZC3-7621 |
TCGGTACCACGATCTGGGGCCTGGGGACGC |
| AhZC3-6933 |
ATCTCGAGTGCTGGCTCTGCCGCGGCGC |
| pGL4-ZNFX1 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AhZFX1-0666 |
CGATAAGCTTTCACTGCCGCCGGCGAGTGC |
| pGL4-X1-Δ1 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AhX1d4 |
GATAAGCTTGGGCGGAGCCGGGCGGGCGGC |
| pGL4-X1-Δ2 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AhX1d3 |
GATAAGCTTGGCCCAGGCACGGCCGGCGCC |
| pGL4-X1-Δ3 | hX1d1 |
TCGGTACCGGGCTTGTCCGCTTCCTCGCC |
| AhZFX1-0666 |
CGATAAGCTTTCACTGCCGCCGGCGAGTGC |
| pGL4-X1-Δ4 | hZNFX1d3 |
CTAGGTACCAGGCCCTCGTGCTCTCCACCC |
| AhZFX1-0666 |
CGATAAGCTTTCACTGCCGCCGGCGAGTGC |
| pGL4-X1-Δ5 | hZNFX1d4 |
ATTGGTACCAGGGTCTGCGGGGAACGGAAA |
| AhZFX1-0666 |
CGATAAGCTTTCACTGCCGCCGGCGAGTGC |
| pGL4-X1-Δ6 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AX1_WT |
CGATAAGCTTCACTTTCGGTTTCCGTTCCCCGCAG |
| pGL4-X1-Δ6mt1 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AX1_mt1 |
CGATAAGCTTCACTTTCGGTTTAAGTTCCCCGCAG |
| pGL4-X1-Δ6mt2 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AX1_mt2 |
CGATAAGCTTCACTTTCGGTTTCCGTTAACCGCAG |
| pGL4-X1-Δ6mt12 | hZX1-1208 |
TCGGTACCCGAAACGCTCTCTTTCCCGCCC |
| AX1_mt12 |
CGATAAGCTTCACTTTCGGTTTAAGTTAACCGCAG |
| pGL4-X1-Δ7 | hZNFX1d3 |
CTAGGTACCAGGCCCTCGTGCTCTCCACCC |
| AX1_WT |
CGATAAGCTTCACTTTCGGTTTCCGTTCCCCGCAG |
| pGL4-X1-Δ7mt1 | hZNFX1d3 |
CTAGGTACCAGGCCCTCGTGCTCTCCACCC |
| AX1_mt1 |
CGATAAGCTTCACTTTCGGTTTAAGTTCCCCGCAG |
| pGL4-X1-Δ7mt2 | hZNFX1d3 |
CTAGGTACCAGGCCCTCGTGCTCTCCACCC |
| AX1_mt2 |
CGATAAGCTTCACTTTCGGTTTCCGTTAACCGCAG |
| pGL4-X1-Δ7mt12 | hZNFX1d3 |
CTAGGTACCAGGCCCTCGTGCTCTCCACCC |
| AX1_mt12 |
CGATAAGCTTCACTTTCGGTTTAAGTTAACCGCAG |
Transient transfection and luc assay
Plasmids (100 ng/1-2×105 cells) were
transfected into HL-60 cells via the DEAE dextran method (26) in 96 well plates. After 24 h of
transfection, the culture medium was changed to RPMI/1640
containing 10% FCS with or without TPA (8.1 nM). Control cells were
treated without TPA and cultured in the medium containing 0.0005%
DMSO. After a further 24 h of incubation at 37°C, cells were
collected and lysed with 100 µl of 1X cell culture lysis
reagent containing 25 mM Tris phosphate (pH 7.8), 2 mM
dithiothreitol, 2 mM
1,2-diaminocyclohexane-N,N,N′,N′,-tetraacetic acid,
10% glycerol and 1% Triton X-100, and then were mixed and
centrifuged at 12,000 x g for 5 sec at 4°C. The supernatant was
stored at -80°C. The Luc assay was performed with a Luciferase
Assay system (Promega Corporation), and relative Luc activities
were normalized by the protein concentration and calculated as
described previously (10,11).
Western blot analysis
Whole proteins were extracted by RIPA buffer (20 mM
Tris-HCl (pH 7.4), containing 0.1% SDS, 1% Triton X100, and 1%
sodium deoxycholate). Proteins were separated via 8/15% SDS-PAGE.
Western blot analysis was carried out as previously described
(10,11) with antibodies against ZNFX1 (Abcam,
cat no. ab179452) and β actin (Sigma Aldrich; Merck KGaA, cat no.
A5441), diluted 1:2,600 and 1:10,000, respectively, followed by the
addition of horse radish peroxidase-conjugated secondary antibody
for ZNFX1 (Sigma Aldrich, cat no. A0545) and β-actin
(Sigma-Aldrich, cat no. A9917) diluted 1:20,000. Signal intensities
of samples at 0, 1, 2, 4, 8, 16, 24, 40 and 48 h following
treatment were quantified with a ChemiDoc and ImageLab System
(BioRad Laboratories, Inc.).
RT-qPCR
RT-qPCR was carried out as described previously
(9-11). Total RNAs were extracted by
GeneElute Mammalian Total RNA Miniprep Kit (Sigma Aldrich),
according to the manufacturer's protocols. First strand cDNAs were
synthesized with ReverTra Ace (Toyobo Life Science), random primers
(Takara Bio, Inc.), and total RNAs extracted from HL-60 cells,
which were treated with TPA (8.1 nM) for 0, 8, 24, 48 and 72 h. The
reaction was carried out by incu bating at 30°C for 10 min, 42°C
for 40 min, and then heating at 94°C for 5 min. The sequences (from
5′-3′) of the primer pairs used to amplify human ZNFX1,
integrin subunit α M (ITGAM; CD11b), and GAPDH
cDNAs were hZX1RT: GGCCCTCAAAAGAAGCCCTG and AhZX1RT:
GATGGGGAACTTCTGGAGGC; hCD11b 459: GGGCTCTGCTTCCTGTTT G and AhCD11b
758: CTGCGTTATTGGCTTCACC; and hGAPDH556: TGC ACC ACC AAC TGC TTA GC
and hGAPDH642: GGCATG GACTGTGGTCATGAG, respectively.
Conditions for PCR, which was performed with BIOTAQ
DNA polymerase (Bioline), were as follows: 96°C for 10 sec, 55°C
for 5 sec, and 72°C for 10 sec, for 30 (ZNFX1), 32
(ITGAM), and 22 (GAPDH) cycles. The PCR products were
electrophoresed on 5% acrylamide gels and stained with ethidium
bromide. The signal intensities were quantified using a ChemiDoc
and ImageLab System.
qPCR analysis of transcripts
cDNAs were amplified using Thunderbird Real time PCR
Master Mix (Toyobo Life Science) and 0.3 µM of each primer
pair. The primer pairs for amplifying human ZNFX1 and
GAPDH transcripts were the same as described above. qPCR was
carried out using an Applied Biosystems 7300 Real Time PCR System
(Thermo Fisher Scientific, Inc.) (9-11).
Amplification was performed initially for 1 min at 95°C, followed
by 40 cycles (95°C for 15 sec and 60°C for 30 sec). Relative gene
expression values were obtained by normalizing CT
(threshold cycle) values of target genes in comparison with
CT values of the GAPDH gene using the
ΔΔCq method (27).
Surveillance of putative TSS and
TF-binding sequences
The putative TSS was identified in 543-bp
ZNFX1 promoter region from the NCBI Gene database
(https://www.ncbi.nlm.nih.gov/gene/57169). The 543 bp
and the 21 bp TPA responsive element were analyzed by JASPAR
database (http://jaspar.genereg.net/) with
threshold 97.5 and 92%, respectively.
Statistical analysis
All statistical analyses were carried out with Excel
software (Microsoft Excel; version 2013; Microsoft Corporation).
The experiments were repeated at least three times. All data are
presented as the mean ± standard deviation. A Student's t-test was
performed to evaluate the significant differences between TPA
treated and non-treated experiments. P<0.05 was considered to
indicate a statistically significant difference. To compare
multiple data sets, one way ANOVA was performed followed by a
Tukey's post hoc test (SPSS; version 19; IBM Corp.).
Results
Isolation and characterization of the 5'
flanking region of the human ZNFX1 gene
The duplicated GGAA motif containing 14 nucleotide
consensus sequences, which are present in the upstream region of
several TPA responding genes (13), are also contained within 500 bp
from the putative TSSs of several ZNF protein encoding genes
(7,8). To examine the response to TPA, we
isolated ~500-bp of the 5′upstream regions of the ZC3HAV1,
ZNF252, ZNF343, ZNF555, ZNF782 and
ZNFX1 genes to construct Luc reporter plasmids. They were
transfected into HL-60 cells (Fig.
1A). As the Luc activity from the control vector transfected
cells was very low, we used the value just for a subtraction.
Notable Luc activity of the pGL4-ZNF343-transfected cells was
detected; however, it was not affected by TPA treatment. Therefore,
we used it for the reference for the easy, reproducible,
cost-effective DEAE dex based multiple transfection assay (28). As the ZNFX1 promoter was
significantly activated by TPA treatment compared without
treatment, we focused on the regulation of the ZNFX1 gene
expression mechanism during the differentiation of the HL 60 cells.
Sequence analysis revealed that the pGL4-ZNFX1 contains a
nucleotide identical to NCBI Sequence ID, NC_018931.2 (nucleotide
from 47799505 47798963) and that it covers the most upstream 5′ end
of the cDNA (Sequence ID: NM_021035.2 of the ZNFX1 mRNA;
GENE ID: 57169 ZNFX1). Interestingly, this 543-bp region also
contains a 5′ upstream end of the ZFAS1 mRNA (NM_021035.2;
GENE ID: 441951ZFAS1) in a reverse orientation to that of the
ZNFX1 gene (Fig. 1B). The
TSS was tentatively set as +1 at the most upstream 5′ end of the
ZNFX1 cDNA that is shown in the NCBI Gene database
(https://www.ncbi.nlm.nih.gov/gene/57169). Sequence
analysis of the 543-bp region with 97.5% relative scores suggested
characteristic recognition sequences of several known TFs (Fig. 1B). Although no particular sequences
similar to the TATA or CCAAT boxes were present, putative binding
sites for E2F1 (-347 to -336 and -111 to -100), E2F4/6 (-347 to
-336), EHF (-158 to -150), ETS1 (-208 to -202, -158 to -150, and
+101 to +106), KLF5/16 and Sp1/2/4 (257 to 241), SPI1 (-245 to
-239, -208 to -202, and -157 to -151), SPIB (-158 to 150), TFAP2A
(-292 to -282 and -52 to -41), and ZNF354C (+25 to +30) are
contained in the 543-bp region (Fig.
1B).
 | Figure 1Promoter activities of the 5′
upstream region of the human ZNF family protein encoding genes. (A)
(Left panel) The 5′ flanking sequence of the human ZNF
protein-encoding genes, which has been ligated upstream of the Luc
gene of the pGL4.10[luc2] vector, is shown. The most upstream
5′regions of the cDNA reported are designated +1. HL 60 cells were
transiently transfected Luc reporter plasmids and they were treated
with or without TPA (8.1 nM) for 24 h. Luc activities were
normalized to that of the pGL4-ZNF343-transfected cells. Results
are presented as the mean ± standard deviation from four
independent experiments. (B) Nucleotide sequence of the 5′ flanking
sequence of the human ZNFX1 gene. The nucleotide sequence of
the 543 bp fragment that was obtained from PCR is shown. The most
upstream 5′ end of the ZNFX1 cDNA (Gene ID: 57169,
NM_021035.2, NC_018931.2, and NC_000020.11), which is typed in
bold, is designated as nucleotide +1. Putative transcription factor
binding sites that are predicted by JASPAR analysis with 97.5%
relative scores are indicated by arrows. The GGAA and TTCC motifs
are doubly underlined. The substituted nucleotide T at 210, which
is identical to the Single Nucleotide Polymorphisms database (rs
3818066 A/G), is indicated by the arrowhead. Underlined characters
represent overlap with the human ZFAS1 gene (GENE ID:
441951), which is transcribed in the opposite direction to that of
the ZNFX1 gene. Statistical analysis was performed with a
Student's t test. *P<0.05. E2F1, eukaryotic
translation termination factor 1; EHF, ETS homologous factor; ETS1,
E26 transformation-specific 1; KLF5, Kruppel like factor 5; Luc,
luciferase; SPI1, spleen focus forming virus proviral integration
oncogene; SPIB, Spi B transcription factor; TFAP2A, transcription
factor AP 2 α; ZC3HAV1, zinc finger CCCH-type containing, antiviral
1; ZNF, zinc finger protein; ZNFX1, zinc finger nfx-1-type
containing 1; ZFAS1, ZNFX1 antisense 1. |
Effects of TPA on ZNFX1 gene expression
and its protein amount in HL-60 cells
To examine whether human ZNFX1 gene
expression is upregulated during the macrophage like
differentiation of HL-60 cells, total RNAs were extracted after TPA
(8.1 nM) treatment. The ZNFX1 gene transcripts were
accumulated from 24-48 h of TPA addition (Fig. 2). This was accompanied with the
induction of ITGAM expression (Fig. 2A), which encodes the adherent
molecule integrin subunit α M/CD11b/Mac1 (29). The relative gene expression of
ZNFX1 compared with that of GAPDH was induced after
24 h of TPA addition (Fig. 2B).
However, the amount of transcripts decreased in response to longer
durations following TPA treatment (Fig. S1). Prior to the upregulation of
transcription, the amount of ZNFX1 protein began to increase from
1-24 h after TPA treatment (Fig.
3), suggesting that the post-transcriptional regulation,
including translation rate or stability of the ZNFX1 protein, may
have been altered following TPA treatment. The accumulation of the
ZNFX1 protein occurred prior to that of CD11b, which was
significantly induced at 48 h after TPA treatment compared with the
other time points following treatment (Fig. S2).
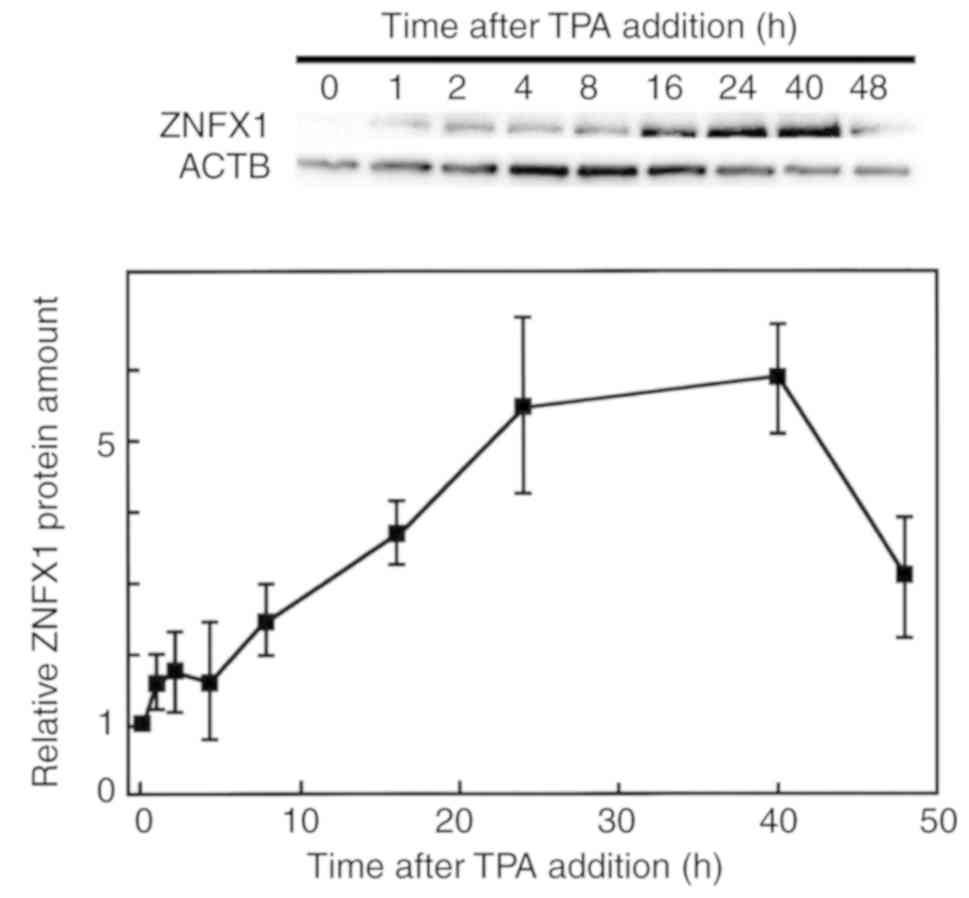 | Figure 3Effects of TPA on ZNFX1 protein
levels in HL-60 cells. The culture medium of HL 60 cells was
replaced with RPMI 1640 medium containing 10% fetal calf serum with
8.1 nM of TPA, and cells were harvested after 0, 2, 4, 8, 16, 24,
40 and 48 h of TPA treatment. The extracted proteins were then
separated by 15% SDS PAGE, and western blotting was performed with
primary antibodies against ZNFX1 and ACTB. Each band was
quantified, and results show the relative ZNFX1/ACTB protein
expression ratio. Results are presented as the mean ± standard
deviation from three independent experiments. ATCB, β actin; TPA,
12 O tetra decanoylphorbol 13 acetate; ZNFX1, zinc finger
nfx-1-type containing 1. |
Effects of TPA on ZNFX1 promoter
activity
To investigate the molecular mechanism as to how the
human ZNFX1 promoter is upregulated by TPA, pGL4-ZNFX1 and
its derivative deletion constructs (Fig. 4A) were transiently transfected into
HL 60 cells. The relative Luc activity of the
pGL4-ZNFX1-transfected cells significantly increased after the
addition of TPA (8.1 nM) to the cell culture compared with the
corresponding control (Fig. 4A).
This induction by TPA was also observed in pGL4-X1-Δ3, Δ4, and Δ5
transfected cells, but not in the pGL4-X1-Δ1 and Δ2 transfected
cells. The Luc activities of the pGL4-X1-Δ1 and Δ2 transfected
cells were reduced to almost background level, implying that they
had lost the essential elements for transcription. In addition, a
notable response to TPA was not observed from the pGL4-X1-Δ1 and Δ2
transfected cells, suggesting that the 97 nucleotides from +87 to
+183 are primarily required for the positive response to TPA in HL
60 cells.
 | Figure 4Deletion and mutation analysis of the
543 bp human ZNFX1 promoter region. The 5′ flanking sequence
of the human ZNFX1 gene, which has been ligated upstream of
the Luc gene of the pGL4.10[luc2] vector, was
presented (left panel). The putative 5′ end of the cDNA is
designated +1. The GGAA and TTCC sequences and the transcription
factor binding elements that were predicted from the JASPAR program
(score >97.5) are schematically shown as shaded and unshaded
rectangles. (A) Luc reporter plasmids were transiently transfected
into HL 60 cells and treated with or without TPA (8.1 nM), and then
they were collected after 24 h incubation. (B) The transient
transfection and Luc assay experiments were performed with
pGL4-ZNFX1, pGL4-X1-Δ6, pGL4-X1-Δ6mt1, pGL4-X1-Δ6mt2,
pGL4-X1-Δ6mt12, pGL4-X1-Δ7, pGL4-X1-Δ7mt1, pGL4-X1-Δ7mt2, and
pGL4-X1-Δ7mt12. Histograms show relative Luc activities of deletion
construct transfected cells; results are presented as the mean ±
standard deviation from at least three independent experiments.
Statistical analysis was performed with a Student's t test.
*P<0.05 vs. TPA. Luc, luciferase; TPA,
12-O-tetra decanoylphorbol-13-acetate; ZNFX1, zinc finger
nfx-1-type containing 1. |
To examine the TPA responding elements in detail,
point mutations were introduced in the pGL4-X1-Δ6 and pGL4-X1-Δ7
plasmids (Fig. 4B). The response
to TPA was completely abolished when both GGAA motifs were
disrupted (pGL4-X1-Δ6mt12 and pGL4-X1-Δ7mt12), suggesting that the
duplicated GGAA motif serves a role in the positive response to
TPA. JASPAR analysis with 92% relative scores suggested that
putative binding elements for ETS1, MZF1, SPI1, STAT1, and STAT2
proteins are involved in the positive response to TPA (Table II). As expected, the disruption of
the duplicated GGAA motif in the pGL4-X1-Δ6mt12 and pGL4-X1-Δ7mt12
constructs excluded the possibilities of the binding of these TFs
to the 21 bp element. Taken together, these results suggested that
the ZNFX1 promoter is regulated by the duplicated GGAA
motif, which is present +50 to +70-bp downstream of the putative
TSS, pertaining to the positive response to TPA in differentiating
HL-60 cells.
 | Table IIPutative transcription factor binding
sites in the TPA-responsive 21-bp element in the ZNFX1
promoter. |
Table II
Putative transcription factor binding
sites in the TPA-responsive 21-bp element in the ZNFX1
promoter.
| Matrix ID | Gene | Score | Relative score | Start | End | Strand | Predicted
sequence |
|---|
| MA0098.1 | ETS1 | 7.79718 | 1 | 11 | 16 | - | TTTCCG |
| MA0056.1 | MZF1 | 8.50962 | 0.973735 | 4 | 9 | + | CGGGGA |
| MA0080.1 | SPI1 | 7.72714 | 0.958724 | 6 | 11 | + | GGGAAC |
| MA0517.1 | STAT1::STAT2 | 16.9526 | 0.927577 | 6 | 20 | - |
TCGGTTTCCGTTCCC |
Discussion
Although, it might be worth to analyze expression of
the selected ZNF family genes, their responses to TPA were not
greater than that of ZNFX1. In addition, as epigenetic or
post transcriptional regulation could serve a role in the
transcription of these genes, expression may not always be
associated with promoter activities. Therefore, in this study, the
543 bp region upstream of the human ZNFX1, which is
head-head oriented with the ZFAS1 gene, was characterized.
IFN treatment is known to affect HL 60 cell differentiation
(30). Additionally, duplicated
GGAA motifs are frequently found in 5′-flanking regions of the
human interferon stimulated genes (ISGs) (9). It should be noted that TPA responding
duplicated GGAA motif is present 0.1 kb downstream of the putative
TSS of the ZNFX1 that belongs to the ISGs (31). We observed upregulation of the
human ZNFX1 gene and protein expression during HL-60
differentiation into macrophage like cells. JASPAR analysis
indicated that ETS1, MZF1, SPI1, and STAT1/STAT2 are candidate
proteins to bind to the TPA responding as its overexpression was
reported to exhibit no effect on TPA-induced monocyte/macrophage
differentiation of HL-60 cells (32). Given that STAT1 and ETS family
proteins, which co operatively regulate transcription of the
CD40 gene in microglia/macrophages (33,34),
they may regulate ZNFX1 gene expression. The JAK/STAT
signaling pathway serves a role in the TPA inducible expression of
2′-5′-oligoadenylate synthetase 1 (OAS1), which belongs to
the ISGs, in HL-60 cells (35). Of
note, the OAS1 promoter possesses a duplicated GGAA motif
responsive to the ETS family TF E74 like ETS transcription factor 1
in HeLa S3 cells (9). The
TPA-responsive 21 bp region harbors similar sequences to the
consensus binding motifs for ETS family type III proteins,
including SPI1, SPIB, and SPIC, and type I proteins, including
ETS1/2 and GABPα (14). GABP α/β
protein targets the GGAA motif in the promoter region of mouse
RB1, which is regulated during myogenesis (36). Spi1 (PU.1) and SPIB proteins occupy
similar regions in the genome, suggesting they co operatively
regulate the expression of genes that regulate B cell development
(37). An electrophoretic mobility
shift assay (EMSA) with antibodies indicated that SPI1 (PU.1) binds
to the TPA responding regions of the human poly(ADP ribose)
glycohydrolase (38) and
eukaryotic translation termination factor 4 (39) gene promoters. Although JASPAR
analysis in our study suggested candidate, it has not been shown
which TFs bind to the TPA-responding sequence. ZNFX1
expression could be induced by combinations or the expression
profiles of the duplicated GGAA-recognizing TFs, which include ETS
family proteins and several DNA binding proteins (8). This should be elucidated in future
experiments, such as EMSA, Footprint analysis, and chromatin
immunoprecipitation assay, along with overexpression/knockdown
experiments.
Binding of transcription suppressor ETS2 repressor
factor to the GGAA motif is hindered by upregulation of the ETS
transcription factor ERG in prostate cancer (40). Occupation or combinations of the
GGAA motif binding proteins near TSSs play a role in both positive
and negative regulation of cellular differentiation regulator
encoding gene expression (8). In
other words, if the binding of strong trans-activating TFs
were replaced by weak ones after TPA treatment, the GGAA motif
could be a suppressive element (8). This may partly explain the reason why
the ZC3HAV1, ZNF343, ZNF252, ZNF555 and
ZNF782 promoters exhibited marked activation in response to
TPA treatment. Alternatively, specific elements near the TSSs could
hinder the positive response of the duplicated GGAA motifs.
Furthermore, TPA inducible microRNAs (41) should be taken into account for
regulation of gene expression.
TPA inducible genes in HL-60 cells have been studied
previously (42). The induction
profile of the expression of the RB1 gene, with a 5′
flanking region has duplicated GGAA motifs (13), resembles that of the ZNFX1
gene. Although, in this study, RB1 gene expression was not
analyzed, it has been reported to be induced at 10 h after TPA
treatment (13). The expression of
these two genes continued to increase after the TPA treatment for
at least for 48 h when almost all cells attach to the culture dish
(13). RB1, in association with
E2F proteins, regulates the expression of various genes to control
cell cycle progression (43,44).
Interestingly, the human ZNFX1 and RB1 genes are
under the control of bi directional promoters, having ZFAS1
and LINC00441as partner genes, respectively. The expression
of the ZFAS1 (ZNFX1-AS1) gene is downregulated in
breast tumors (45), and its
overexpression inhibits proliferation to induce the apoptosis of
cancer cells (46), implying that
ZFAS1 serves as a tumor suppressor. ZNFX1 protein expression
increased until 40 h after treatment with TPA I our study,
suggesting that it plays a role in sustaining the macrophage like
state of the HL-60 cells. Certain ZNFs, including zinc finger E-box
binding homeobox 1, ZFP36, broad-complex, tramtrack and
bric-à-brac-zinc finger, and ectopic virus integration site 1 are
known to regulate cell differentiation (5,47-49).
Additionally, epigenetic regulation should be considered in this
process. In Caenorhabditis elegans, ZNFX1 functions as an
RNA helicase that directs transgenerational epigenetic inheritance
(50).
A potential cancer therapy with low toxic effect, or
differen tiation/apoptosis inducing therapy, has been proposed
(51,52). Providing that upregulation of both
ZNFX1 and ZFAS1 genes affect the differentiation
process, further investigations into the regulation of their
expression may contribute to the estab lishment of differentiation
inducing gene therapy.
Supplementary Data
Abbreviations:
|
FCS
|
fetal calf serum
|
|
IFN
|
interferon
|
|
Luc
|
luciferase
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TPA
|
12-O-tetradecanoylphorbol-13-acetate
|
|
TSS
|
transcription start site
|
Acknowledgments
The authors are grateful to Ms. Sayaka Ishibashi and
Ms. Chihiro Katsuda (Department of Gene Regulation, Faculty of
Pharmaceutical Sciences, Tokyo University of Science), for their
outstanding technical assistance.
Funding
The present study was supported in part by JSPS
KAKENHI (grant no. 24510270) and a Research Fellowship from the
Research Center for RNA Science, RIST, Tokyo University of
Science.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HH, MY, HO, YK, and KN constructed the Luc reporter
plasmids. HH and HO performed experiments and analyzed the data
(transfection assay, RT qPCR, western blotting, and statistical
analysis). YM performed ANOVA. FU interpreted the data and wrote
the manuscript. ST collected and analyzed/interpreted the data. TO
and MA interpreted the data and edited the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hasegawa A, Kaneko H, Ishihara D, Nakamura
M, Watanabe A, Yamamoto M, Trainor CD and Shimizu R: GATA1 binding
kinetics on conformation-specific binding sites elicit differential
transcriptional regulation. Mol Cell Biol. 36:2151–2167. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Q, Niu N, Wada Y and Liu J: The role
of Cdkn1A interacting zinc finger protein 1 (CIZ1) in DNA
replication and pathophysi ology. Int J Mol Sci. 17:2122016.
View Article : Google Scholar
|
|
3
|
Tetreault MP, Weinblatt D, Shaverdashvili
K, Yang Y and Katz JP: KLF4 transcriptionally activates non
canonical WNT5A to control epithelial stratification. Sci Rep.
6:261302016. View Article : Google Scholar
|
|
4
|
Liu Y, Ma D and Ji C: Zinc fingers and
homeoboxes family in human diseases. Cancer Gene Ther. 22:223–226.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Prenzler F, Fragasso A, Schmitt A and Munz
B: Functional analysis of ZFP36 proteins in keratinocytes. Eur J
Cell Biol. 95:277–284. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uchiumi F, Larsen S and Tanuma S:
Transcriptional regulation of the human genes that encode DNA
repair and mitochondrial function associated proteins. DNA Repair.
Chen C: InTechOpen; London, UK: pp. 129–167. 2015
|
|
7
|
Uchiumi F, Larsen S and Tanuma S: Possible
roles of a duplicated GGAA motif as a driver cis element for cancer
associated genes. Understand Cancer-research and treatment.
iConcept: iConcept Press Ltd.; Hong Kong: pp. 1–25. 2016
|
|
8
|
Uchiumi F, Miyazaki S and Tanuma S: The
possible functions of duplicated ets (GGAA) motifs located near
transcription start sites of various human genes. Cell Mol Life
Sci. 68:2039–2051. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Larsen S, Kawamoto S, Tanuma S and Uchiumi
F: The hematopoietic regulator, ELF 1, enhances the transcriptional
response to Interferon β of the OAS1 anti viral gene. Sci Rep.
5:174972015. View Article : Google Scholar
|
|
10
|
Uchiumi F, Shoji K, Sasaki Y, Sasaki M,
Sasaki Y, Oyama T, Sugisawa K and Tanuma S: Characterization of the
5′-flanking region of the human TP53 gene and its response to the
natural compound, resveratrol. J Biochem. 159:437–447. 2016.
View Article : Google Scholar
|
|
11
|
Uchiumi F, Arakawa J, Iwakoshi K,
Ishibashi S and Tanuma S: Characterization of the 5′-flanking
region of the human DNA helicase B (HELB) gene and its response to
trans-Resveratrol. Sci Rep. 6:245102016. View Article : Google Scholar
|
|
12
|
Uchiumi F, Arakawa J, Takihara M, Akui M,
Ishibashi S and Tanuma S: The effect of trans resveratrol on the
expression of the human DNA-repair associated genes. Int Mol Med.
3:783–792. 2016.
|
|
13
|
Uchiumi F, Watanabe T and Tanuma S:
Characterization of various promoter regions of the human DNA
helicase encoding genes and identification of duplicated ets (GGAA)
motifs as an essential transcription regulatory element. Exp Cell
Res. 316:1523–1534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Perdomo Sabogal A, Nowick K, Piccini I,
Sudbrak R, Lehrach H, Yaspo ML, Warnatz HJ and Querfurth R: Human
lineage-specific transcriptional regulation through GA binding
protein transcription factor alpha (GABPa). Mol Biol Evol.
33:1231–1244. 2016. View Article : Google Scholar
|
|
15
|
Oikawa T and Yamada T: Molecular biology
of the Ets family of transcription factors. Gene. 303:11–34. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu T, Trojanowska M and Watson DK: Ets
proteins in biological control and cancer. J Cell Biochem.
91:896–903. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sevinsky JR, Whalen AM and Ahn NG:
Extracellular signal-regulated kinase induces the megakaryocyte
GPIIb/CD41 gene through MafB/Kreisler. Mol Cell Biol. 24:4534–4545.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cho HY and Lee SW, Seo SK, Choi IW, Choi I
and Lee SW: Interferon sensitive response element (ISRE) is mainly
respon sible for IFN-α-induced upregulation of programmed death 1
(PD 1) in macrophages. Biochim Biophys Acta. 1779:811–819. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Champhekar A, Damle SS, Freedman G,
Carotta S, Nutt SL and Rothenberg EV: Regulation of early T lineage
gene expression and developmental progression by the progenitor
cell transcription factor PU.1. Genes Dev. 29:832–848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akagi T, Kuure S, Uranishi K, Koide H,
Costantini F and Yokota T: ETS related transcription factors ETV4
and ETV5 are involved in proliferation and induction of
differentiation-associated genes in embryonic stem (ES) cells. J
Biol Chem. 290:22460–22473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang C, Kam RK, Shi W, Xia Y, Chen X, Cao
Y, Sun J, Du Y, Lu G, Chen Z, et al: The proto-oncogene
transcription factor Ets 1 regulates neural crest development
through histone deacetylase 1 to mediate output of bone
morphogenetic protein signaling. J Biol Chem. 290:21925–21938.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kar A and Gutierrez-Hartmann A: Molecular
mechanisms of ETS transcription factor mediated tumorigenesis. Crit
Rev Biochem Mol Biol. 48:522–543. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Komura S, Semi K, Itakura F, Shibata H,
Ohno T, Hotta A, Woltjen K, Yamamoto T, Akiyama H and Yamada Y: An
EWS FLI1 induced osteosarcoma model unveiled crucial role of
impaired osteogenic differentiation on osteosarcoma development.
Stem Cell Reports. 6:592–606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dean KC, Huang L, Chen Y, Lu X and Liu Y:
An Rb1-dependent amplification loop between Ets1 and Zeb1 is
evident in thymo cyte differentiation and invasive lung
adenocarcinoma. BMC Mol Biol. 16:82015. View Article : Google Scholar
|
|
25
|
Gulick T: Transfection using DEAE-dextran.
Curr Protoc Mol Biol. Chapter 20: Unit 20.4. 2003. View Article : Google Scholar
|
|
26
|
Katagiri K, Katagiri T, Koyama Y, Morikawa
M, Yamamoto T and Yoshida T: Expression of src family genes during
monocytic differentiation of HL-60 cells. J Immunol. 146:701–707.
1991.PubMed/NCBI
|
|
27
|
Schmittgen TD and Livak KJ: Analyzing real
time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
28
|
Uchiumi F, Ohi H and Tanuma S: Application
of DEAE-dextran to an efficient gene transfer system. Seikagaku.
86:532–537. 2014.In Japanese. PubMed/NCBI
|
|
29
|
White SL, Belov L, Barber N, Hodgkin PD
and Christopherson RI: Immunophenotypic changes induced on human
HL60 leukemia cells by 1 alpha, 25-dihydroxyvitamin D3 and
12-O-terradecanoyl phorbol 13 acetate. Leuk Res. 29:1141–1151.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grant S, Bhalla K, Weinstein B, Pestka S,
Mileno MD and Fisher PB: Recombinant human interferon sensitizes
resistant myeloid leukemic cells to induction of terminal
differentiation. Biochem Biophys Res Commun. 130:379–388. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Forde N, Duffy GB, McGettigan PA, Browne
JA, Mehta JP, Kelly AK, Mansouri Attia N, Sandra O, Loftus BJ,
Crowe MA, et al: Evidence for an early endometrial response to
pregnancy in cattle: Both dependent upon and independent of
interferon tau. Physiol Genomics. 44:799–810. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robertson KA, Hill DP, Kelley MR, Tritt R,
Crum B, Van Epps S, Srour E, Rice S and Hromas R: The myeloid zinc
finger gene (MZF-1) delays retinoic acid-induced apoptosis and
differentiation in myeloid leukemia cells. Leukemia. 12:690–698.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu H, Yang G, Lu F, Wang R, Yao L and Lu
Z: Transcription up regulation of restin by all-trans retinoic acid
through STAT1 in cancer cell differentiation process. Biochem
Biophys Res Commun. 343:1009–1016. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nguyen VT and Benveniste EN: Involvement
of STAT-1 and Ets family members in interferon gamma induction of
CD40 transcription in microglia/macrophages. J Biol Chem.
275:23674–23684. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cohen S, Dovrat S, Sarid R, Huberman E and
Salzberg S: JAK-STAT signaling involved in phorbol 12 myristate 13
acetate and dimethyl sulfoxide induced 2′-5′ oligoadenylate
synthetase expression in human HL 60 leukemia cells. Leuk Res.
29:923–931. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deléhouzée S, Yoshioka T, Sawa C, Sawada
J, Ito T, Omori M, Wada T, Yamaguchi Y, Kabe Y and Handa H: GABP,
HCF-1 and YY1 are involved in Rb gene expression during myogenesis.
Genes Cells. 10:717–731. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solomon LA, Li SK, Piskorz J, Xu LS and
DeKoter RP: Genome wide comparison of PU.1 and Spi-B binding sites
in a mouse B lymphoma cell line. BMC Genomics. 16:762015.
View Article : Google Scholar
|
|
38
|
Uchiumi F, Sakakibara G, Sato J and Tanuma
S: Characterization of the promoter region of the human parg gene
and its response to PU.1 during differentiation of HL-60 cells.
Genes Cells. 13:1229–1247. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hamada H, Goto Y, Arakawa J, Murayama E,
Ogawa Y, Konno M, Oyama T, Asai M, Sato A, Tanuma SI and Uchiumi F:
Characterization of the human E2F4 promoter region and its response
to 12-O-tetradecanoylphorbol 13 acetate. J Biochem. pii: mvz047.
2019. View Article : Google Scholar : Epub ahead of
print.
|
|
40
|
Bose R, Karthaus WR, Armenia J, Abida W,
Iaquinta PJ, Zhang Z, Wongvipat J, Wasmuth EV, Shah N, Sullivan PS,
et al: ERF mutations reveal a balance of ETS factors controlling
pros tate oncogenesis. Nature. 546:671–675. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kasashima K, Nakamura Y and Kozu T:
Altered expression profiles of microRNAs during TPA-induced
differentiation of HL-60 cells. Biochem Biophys Res Commun.
322:403–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zheng X, Ravatn R, Lin Y, Shih WC, Rabson
A, Strair R, Huberman E, Conney A and Chin KV: Gene expression of
TPA induced differentiation in HL 60 cells by DNA microarray
analysis. Nucleic Acids Res. 30:4489–4499. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dick FA and Rubin SM: Molecular mechanisms
underlying RB protein function. Nat Rev Mol Cell Biol. 14:297–306.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bagchi S, Weinmann R and Raychaudhuri P:
The retinoblastoma protein copurifies with E2F-1 an E1A-regulated
inhibitor of the transcription factor E2F. Cell. 65:106310721991.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Askarian Amiri MN, Crawford J, French JD,
Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani
SR, et al: SNORD-host RNA Zfas1 is a regulator of mammary
development and a potential marker for breast cancer. RNA.
17:878–891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang T, Ma S, Qi X, Tang X, Cui D, Wang Z,
Chi J, Li P and Zhai B: Long noncoding RNA ANFX1-AS1 suppresses
growth of hepatocellular carcinoma cells by regulating the
methylation of miR-9. Onco Targets Ther. 9:5005–5014. 2016.
View Article : Google Scholar :
|
|
47
|
Chevier A and Corcoran LM: BTB-ZF
transcription factors, a growing family of regulators of early and
late B-cell develop ment. Immunol Cell Biol. 92:481–488. 2014.
View Article : Google Scholar
|
|
48
|
Singh S, Howell D, Trivedi N, Kessler K,
Ong T, Rosmaninho P, Raposo AA, Robinson G, Roussel MF, Castro DS
and Solecki DJ: Zeb1 controls neuron differentiation and germinal
zone exit by a mesenchymal epithelial like transition. Elife.
5:pii: e12717. 2016. View Article : Google Scholar
|
|
49
|
Glass C, Wilson M, Gonzalez R, Zhang Y and
Perkins AS: The role of EVI1 in myeloid malignancies. Blood Cells
Mol Dis. 53:67–76. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wan G, Fields BD, Spracklin G, Shukla A,
Phillips CM and Kennedy S: Spatiotemporal regulation of liquid like
condensates in epigenetic inheritance. Nature. 557:679–683. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Leszczyniecka M, Roberts T, Dent P, Grant
S and Fisher PB: Differentiation therapy of human cancer: Basic
science and clinical applications. Pharmacol Ther. 90:105–156.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Luo H, Liu WH, Liang HY, Yan HT, Lin N, Li
DY, Wang T and Tang LJ: Differentiation inducing therapeutic effect
of Notch inhibition in reversing malignant transformation of liver
normal stem cells via MET. Oncotarget. 9:18885–18895. 2018.
View Article : Google Scholar : PubMed/NCBI
|


















