Introduction
CD46, a transmembrane protein, plays pivotal roles
in various biological processes, including immune activation and
the modulation of adaptive immunity. Notably, it regulates
complement activity, a crucial component of the immune system that
targets invading pathogens and damaged cells (1). CD46 functions as a cofactor in the
degradation of complement components, thereby inhibiting complement
activation. This inhibition is essential to prevent excessive or
inappropriate activation of the complement system, which could
result in tissue damage and autoimmune disorders. CD46 is widely
distributed in tissues, being expressed by almost all human cell
populations, with the exception of erythrocytes (2). It is commonly expressed in four
distinct isoforms, resulting from alternative mRNA splicing of a
single gene transcript (3).
Beyond its role in complement regulation, CD46 has
been linked to several types of cancer and other diseases (4-9). A
number of tumor types exhibit CD46 overexpression. For instance, in
hepatocellular carcinoma, CD46 expression is ~6 times higher than
in liver cirrhosis and chronic hepatitis (10). High concentrations of CD46 have
been observed in the sera of cancer patients (11). CD46 expression is an unfavorable
prognostic factor in ovarian cancer, breast cancer and
hepatocellular carcinoma (12-14). Additionally, CD46 contributes to
the tumorigenesis and development of bladder cancer (15). Our previous research demonstrated
that restoring CD46 expression in bladder cancer cells conferred
protection against cetuximab-mediated inhibition of
phosphoinositide-3-kinase (PI3K)/protein kinase B (AKT) and
extracellular signal-regulated kinase (ERK) phosphorylation
(16). This restoration also
protected cells from complement-dependent cytotoxicity and
antibody-dependent cellular cytotoxicity induced by cetuximab.
Thus, CD46 shows a protective effect in cancer cells against both
direct (via involvement of PBMC or complement) and indirect
cytotoxic activities of cetuximab in bladder cancer cells.
Studies have underscored the importance of matrix
metalloproteinases (MMPs) in tumor growth and metastasis, with a
focus on MMP2 and MMP9 (17,18). Researchers have characterized
promoter region of MMP9, identifying binding sites for
several transcription factors such as activator protein 1 (AP-1),
specificity protein 1 (SP-1), E-twenty six (Ets) and nuclear factor
kappa B (NF-κB) (19). Further
research indicates that AP-1 activation is essential for sustaining
cancer cell invasion and aiding epithelial wound healing (20-22). This activation predominantly
occurs through the mitogen-activated protein kinase (MAPK)
cascades. The MAPK family is divided into three primary groups:
ERK, c-Jun N-terminal kinase (JNK) and p38 MAPK. Certain MAPK
family members, including ERK1/2 and p38 MAPK, along with PI3K/AKT,
play a pivotal role in upregulating MMP9 expression (23-27). Additionally, AP-1 activation is
primarily facilitated by the PI3K/AKT and JNK pathways, working in
synergy (28).
Despite significant progress in understanding CD46,
a number of aspects of its functions and the molecular mechanisms
involved in cancer cell metastasis remain unclear. Our previous
research demonstrated that CD46 is highly expressed in bladder
cancers, where it helps protect cancer cells from various cytotoxic
insults (16,29). Moreover, CD46 appears to influence
cell migration by altering the expression of several cytoskeletal
proteins (30). Given the strong
correlation between MMPs, particularly MMP9, and the progression
and recurrence of bladder tumors (18,31,32), the present study aimed to
investigate the impact of CD46 on MMP9 in bladder cancer and to
elucidate the specific mechanisms related to CD46 and MMP9 in
bladder cancer cells.
Materials and methods
Cell lines and cell culture
Human bladder cancer cell lines [J82, 5637, HT-1376
(cat. no. CRL-1472), UM-UC-3], melanoma cells (M010119), prostate
cancer cells (LNCaP), colon cancer cells (HCT116) and lung cancer
cells (A549) were procured from the American Type Culture
Collection. 253J cells were kindly provided by Dr Wun-Jae Kim
(Chungbuk National University, Korea). J82, 5637, HT-1376, UM-UC-3,
253J, M010119 and LNCaP cells were cultured in RPMI 1640 medium
(Welgene, Inc.) supplemented with 5% heat-inactivated fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
HCT116 and A549 cells were cultured in Dulbecco's modified Eagle's
medium (DMEM; Welgene, Inc.) supplemented with 5% heat-inactivated
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
Cultivation was at 37°C in an atmosphere containing 5%
CO2, with media changes every 2-3 days. To establish
cell lines with CD46 overexpression, the lentiviral vector
pBlasti-eGFP-CD46 kindly provided by Dr Silvio Hemmi (Institute of
Molecular Life Sciences, University of Zurich, Zurich, Switzerland)
was co-transfected with helper in the 2nd generation transfection
system into 293T cells, as detailed in a previous study (29). The 293T cells were cultured in
DMEM supplemented with 10% FBS at a temperature of 37°C and 5%
CO2. Following a 48 h incubation period at 37°C; the
supernatant of the 293T cells were collected and the lentivirus was
concentrated by subjecting to centrifugation at 2,100 x g for 5 min
at 4°C and filtering the supernatant through a 0.45 µm
filter. The harvested lentiviral vectors were then introduced into
the target cells for transduction. Subsequently, cell lines were
plated into a 6-well plate and the cells were cultured until they
reached 80% confluence. To generate stable cell lines, cells were
transduced with these lentiviral vectors in a milieu containing
polybrene at a concentration of 8 µg/ml. Then, lentivirus
was added and co-cultured with the cells at 37°C for 24 h
(Multiplicity of infection, 30). After that, the medium was
replaced, and then culture continued in a 5% CO2 and
37°C incubator for another 48 h. Following a 72 h incubation
period, cells underwent a selection process using 10 µg/ml
blasticidin (MilliporeSigma), over a course of 3 days.
Western blotting
For the cancer cells, protein extraction was
performed using Pro-Prep (cat. no. 17081; Intron Biotechnology,
Inc.). The quantification of proteins was conducted using the BCA
Protein Assay kit (cat. no. 23227; Thermo Fisher Scientific, Inc.).
Proteins (15 µg) were subjected to separation on a 7.5%
SDS-polyacrylamide gel, using the Bio-Rad electroporation system
(Bio-Rad Laboratories, Inc.), followed by transfer to PVDF
membranes (cat. no. IPVH00010; MilliporeSigma). After the membranes
were blocked with 3% BSA (BioShop) in tris-buffered saline
containing 0.1% Tween-20 (TBST) at room temperature for 1 h and
then incubated at 4°C overnight with primary antibodies. The
primary antibodies used included: CD46 (1:2,000; cat. no. ab108307;
Abcam); MMP9 (1:1,000; cat. no. 7-11C; Santa Cruz Biotechnology,
Inc.), phosphorylated (p-)AKT (1:1,000; cat. no. 05-669;
MilliporeSigma), p38α (1:1,000; cat. no. sc-535; Santa Cruz
Biotechnology, Inc.), JNK1/3 (1:1,000; cat. no. sc-474; Santa Cruz
Biotechnology, Inc.), p-JNK (1:1,000; cat. no. sc-6254 Santa Cruz
Biotechnology, Inc.), ERK1 (1:1,000; cat. no. sc-271269 Santa Cruz
Biotechnology, Inc.), c-Fos (1:1,000; cat. no. sc-166940; Santa
Cruz Biotechnology, Inc.), p-c-Fos (1:1,000; cat. no. sc-81485;
Santa Cruz Biotechnology, Inc.), c-Jun (1:1,000; cat. no. sc-1694;
Santa Cruz Biotechnology, Inc.), p-c-Jun (1:1,000; cat. no. sc-822;
Santa Cruz Biotechnology, Inc.); p-p38 (1:2,000; cat. no. 05-1059;
MilliporeSigma), AKT (1:2,000; cat. no. 9272; Cell Signaling
Technology, Inc.); MMP2 (1:2,000; cat. no. NB200-114; Novus
Biologicals; Bio-Techne); p-ERK (1:2,000; cat. no. 05-797R;
MilliporeSigma); and β-actin (1:2,000; cat. no. 26628-22-8;
MilliporeSigma). After washing three times with TBST, the membranes
were incubated at room temperature for 1 h with the secondary
antibody (Anti-mouse IgG HRP-linked antibody; 1:5,000; cat. no.
7076S; Anti-rabbit IgG HRP-linked antibody; 1:10,000; cat. no.
7074; Cell Signaling Technology, Inc.). Following secondary
antibody incubation, the membranes were washed four times again and
then completely covered with a 1:1 mixture of HRP substrate
peroxide solution/HRP substrate Luminol reagent (cat. no.
WSKLS0500; MilliporeSigma). Band visualization was conducted using
the Fusion Solo system (Vilber Lourmat Deutschland GmbH) and
analyzed with ImageJ software (version 1.53m; National Institutes
of Health, USA).
Reverse transcription-quantitative (RT-q)
PCR
Total RNA was extracted from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manual instructions. cDNA was generated with M-MLV
reverse transcriptase (cat. no. M170B; Promega Corporation). mRNA
expression was determined by qPCR on a CFX96 real-time PCR System
(Bio-Rad Laboratories, Inc.) using TOPreal SYBR Green qPCR premix
kit (cat. no. RT500M; Enzynomics), using GAPDH as the internal
loading control. The primer sequences for PCR were as follows:
human CD46 (Forward: 5′-CCACGACCATTTGAAGCTAT-3′, Reverse:
5′-TCCAGGTGCTGGATCACAC-3′), human MMP-9 (Forward:
5′-CATCGTCATCCAGTTTGG-3′, Reverse: 5′-GATGGATTGGCCTTGGAA-3′) and
human GAPDH (Forward: 5′-GAAGGTGAAGGTCGGAGTC-3′, Reverse:
5′-GAAGATGGTGATGGGATTTC-3′). PCR cycling conditions for all samples
included: 10 min at 95°C for enzyme activation, followed by 40
cycles consisting of melting (95°C; 15 sec) and annealing/extension
(72°C; 30 sec) phases. The data were analyzed as previously
described (33). Briefly, using
the comparative CT method, the Ct values obtained were converted to
picogram quantities based on each gene's standard curve from target
cDNA. The quantity of CD46 and MMP9 were normalized to GAPDH and
adjusted by subtracting values from no reverse transcriptase
controls, averaging the result for each triplicate (34).
Transient of CD46 short interfering
(si)RNA
Two variants of CD46 siRNA (CCAAAACCCUACUAUGAGA,
GGAUACUUCUAUAUACCUCUU) and negative control siRNA
(UGCAGGUUUAUAGUCCACAUU) were procured from Bioneer Corporation.
5637 cells underwent transfection with these siRNAs using
Lipofectamine® RNAiMAX transfection reagent (cat. no.
13778-075; Invitrogen; Thermo Fisher Scientific, Inc.), adhering to
the manufacturer's protocol. Cells were cultured in six-well plates
until they reached 60% confluence. To prepare the transfection mix,
7.5 µl Lipofectamine® RNAiMAX was diluted in 150
µl Opti-MEM Medium (Gibco; Thermo Fisher Scientific, Inc.)
and mixed. DNA siRNA (25 pmol) was diluted in 150 µl
Opti-MEM Medium and mixed. The diluted DNA was then combined with
the previously diluted Lipofectamine® RNAiMAX, at a 1:1
ratio, to form a DNA-lipid complex. This complex was allowed to
incubate for 15 min at room temperature to a stable DNA-lipid
complex. Then, the complex was administered to the cultured cells.
Following transfection, the cells were incubated at 37°C. At 6 h
post-transfection, the cells were chanced and incubated at 37°C
with media containing 5% FBS. At 48 h post-transfection, the cells
were lysed for western blot analysis as described earlier.
Wound migration assay
5637, J82 and 253J cells were cultured in six-well
plates until they reached 90% confluence in 2 ml of growth medium.
The cells were then washed and incubated in RPMI containing 0.5%
FBS for 16 h. A sterile 200 µl pipette tip was used to
scrape the cell monolayer. Subsequently, the cells were gently
washed thrice with PBS and FBS-free medium was added for starvation
culture at 37°C. Images were captured at 0, 12 and 24 h
post-scraping using an inverted microscope (Olympus IX51; Olympus
Corporation). The open wound area percentages were measured and
calculated with ImageJ software (version 1.53m; National Institutes
of Health).
Transwell assays
The migration devices (cat. no. 3422; Corning, Inc.)
and invasion devices (cat. no. 354483; Corning, Inc.) were
subjected to room temperature equilibration for 10 min, then the
two devices were incubated for 2 h in a 37°C incubator, and
300-µl serum-free RPMI was added in the upper chamber and
600 µl in the lower chamber. Cells at 80-90% confluence were
washed and incubated in RPMI with 0.1% FBS for 16 h prior to their
application to the chambers. Subsequently, 5637 and J82 cells
(5×104) in 200 µl of serum-free medium were
seeded into the upper chamber and the lower chambers were filled
with 600 µl of RPMI media containing 10% fetal bovine serum.
After a 24-h incubation, cells on the bottom membrane of the
chamber were gently wiped with a cotton swab and migrating cells
were stained with Diff-Quik stain solution (cat. no. 38721; Sysmex
Corporation) at room temperature. After drying, images were
captured under a light microscope (Olympus IX51; Olympus
Corporation) and cells were counted in four fields per chamber.
Cell proliferation assay
Cell proliferation was assessed using the WST-1
assay (EZ-Cytox Cell Viability Assay kit; ITSBio, Inc.), following
the manufacturer's instructions. Briefly, cells were seeded in
96-well plates with 100 µl of RPMI media containing 10% FBS
and cultured at 37°C. Then, at the indicated time points (0, 24, 48
and 72 h), 10 µl of the kit solution was added to the cells,
followed by a 30-min incubation at 37°C. Cell viability was
measured at 450 nm using an Epoch Microplate Spectrophotometer
(BioTek Instruments, Inc.).
Gelatin zymography
Cells at 80-90% confluence were washed and incubated
in RPMI without FBS for 24 h in 60 mm plates. The samples were then
electrophoresed on a 7.5% SDS-polyacrylamide gel containing 1 mg/ml
gelatin. Post-electrophoresis, the gel was washed twice for 2 h at
room temperature using a washing buffer [50 mM Tris-HCl (pH 7.5), 5
mM CaCl2, 1 µM ZnCl2 and 2.5% Triton
X-100] and then briefly rinsed with H2O. Subsequently,
the gel was incubated with 1X of Zymogram developing buffer 10X, pH
7.45 (cat. no. 42620000-2; Bioworld Technology, Inc.) at 37°C for
24 h. After incubation, the gels were stained with Coomassie
Brilliant Blue R-250 Staining Solution (cat. no. 1610436; Bio-Rad
Laboratories, Inc.) and destained with Coomassie Brilliant Blue
R-250 Destaining Solution (cat. no. 1610438; Bio-Rad Laboratories,
Inc.).
Luciferase assay
Transcriptional activity was assessed using a
luciferase assay system. The pGL4.17[luc 2/Neo] vector, containing
the MMP9 promoter region (−924/+13), known as the MMP9-luc
promoter, was used. The MMP9-luc vector was kindly provided by Dr
Young-Han Lee (Konkuk University, Korea). The AP-1-luc vector was
obtained from Stratagene (Agilent Sumitomo Dainippon Pharma Co.,
Ltd.). CD46-overexpressing and control cells were seeded in a
24-well plate at a density of 1×105 cells per well and
cultured for 24 h. The cells were transfected with 0.1 µg of
the reporter vector and 2 ng of Renilla using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions. At 6 h
post-transfection, the cells were washed and cultured with media
containing 5% FBS at 37°C for 18 h. The addition of SB202190 or
LY294002 was at 24 h post-transfection. 24 h following addition of
inhibitor, luciferase and Renilla activities were measured.
Light signals were detected using the Dual-Luciferase Reporter
Assay System (Promega Corporation) and a Glomax Navigator
instrument (Promega Corporation), in accordance with the
manufacturer's instructions.
MMP9 ELISA
Cell media or mouse sera were collected under
sterile conditions, immediately frozen and stored at −20°C until
use. MMP9 concentration was determined using the human MMP9
Quantikine ELISA Kit (cat. no. DMP900; R&D Systems), following
the manufacturer's instructions.
Xenograft model and lung metastasis
model
A total of 28 female BALB/c nude mice (6-weeks-old;
17-22 g; Orient Bio, Inc.) were housed under specific pathogen-free
conditions in the animal facility at the Hwasun Biomedical
Convergence Center, with a 12-h light/dark cycle (light from 7:00
am to 7:00 pm) and controlled temperature maintained at 24±2 °C for
optimal growth, while maintaining a relative humidity range of
50±10%. The mice were allowed ad libitum access to water and
food pellets (PicoLab 5053). All procedures performed in studies
were approved by the Animal Use and Care Committees at Chonnam
National University Medical School (approval no. H2022-70). Mice
were used to establish lung metastasis models through tail vein
inoculation with 1.5×106 UM-UC-3 and 5637 cells. For
each cell line, mice were randomly assigned to the experimental
group (overexpressing CD46) and the control group (7 mice per
group). Overall survival and physiological signs of mice in the
groups were monitored daily (for a period of 6 weeks). If the
animals reached any of the following humane endpoints, they were
sacrificed: Animal weight loss >20%; severely decreased
mobility/activity, moribund behavior or wasting syndrome. Notably,
none of the mice suffered from those signs of illness or mortality
during the experimental process. At the end of the experiment, the
mice were deeply anesthetized with 5% isoflurane (Ifran; Hana
Pharm, Co., Ltd.) to minimize any potential pain or distress during
the procedure, followed by cervical dislocation to sacrifice the
mice. Their blood was collected via the left ventricle using a
23-25 gauge needle. The lung metastasis tumors' fluorescence was
detected using the FOBI Fluorescence In Vivo Imaging System
(Cellgentek, Co., Ltd.). Additionally, lung and liver tissues were
fixed with 10% buffered formalin at 4°C for 48 h. Then tissues were
washed three times with PBS 1X and dehydrated with increasing
alcohol concentration from 30 to 100% within 4 h and xylene within
1 h, embedded in paraffin and sliced into 5-µm serial
sections and subsequently analyzed through H&E staining at room
temperature for 1 min 40 and 15 sec and immunohistochemistry.
Immunohistochemical staining
Tumor paraffin (5 µm) sections from the mice
were deparaffinized and rehydrated in graded alcohols and distilled
water. Then they initially treated with 3%
H2O2 in 60% methanol for 10 min and then
incubated with Immuno-Block reagent (BioSB, Santa Barbara, CA) for
30 min at room temperature. These sections were further incubated
overnight at 4°C with primary antibodies against MMP9 (1:200; cat.
no. HPA001238; Atlas Antibodies AB). For the secondary antibody,
anti-rabbit immunoglobulin IgG (1:500; cat. no. 30014; Vector
Laboratories, Inc.) was used and incubated for 1 h at room
temperature. Immunoreactivity was then visualized using an enhanced
DAB kit (Abcam). The results were observed using a light
microscope.
Statistical analysis
GraphPad Prism software (Dotmatics) was employed for
all statistical analyses. Comparison of two data sets was done with
two-tailed unpaired Student's t-test in case of normal distributed
data, or with U-Mann Whitney test in case of non-parametric data,
considering P-values <0.05 as statistically significant.
Multiple groups were analyzed by one-way ANOVA followed by Tukey's
post hoc test or two-way ANOVA followed by Bonferroni's post hoc
test. The results are presented as mean ± standard deviation.
Results
Forced expression of CD46 alters
expression of MMP9 in bladder cancer cells
The observations indicated that ~50% of bladder
cancers exhibit an overexpression of CD46 compared with normal
urothelia (29). CD46 typically
acts as a protector against complement-mediated cytotoxicity in
cells. Previous studies reported that CD46 also shields cancer
cells from the indirect cytotoxic effects of cetuximab (an EGFR
inhibitor), by modulating AKT and ERK phosphorylation in bladder
cancer cells (16). The direct
role of intracellular CD46 in cancer cells, however, remains poorly
understood. Recently, we found that overexpression and suppression
of CD46 in bladder cancer cells regulated several genes associated
with migration and invasion, as identified by DNA microarray
analysis (30). These genes
include matrix Gla protein (MGP) and keratin 13 (KRT13) (30). Therefore, the present study
explored whether CD46 affected the expression and activity of MMPs,
which are pivotal in cancer cell invasion and metastasis across
various types of cancer. Focusing on MMP2 and MMP9, known to play
significant roles in bladder cancer metastasis (35), the present study examined if
forced CD46 expression could alter their expression levels.
Previously described stable transfectants overexpressing CD46 in
bladder cancer cells (5637, J82, UM-UC-3, 253J and HT1376) and
melanoma cells (M010119) were used (29,36). Additionally, LNCaP prostate cancer
cells, HCT116 colon cancer cells and A549 lung cancer cells were
transfected with pBlasti-CD46 vectors, using pBlasti as control
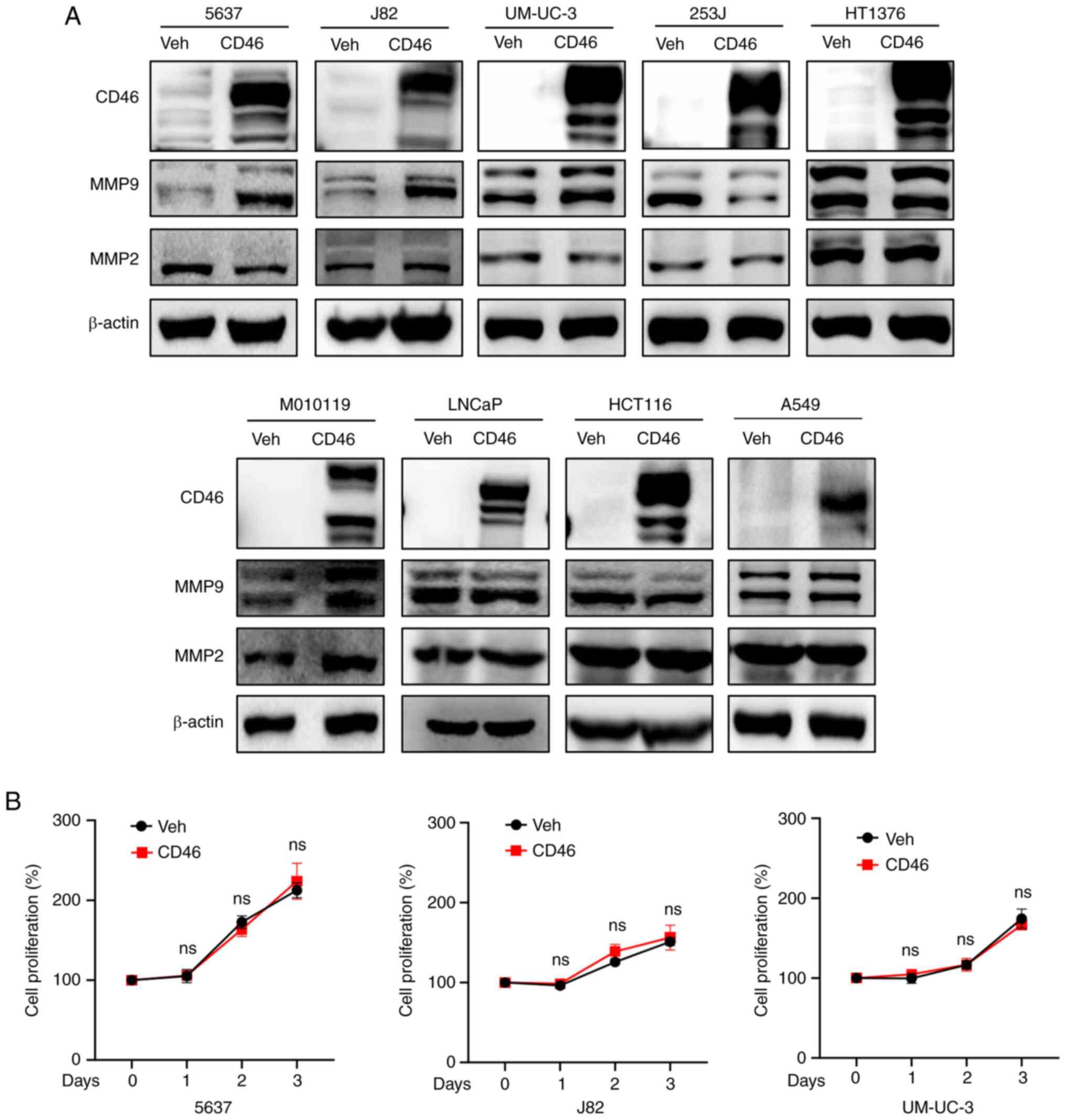
(vehicle group). Western blotting analysis revealed that CD46
overexpression stimulated MMP9 expression in 5637, J82 and UM-UC-3
cells, but not evidently in 253J cells (Fig. 1A). In HT1376 cells, where CD46
previously showed a stimulatory effect on migration (30), MMP9 expression remained unchanged.
By contrast, most non-bladder cancer cells (LNCaP, HCT116 and A549)
did not exhibit enhanced MMP9 upregulation with CD46, except for
M010119 melanoma cells, which showed a marked increase in MMP9
expression. In all tested cells, CD46 stimulation of MMP2
expression was not observed, with the exception of M010119. Given
the consistent upregulation of MMP9 mediated by CD46, the present
study focused on three bladder cancer cell lines that exhibited
increased MMP9 expression (5637, J82 and UM-UC-3). To determine the
effect of CD46 overexpression on cancer cell proliferation, a cell
proliferation assay was conducted over three days. The results
indicated that CD46 overexpression did not significantly affect
cell proliferation in these three bladder cancer cells (Fig. 1B; P>0.05 by Mann-Whitney U
test), thus underscoring the CD46-induced MMP9 overexpression.
These findings suggested that CD46 selectively enhances the
expression of MMP9, a key factor in cancer metastasis, in bladder
cancer cells.
CD46 promotes phosphorylation of p38
MAPK, PI3K/AKT and AP-1
To elucidate the mechanism behind CD46's role in
elevating MMP9 expression, the present study examined several key
signaling pathways that govern MMP9 regulation. There is mounting
evidence indicating the involvement of both MAPK and PI3K/AKT
cascades in MMP9 regulation (27,37,38). The primary constituents of the
MAPK family include ERK, p38 and JNK, while PI3K/AKT is crucial for
cell survival and growth (39).
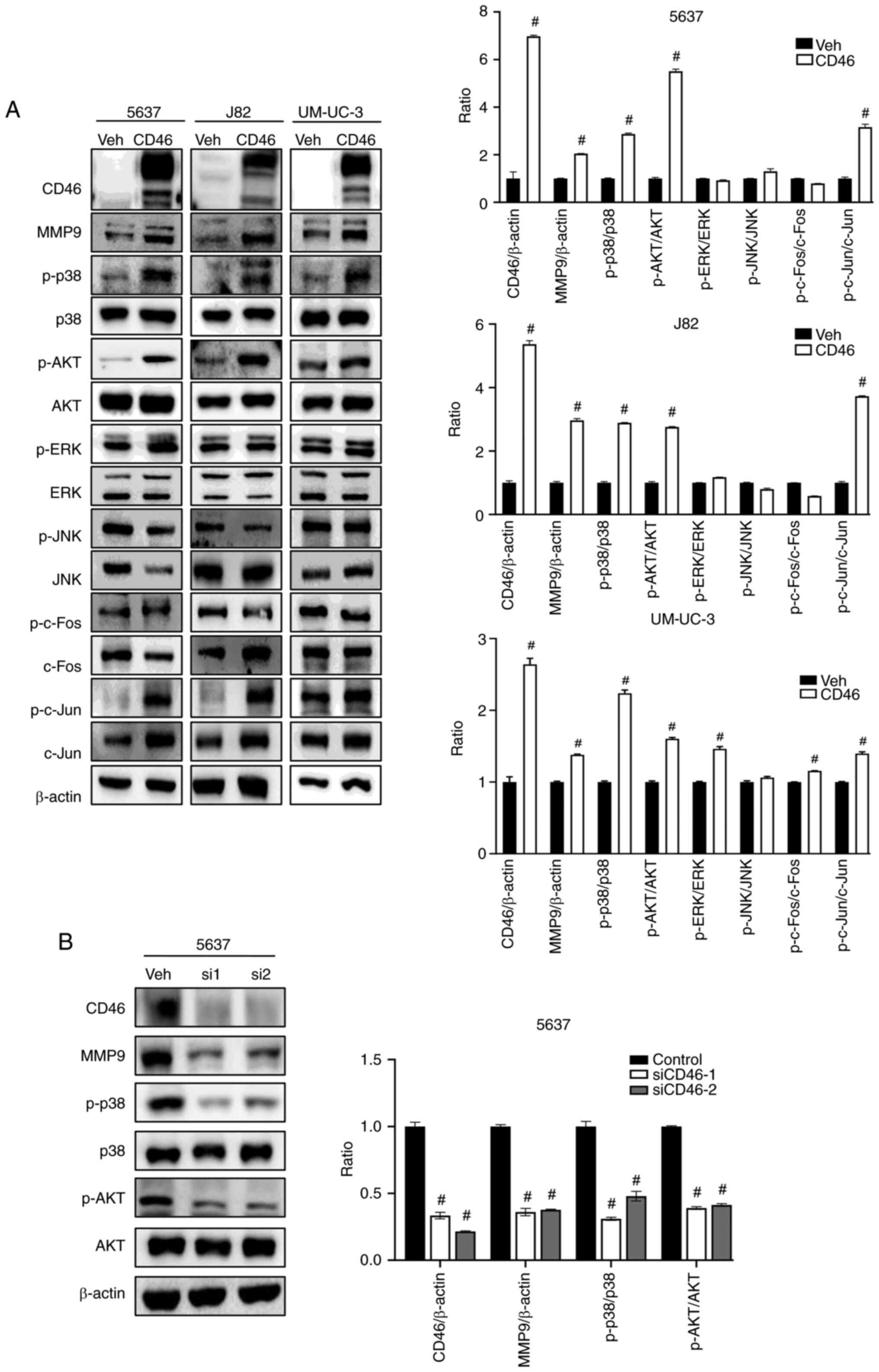
Investigating the potential link between CD46 and MMP9, the present
study assessed alterations in the MAPK and PI3K/AKT pathways in
bladder cancer cells exhibiting CD46-induced MMP9 expression. As
depicted in Fig. 2A, CD46
stimulation not only enhanced MMP9 expression but also increased
phosphorylation levels of p38 MAPK and AKT proteins in all three
cell lines examined. The right panel of Fig. 2A presents a quantitation graph
corresponding to the left panel. No significant changes were
observed in JNK and ERK phosphorylation across the cell lines, with
the exception of p-ERK in UM-UC-3 cells (Student's t-test;
P<0.0001). To minimize potential nonspecific effects of
overexpression via viral vectors, we introduced CD46 siRNA (siCD46)
into 5637 cells through transient transfection. CD46 suppression
effectively reduced MMP9 expression and decreased phosphorylation
of p38 and AKT (Fig. 2B;
Student's t-test; P<0.0001). Jun and Fos family proteins form a
dimeric complex of activator protein-1 (AP-1), influenced by both
MAPK and PI3K/AKT pathways (37,38,40,41). Fig.
2A illustrated that CD46 overexpression also led to increased
phosphorylation of c-Jun, but not c-Fos, in all three cell lines
tested (Student's t-test; P<0.0001). These results indicated
that CD46 enhanced the activation of p38 MAPK and PI3K/AKT in
bladder cancer cells, which appears to be associated with increased
MMP9 expression, suggesting a potential role of CD46-mediated
activation in bladder cancer metastasis through the p38 MAPK and
PI3K/AKT pathways.
CD46 activates p38 MAPK and PI3K/AKT
through phosphorylation of c-Jun to promote expression of MMP9
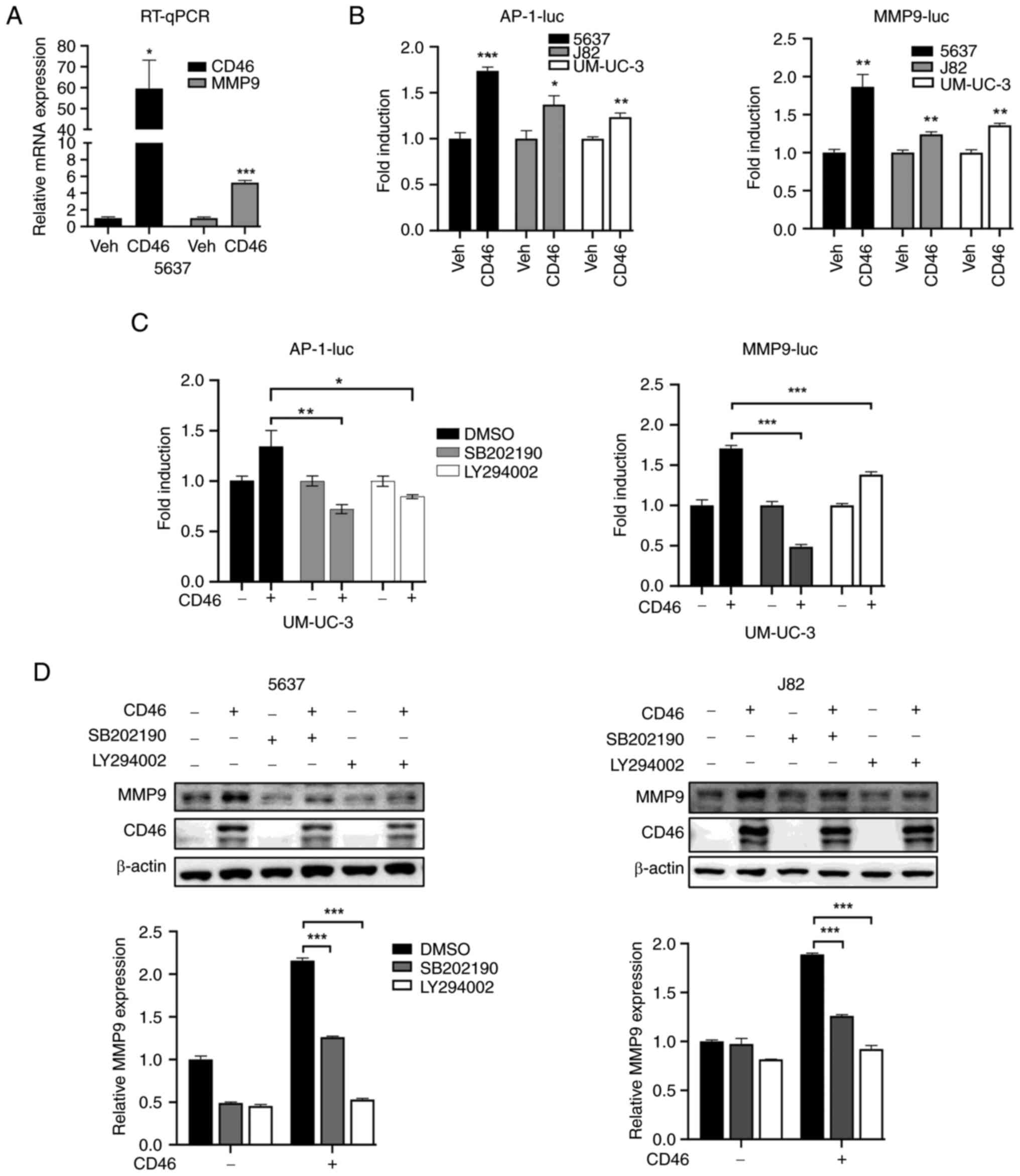
To examine the influence of CD46 on MMP9
transcription, the present study conducted RT-qPCR in 5637 cells.
The results showed that CD46 overexpression significantly increased
MMP9 mRNA levels (Fig. 3A;
P<0.01). Additionally, reporter gene transcription assays with
MMP9 or AP-1 promoters revealed CD46-induced upregulation of both
MMP-9 and AP-1 promoter activities in all three cell types examined
(Fig. 3B; P<0.01).
Subsequently, the present study explored if CD46-mediated MMP9
overexpression involved p38 MAPK and PI3K/AKT pathway activation.
UM-UC-3 cells were treated with specific inhibitors: p38 inhibitor
SB202190 (20 µM) and AKT inhibitor LY294002 (10 µM)
for 24 h. The results, depicted in Fig. 3C, demonstrated that both
inhibitors significantly reduced CD46-induced MMP9 promoter
activity (P<0.0001) and AP-1 activity (P<0.01). Moreover,
these inhibitors also decreased CD46-stimulated MMP-9 protein
overexpression in 5637 and J82 cells, as confirmed by western blot
analysis (Fig. 3D; P<0.0001).
Collectively, these findings suggested that CD46 enhanced MAPK and
AKT signaling, leading to AP-1-mediated transcription and,
consequently, increased MMP9 expression in bladder cancer
cells.
CD46-stimulated MMP9 promotes migratory
and invasive potential of bladder cancer cells in vitro
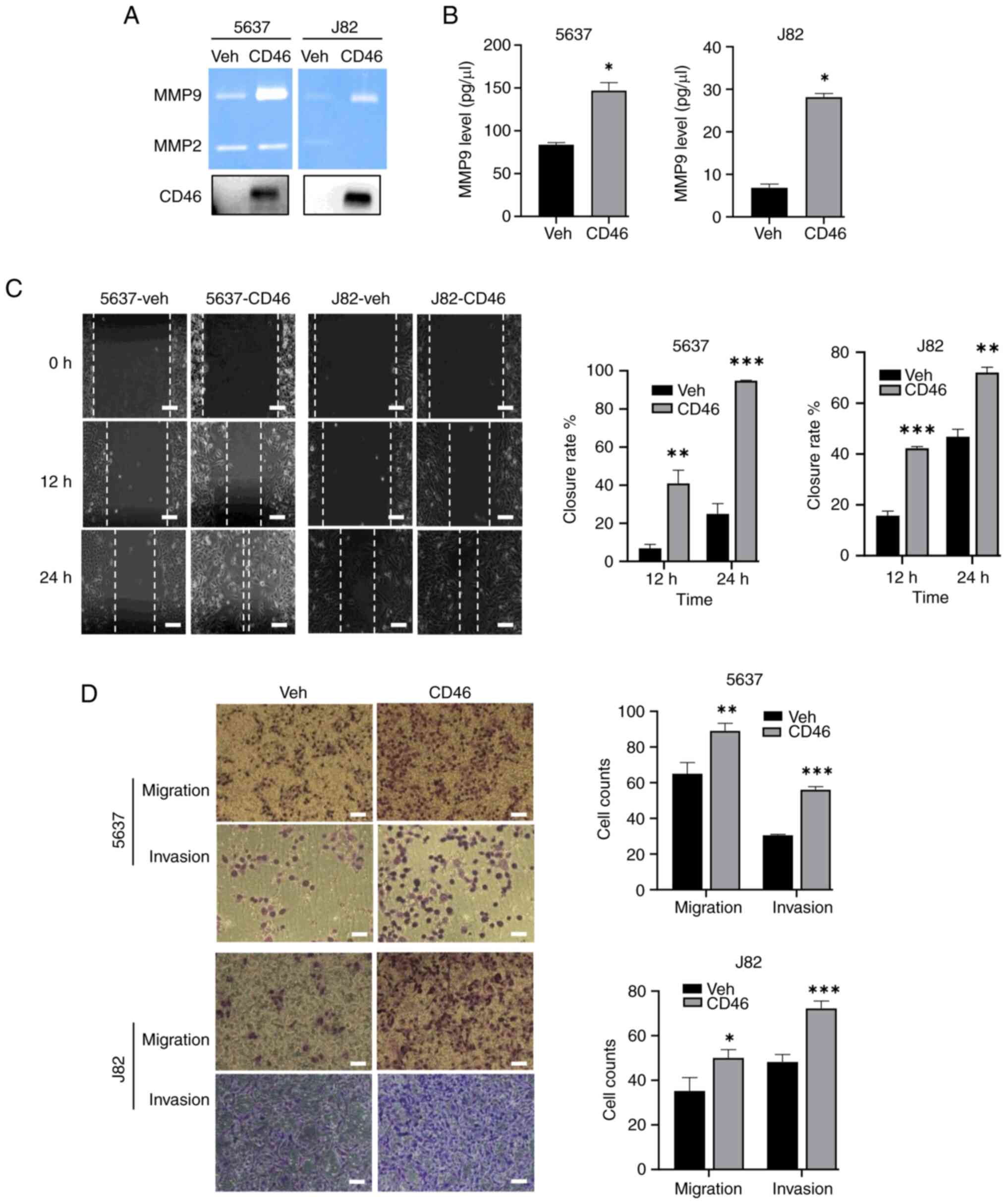
To explore the functional impact of CD46-stimulated
MMP9 expression, the bioactivity of MMP9 secreted by 5637 and J82
cells was analyzed using gelatin zymography. This revealed that
CD46 markedly elevated the gelatinase activity of MMP9 in cells
overexpressing CD46 (Fig. 4A). It
was also observed that CD46 is actively secreted into the cell
culture media in CD46-overexpressed cancer cells (bottom panel of
Fig. 4A). MMP9 is known to
actively cleave membrane CD46 to increase soluble CD46 (42,43). Additionally, MMP9 levels secreted
into the media were quantified by ELISA, finding significantly
elevated MMP9 levels in the media of both cell types (Fig. 4B; P<0.01 for both 5637 and
J82). The influence of CD46 on cell migration and wound healing was
further investigated using wound-healing assays. Here, the width of
the healed gap was measured after a specified incubation period. As
depicted in Fig. 4C, CD46
overexpression significantly accelerated wound repair in 5637 and
J82 cells at each time point (P<0.001). In 253J cells in which
CD46 did not upregulate MMP9 expression, CD46 did not affect the
wound repair capability of cells (Fig. S1). Furthermore, to confirm the
role of CD46 in promoting bladder cancer cell migration and
invasion, Transwell migration and invasion assays were conducted.
Post-incubation, filters were stained and examined under a
microscope at randomly chosen areas to count cells. CD46
overexpression substantially increased both migration and invasion
in 5637 and J82 cells (Fig. 4D;
P<0.01). Correspondingly, suppression of CD46 by siRNA caused
5637 cells to slow wound repair and decreased migration and
invasion (Fig. S2). These
findings suggested that the upregulation of MMP9 expression induced
by CD46 was linked with enhanced bioactivity and secretion of MMP9
in bladder cancer cells.
CD46 promotes bladder cancer metastasis
in vivo
To explore the effect of CD46 overexpression in a
live experimental metastasis model, 5637 cells were injected into
the tail vein of nude mice, with seven mice in each group. Over a
period of six weeks following the injection, the weight of the mice
was monitored. The results indicated that CD46 overexpression did
not significantly alter the weight of the mice groups (Fig. S3A; P>0.05; Mann-Whitney U
test). At the end of the six-week period, the mice were sacrificed
and examined for metastatic tumor nodules in the lung and liver.
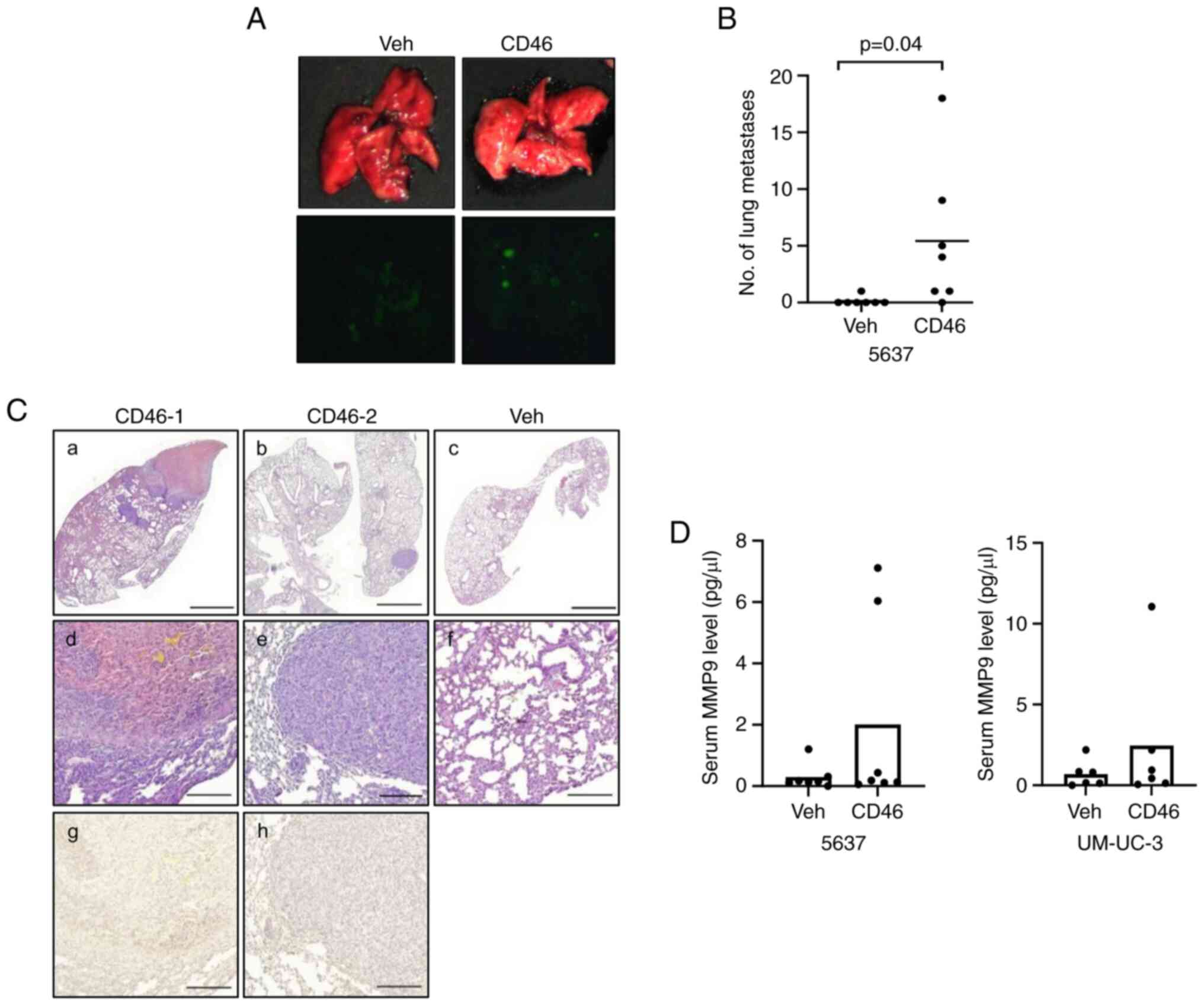
Gross and fluorescent examinations of the lung surfaces were
conducted (Fig. S3B and C). The
lungs of the mice injected with 5637 cells displayed multiple
positive fluorescence foci, as shown in a representative image
(Fig. 5A). The lung tissues were
fixed and embedded in paraffin to identify metastatic tumor nodules
within the lung parenchyma. Serial lung tissue sections, taken
every 1-2 mm up to 5 sections, were stained with H&E and the
tumor nodules counted. The number of lung metastases was
significantly higher in the CD46-5637 cells group, with 6 out of 7
mice showing more than one micrometastatic nodule, in contrast to
almost none in the control group (Fig. 5B; P=0.04). The average tumor area
in lung metastatic nodules of the CD46-5637-injected mice was
11.99±0.19 mm2, compared with >1 mm2 in
control cell-injected mice (data not shown). The entire microscopic
examination of the lung tissues is shown in Fig. S3D. Fig. 5C (a-f) shows a representative lung
tissue with a tumor nodule. Immunohistochemical staining revealed
that the metastasized tumor cells did not significantly overexpress
MMP9 (Fig. 5C, g-h). No tumor
nodules were detected in the liver of any mice. Additionally, serum
analysis during sample collection showed that mice injected with
CD46-overexpressing cells had higher blood levels of MMP9 compared
with those injected with control cells, as determined by ELISA
(Fig. 5D). These results
demonstrated a significant increase in lung metastasis in mice due
to CD46 overexpression. While overexpression of MMP9 in these
metastatic tumor nodules was not definitively positive, the higher
MMP9 levels in the blood of mice injected with CD46-overexpressing
cells suggested a potential association between CD46, MMP9
expression and tumor metastasis.
Discussion
CD46 is a protein with diverse roles in various
biological processes, notably in inhibiting complement activation.
This function is essential for preventing excessive or
inappropriate activation of the complement system, which could lead
to tissue damage and autoimmune disorders. Apart from complement
regulation, CD46 has been implicated in several diseases, including
different types of cancer. Our previous research established that
restoring CD46 expression in bladder cancer cells offers protection
against the inhibitory effects of cetuximab on AKT and ERK
phosphorylation. Furthermore, this restoration shields cells from
complement-dependent cytotoxicity and antibody-dependent cellular
cytotoxicity induced by cetuximab. Consequently, CD46 plays a
protective role in cancer, counteracting direct cytotoxicity
(through peripheral blood mononuclear cells or the complement
system) and indirect cytotoxicity exerted by cetuximab in bladder
cancer cells. We have also shown that CD46 target gene expression
profiling in HT1376 bladder cancer cells using DNA microarray
identified MGP and keratin 13 (KRT13) as CD46 responsive proteins;
both are associated with cell migration and invasion.
Overexpression of CD46 was found to enhance the migratory potential
of bladder cancer cells (30).
The present study aimed to investigate the effect of CD46 on MMP9
expression and bladder cancer progression. The findings indicated
that CD46 facilitates bladder cancer cell migration and invasion
via MMP9 expression. In a mouse model of cancer metastasis,
achieved through tail vein injection, CD46 overexpression led to a
significant increase in bladder cancer lung metastatic lesions.
Notably, while metastatic tumors did not markedly overexpress MMP9,
elevated levels of MMP9 were detected in the serum of mice injected
with CD46 overexpressing 5637 bladder cancer cells. Although a
number of studies are focused on the role of CD46 as a protector
for complement-mediated cytotoxicity and a viral receptor for
cancer therapeutics, there are several studies reporting CD46
expression as a cancer progression and recurrence indicator. In
breast cancers, expression of CD46 was positively associated with
tumor grade and tumor recurrence and also indicative of decreased
progression-free time and overall survival time (9,12).
CD46 was also highly correlated with shorter revival-free time in
ovarian cancers and increased liver metastasis in colorectal
cancers (13,42). Altogether, this suggests that
upregulated MMP9 can be rapidly secreted into the extracellular
matrix and bloodstream in a tumor microenvironment. Most MMPs,
including MMP9, are secreted as inactive proproteins and are
activated upon cleavage by extracellular proteinases (43).
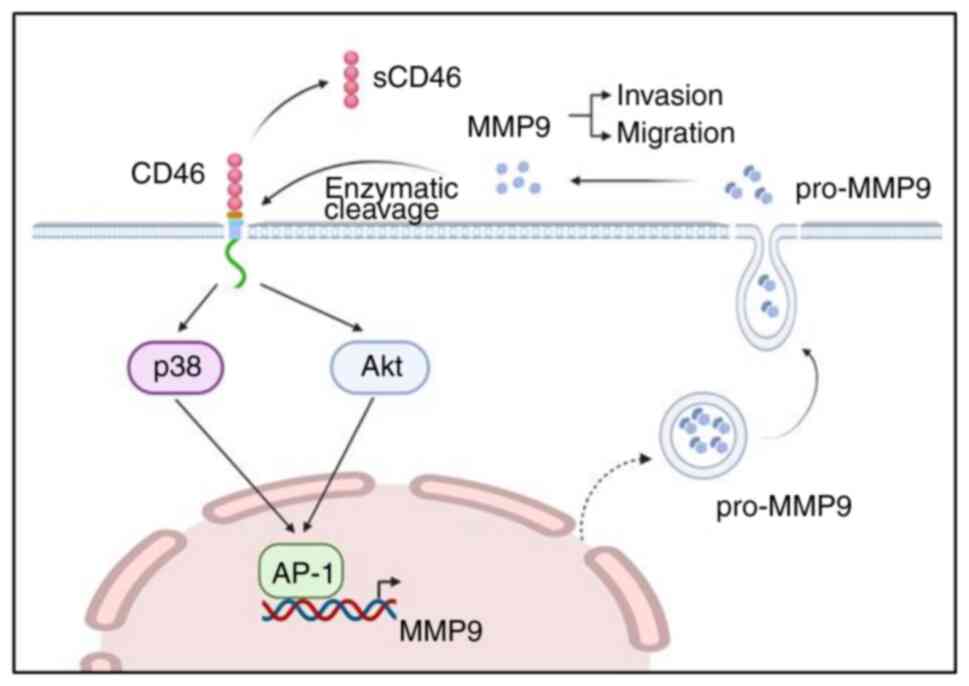
The present study also demonstrated that
CD46-mediated MMP9 activation occurs through p38 MAPK and PI3K/AKT,
which further stimulates AP-1, particularly the activation of c-Jun
protein. Currently, the exact mechanism by which CD46 activates the
p38 MAPK or PI3K/AKT pathways remains to be elucidated.
Nonetheless, substantial evidence links the transmembrane protein
CD46 to cytoskeletal proteins and the MAPK pathway. CD46 has been
long associated with the maintenance of intestinal epithelial
barrier integrity (44). It
directly interacts with Ste20/SPS-1-related kinase (SPAK) and the
cytoplasmic part of E-cadherin (45). SPAK is also involved in the MAPK
pathway. E-cadherin, a transmembrane protein, plays an essential
role in cell-to-cell adhesion. Activation of CD46 on intestinal
epithelial cells leads to a rapid decrease in tight junction
integrity (44). Furthermore,
CD46 activation results in cell proliferation and migration,
aligning with the effects observed following reduced contact
inhibition (46). MMPs play a
role in various physiological processes, including embryonic
development, reproduction, tissue remodeling and carcinogenesis.
MMP9, a key MMP member, primarily degrades collagen type IV in the
basement membrane. MMP9 expression, observed in various cancers, is
thought to aid tumor cell metastasis (47). Activation of EGFR stimulates the
phosphorylation of AKT, MAPK and JNK (37). The MMP9 promoter region
contains multiple transcription factor binding sites, including
those for NF-κB, SP-1, Ets, AP-1 and retinoblastoma (19). Previous research has shown that
MMP9 is upregulated in bladder cancer cells through the activation
of p38 MAP kinase pathways (48)
and the AKT pathway (49).
The present study focused on activating AP-1 to
understand the CD46-mediated activation of p38 MAPK and/or AKT and
its role in regulating MMP9. AP-1 induction, mostly driven by the
JNK and p38MAPK pathways in response to various stimuli, is
well-documented (50). Upon
activation, JNKs move to the nucleus, phosphorylating c-Jun, which
enhances its transcriptional activation capabilities through
homodimerization or heterodimerization with c-Fos (51). The findings of the present study
indicated that the upregulation of MMP9, stimulated by CD46, is
primarily facilitated through the activation of c-Jun
phosphorylation, rather than c-Fos. It is plausible that other
transcription factors, such as NF-κB or SP-1, are also influenced
by CD46 in regulating MMP9. The presence of AP-1 sites ~70 bp
upstream from the transcriptional start site of the MMP9
gene is considered to play a crucial role in the transcriptional
activation of MMP promoters (52). Moreover, the transactivation of
the MMP9 promoter necessitates the specific interaction of AP-1
with other cis-acting elements and certain transcription factors
binding to these sequences (53).
Further research is required to delineate the intricate
transcriptional regulation of the MMP9 promoter and enhancer
regions.
The major limitation of the present study is that it
did not survey MMP9 and CD46 expressions in bladder cancer
patients, which is a potent tool for studying CD46 progression and
development. In the future, it is planned to assess the correlation
of MMP9 and CD46 expressions with stages of bladder cancer
patients.
In conclusion, the results suggested that CD46
enhanced bladder cancer cell migration, invasion and metastasis.
The present study also underscored the potential role of MMP9 in
the metastatic process via the MAPK and AKT pathways activation
(Fig. 6). CD46 could be integral
in regulating these biological processes and may serve as a viable
target for therapeutic intervention in bladder cancer
metastasis.
Supplementary Data
Availability of data and materials
The data generated in the present study are included
in the figures of this article.
Authors' contributions
TNT performed the experiments, data analysis and
preparation of the original draft. HDT, VTN and SYK contributed to
the investigation, methodology, review and editing of the final
manuscript. CM assisted in the animal studies. ECH aided the
clinical background of study and final version of the manuscript.
CJ contributed to the conceptualization, supervision, funding
acquisition, review, revision and approval of the manuscript. TNT
and CJ confirm the authenticity of all the raw data. All authors
have read and approved the final version of this manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Use and Care Committees at Chonnam National University Medical
School (approval no. H2022-70).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
AKT
|
protein kinase B
|
|
AP-1
|
activator protein 1
|
|
EGFR
|
epidermal growth factor receptor
|
|
ELISA
|
enzyme-linked immunoabsorbent
assays
|
|
ERK
|
extracellular signal-regulated
kinases
|
|
Ets
|
E-twenty six
|
|
FBS
|
fetal bovine serum
|
|
H&E
|
hematoxylin and eosin
|
|
JNK
|
c-Jun N-terminal kinases
|
|
KRT13
|
keratin 13
|
|
MAPK
|
mitogen-activated protein kinase
|
|
MGP
|
matrix Gla protein
|
|
MMLV
|
moloney murine leukemia virus
|
|
MMP2
|
matrix metalloproteinase 2
|
|
MMP9
|
matrix metalloproteinase 9
|
|
MMPs
|
matrix metalloproteinases
|
|
NF-κB
|
nuclear factor kappa B
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
SP-1
|
specificity protein 1
|
|
SPAK
|
Ste20/SPS-1-related kinase
|
Acknowledgements
Not applicable.
Funding
The present study was supported by a National Research
Foundation of Korea grant funded by the Korean government (Ministry
of Science and ICT; grant no. 2022R1A2C1003206).
References
|
1
|
Yamamoto H, Fara AF, Dasgupta P and Kemper
C: CD46: The 'multitasker' of complement proteins. Int J Biochem
Cell Biol. 45:2808–2820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Andrews PW, Knowles BB, Parkar M, Pym B,
Stanley K and Goodfellow PN: A human cell-surface antigen defined
by a monoclonal antibody and controlled by a gene on human
chromosome 1. Ann Hum Genet. 49:31–39. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hourcade D, Garcia AD, Post TW,
Taillon-Miller P, Holers VM, Wagner LM, Bora NS and Atkinson JP:
Analysis of the human regulators of complement activation (RCA)
gene cluster with yeast artificial chromosomes (YACs). Genomics.
12:289–300. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sherbenou DW, Aftab BT, Su Y, Behrens CR,
Wiita A, Logan AC, Acosta-Alvear D, Hann BC, Walter P, Shuman MA,
et al: Antibody-drug conjugate targeting CD46 eliminates multiple
myeloma cells. J Clin Invest. 126:4640–4653. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu YQ, Gao YD, Yang J and Guo W: A defect
of CD4+CD25+ regulatory T cells in inducing interleukin-10
production from CD4+ T cells under CD46 costimulation in asthma
patients. J Asthma. 47:367–373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Le Buanec H, Gougeon ML, Mathian A, Lebon
P, Dupont JM, Peltre G, Hemon P, Schmid M, Bizzini B, Künding T, et
al: IFN-α and CD46 stimulation are associated with active lupus and
skew natural T regulatory cell differentiation to type 1 regulatory
T (Tr1) cells. Proc Natl Acad Sci USA. 108:18995–19000. 2011.
View Article : Google Scholar
|
|
7
|
Fishelson Z, Donin N, Zell S, Schultz S
and Kirschfink M: Obstacles to cancer immunotherapy: Expression of
membrane complement regulatory proteins (mCRPs) in tumors. Mol
Immunol. 40:109–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blok VT, Daha MR, Tijsma OM, Weissglas MG,
van den Broek LJ and Gorter A: A possible role of CD46 for the
protection in vivo of human renal tumor cells from
complement-mediated damage. Lab Invest. 80:335–344. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Madjd Z, Durrant LG, Pinder SE, Ellis IO,
Ronan J, Lewis S, Rushmere NK and Spendlove I: Do poor-prognosis
breast tumours express membrane cofactor proteins (CD46)? Cancer
Immunol Immunother. 54:149–156. 2005. View Article : Google Scholar
|
|
10
|
Kinugasa N, Higashi T, Nouso K,
Nakatsukasa H, Kobayashi Y, Ishizaki M, Toshikuni N, Yoshida K,
Uematsu S and Tsuji T: Expression of membrane cofactor protein
(MCP, CD46) in human liver diseases. Br J Cancer. 80:1820–1825.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seya T, Hara T, Iwata K, Kuriyama S,
Hasegawa T, Nagase Y, Miyagawa S, Matsumoto M, Hatanaka M, Atkinson
JP, et al: Purification and functional properties of soluble forms
of membrane cofactor protein (CD46) of complement: Identification
of forms increased in cancer patients' sera. Int Immunol.
7:727–736. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maciejczyk A, Szelachowska J,
Szynglarewicz B, Szulc R, Szulc A, Wysocka T, Jagoda E, Lage H and
Surowiak P: CD46 Expression is an unfavorable prognostic factor in
breast cancer cases. Appl Immunohistochem Mol Morphol. 19:540–546.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Surowiak P, Materna V, Maciejczyk A,
Kaplenko I, Spaczynski M, Dietel M, Lage H and Zabel M: CD46
expression is indicative of shorter revival-free survival for
ovarian cancer patients. Anticancer Res. 26:4943–4948. 2006.
|
|
14
|
Lu Z, Zhang C, Cui J, Song Q, Wang L, Kang
J, Li P, Hu X, Song H, Yang J and Sun Y: Bioinformatic analysis of
the membrane cofactor protein CD46 and microRNA expression in
hepatocellular carcinoma. Oncol Rep. 31:557–564. 2014. View Article : Google Scholar :
|
|
15
|
Zeng J, Xu H, Huang C, Sun Y, Xiao H, Yu
G, Zhou H, Zhang Y, Yao W, Xiao W, et al: CD46 splice variant
enhances translation of specific mRNAs linked to an aggressive
tumor cell phenotype in bladder cancer. Mol Ther Nucleic Acids.
24:140–153. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Do M, Thanh HD, To PK, Kim MS, Moon C and
Jung C: CD46 protects the bladder cancer cells from
cetuximab-mediated cytotoxicity. Sci Rep. 12:224202022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Merdad A, Karim S, Schulten HJ, Dallol A,
Buhmeida A, Al-Thubaity F, Gari MA, Chaudhary AG, Abuzenadah AM and
Al-Qahtani MH: Expression of matrix metalloproteinases (MMPs) in
primary human breast cancer: MMP-9 as a potential biomarker for
cancer invasion and metastasis. Anticancer Res. 34:1355–1366.
2014.PubMed/NCBI
|
|
18
|
Hara I, Miyake H, Hara S, Arakawa S and
Kamidono S: Significance of matrix metalloproteinases and tissue
inhibitors of metalloproteinase expression in the recurrence of
superficial transitional cell carcinoma of the bladder. J Urol.
165:1769–1772. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sato H and Seiki M: Regulatory mechanism
of 92 kDa type IV collagenase gene expression which is associated
with invasiveness of tumor cells. Oncogene. 8:395–405.
1993.PubMed/NCBI
|
|
20
|
Folkman J: Angiogenesis and c-Jun. J Natl
Cancer Inst. 96:6442004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ibrahim SAE, Abudu A, Johnson E, Aftab N,
Conrad S and Fluck M: The role of AP-1 in self-sufficient
proliferation and migration of cancer cells and its potential
impact on an autocrine/paracrine loop. Oncotarget. 9:34259–34278.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu G, Cheng Z, Huang Y, Zheng W, Yang S,
Lin C and Ye J: MyD88 mediates colorectal cancer cell
proliferation, migration and invasion via NF-κB/AP-1 signaling
pathway. Int J Mol Med. 45:131–140. 2020.
|
|
23
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng CY, Hsieh HL, Hsiao LD and Yang CM:
PI3-K/Akt/JNK/NF-κB is essential for MMP-9 expression and outgrowth
in human limbal epithelial cells on intact amniotic membrane. Stem
Cell Res. 9:9–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jian H, Zhao Y, Liu B and Lu S: SEMA4b
inhibits MMP9 to prevent metastasis of non-small cell lung cancer.
Tumour Biol. 35:11051–11056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin CC, Kuo CT, Cheng CY, Wu CY, Lee CW,
Hsieh HL, Lee IT and Yang CM: IL-1 beta promotes A549 cell
migration via MAPKs/AP-1- and NF-kappaB-dependent matrix
metalloproteinase-9 expression. Cell. Signal. 21:1652–1662. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ellerbroek SM, Halbleib JM, Benavidez M,
Warmka JK, Wattenberg EV, Stack MS and Hudson LG:
Phosphatidylinositol 3-kinase activity in epidermal growth
factor-stimulated matrix metalloproteinase-9 production and cell
surface association. Cancer Res. 61:1855–1861. 2001.PubMed/NCBI
|
|
28
|
Funakoshi-Tago M, Tago K, Sonoda Y,
Tominaga S and Kasahara T: TRAF6 and C-SRC induce synergistic AP-1
activation via PI3-kinase-AKT-JNK pathway. Eur J Biochem.
270:1257–1268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Do MH, To PK, Cho YS, Kwon SY, Hwang EC,
Choi C, Cho SH, Lee SJ, Hemmi S and Jung C: Targeting CD46 enhances
anti-tumoral activity of adenovirus type 5 for bladder cancer. Int
J Mol Sci. 19:26942018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nguyen TT, Thanh HD, Do MH and Jung C:
Complement regulatory protein CD46 manifests a unique role in
promoting the migration of bladder cancer cells. Chonnam Med J.
59:160–166. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Davies B, Waxman J, Wasan H, Abel P,
Williams G, Krausz T, Neal D, Thomas D, Hanby A and Balkwill F:
Levels of matrix metalloproteases in bladder cancer correlate with
tumor grade and invasion. Cancer Res. 53:5365–5369. 1993.PubMed/NCBI
|
|
32
|
Gerhards S, Jung K, Koenig F, Daniltchenko
D, Hauptmann S, Schnorr D and Loening SA: Excretion of matrix
metalloproteinases 2 and 9 in urine is associated with a high stage
and grade of bladder carcinoma. Urology. 57:675–679. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jung C, Kim RS, Zhang HJ, Lee SJ and Jeng
MH: HOXB13 induces growth suppression of prostate cancer cells as a
repressor of hormone-activated androgen receptor signaling. Cancer
Res. 64:9185–9192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Papathoma AS, Petraki C, Grigorakis A,
Papakonstantinou H, Karavana V, Stefanakis S, Sotsiou F and Pintzas
A: Prognostic significance of matrix metalloproteinases 2 and 9 in
bladder cancer. Anticancer Res. 20:2009–2013. 2000.PubMed/NCBI
|
|
36
|
Cho YS, Do MH, Kwon SY, Moon C, Kim K, Lee
K, Lee SJ, Hemmi S, Joo YE, Kim MS and Jung C: Efficacy of
CD46-targeting chimeric Ad5/35 adenoviral gene therapy for
colorectal cancers. Oncotarget. 7:38210–38223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Braicu C, Buse M, Busuioc C, Drula R,
Gulei D, Raduly L, Rusu A, Irimie A, Atanasov AG, Slaby O, et al: A
comprehensive review on MAPK: A promising therapeutic target in
cancer. Cancers (Basel). 11:16182019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Datta SR, Brunet A and Greenberg ME:
Cellular survival: A play in three akts. Genes Dev. 13:2905–2927.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park SL, Won SY, Song JH, Kambe T, Nagao
M, Kim WJ and Moon SK: EPO gene expression promotes proliferation,
migration and invasion via the p38MAPK/AP-1/MMP-9 pathway by
p21WAF1 expression in vascular smooth muscle cells. Cell Signal.
27:470–478. 2015. View Article : Google Scholar
|
|
41
|
Shaulian E and Karin M: AP-1 in cell
proliferation and survival. Oncogene. 20:2390–2400. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meng Q, Xu J, Wang J, Zhang X, Yang H, Sun
H, Wu S, Aschner M, Li X, Zhang L, et al: Investigation of the
enhanced antitumour potency of CD46-specific chimeric antigen
receptor-T cells in human colorectal cancer liver metastases after
combination with nanotherapeutics. Nano Today. 52:1019852023.
View Article : Google Scholar
|
|
43
|
Van den Steen PE, Dubois B, Nelissen I,
Rudd PM, Dwek RA and Opdenakker G: Biochemistry and molecular
biology of gelatinase B or matrix metalloproteinase-9 (MMP-9). Crit
Rev Biochem Mol Biol. 37:375–536. 2002. View Article : Google Scholar
|
|
44
|
Cardone J, Al Shouli S and Kemper C: A
novel role for CD46 in wound repair. Front Immunol. 2:282011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yan Y and Merlin D: Ste20-related
proline/alanine-rich kinase: A novel regulator of intestinal
inflammation. World J Gastroenterol. 14:6115–6121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guillot C and Lecuit T: Mechanics of
epithelial tissue homeostasis and morphogenesis. Science.
340:1185–1189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Laronha H and Caldeira J: Structure and
function of human matrix metalloproteinases. Cells. 9:10762020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee SJ, Park SS, Lee US, Kim WJ and Moon
SK: Signaling pathway for TNF-alpha-induced MMP-9 expression:
Mediation through p38 MAP kinase, and inhibition by anti-cancer
molecule magnolol in human urinary bladder cancer 5637 cells. Int
Immunopharmacol. 8:1821–1826. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen YT, Yang CC, Shao PL, Huang CR and
Yip HK: Melatonin-mediated downregulation of ZNF746 suppresses
bladder tumorigenesis mainly through inhibiting the AKT-MMP-9
signaling pathway. J Pineal Res. 66:e125362019. View Article : Google Scholar
|
|
50
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pulverer BJ, Hughes K, Franklin CC, Kraft
AS, Leevers SJ and Woodgett JR: Co-purification of
mitogen-activated protein kinases with phorbol ester-induced c-Jun
kinase activity in U937 leukaemic cells. Oncogene. 8:407–415.
1993.PubMed/NCBI
|
|
52
|
Benbow U and Brinckerhoff CE: The AP-1
site and MMP gene regulation: What is all the fuss about? Matrix
Biol. 15:519–526. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gum R, Lengyel E, Juarez J, Chen JH, Sato
H, Seiki M and Boyd D: Stimulation of 92-kDa gelatinase B promoter
activity by ras is mitogen-activated protein kinase kinase
1-independent and requires multiple transcriptin factor binding
sites including closely spaced PEA3/ets and AP-1 sequences. J Biol
Chem. 271:10672–10680. 1996. View Article : Google Scholar : PubMed/NCBI
|