Gastric cancer (GC) stands as one of the most
prevalent types of cancer worldwide. As outlined in the 2020
GLOBOCAN report, GC ranks fifth globally in incidence and fourth in
mortality (1). Notably, East
Asia, Eastern Europe and South America exhibit particularly
elevated rates of both incidence and mortality associated with GC
(2). As a highly heterogeneous
disease, personalized treatment approaches are essential for GC
(3). In the early stages of GC
development, due to limitations in detection technologies, the most
common classification systems were based on morphology; these
included the World Health Organization (WHO) classification system
(tubular, papillary, mucinous and poorly cohesive) (4), the Lauren classification system
(diffuse, intestinal, and mixed) (5), and the Nakamura classification
system (differentiated and undifferentiated) (6). Early-stage and advanced-stage GCs
are characterized by extensive morphological differences, leading
to an increasing number of classification systems (2). However, relying solely on
histological classification is insufficient for effectively
stratifying patients for individualized treatment and improving
clinical outcomes (7).
With technological advancements, omics techniques
such as next-generation sequencing (NGS), metabolomics and
proteomics have facilitated significant breakthroughs in the
medical field, providing direct evidence at the microscopic level
to understand the heterogeneity of GC. In 2011, Shah et al
(14) differentiated GC subtypes
with epidemiological and histological differences using gene
expression data, demonstrating favorable consistency and enhancing
understanding of tumor biology. In the same year, Tan et al
(15) identified two major
intrinsic subgroups of GC (G-INT and G-DIF) through gene expression
analysis of 37 GC cell lines, laying a solid foundation for
subsequent clinical genomic classification research. The use of
various omics approaches contributes to a deeper understanding of
the detailed biological characteristics of GC, providing a
foundation for personalized treatment and drug development and
improving the effectiveness of cancer treatment and patient
survival rates. The present review summarizes the latest research
on GC omics classification, elucidating its potential clinical
applications in diagnosis, prognosis and treatment response
prediction, and drug design.
Research on the cluster-based classification of GC
has gained significant momentum in the past decade. To date, 27
articles related to the cluster classification of GC patient
samples have been published, 14 of which provided further
validation of the studies (Table
I). A milestone study was reported in 2014 by The Cancer Genome
Atlas (TCGA) research network, which included the most
comprehensive molecular characteristics of gastric adenocarcinoma
at that time (16). Numerous
subsequent cluster-based subtyping studies have been conducted
based on the TCGA database (17-28). Apart from the TCGA research, a
major gene expression profile study conducted by the Asian Cancer
Research Group (ACRG) in 2015 revealed fresh expression subtypes of
GC. This classification scheme is intricately linked to diverse
molecular alterations, disease progression and prognosis patterns
(29). An increasing number of
countries and regions, such as Singapore (3,30),
South Korea (29,31-33) and China (34-38), have started similar research.
These studies commonly employ mainstream classification methods
such as consensus clustering (3,18-20,33,34), K-value clustering (24,39) and non-negative matrix
factorization (21,26,36,37,40), among others, to conduct
cluster-based subtyping of GC patient samples. This finding not
only correlates with different genomic alteration patterns but also
with GC recurrence patterns, prognosis and drug sensitivity.
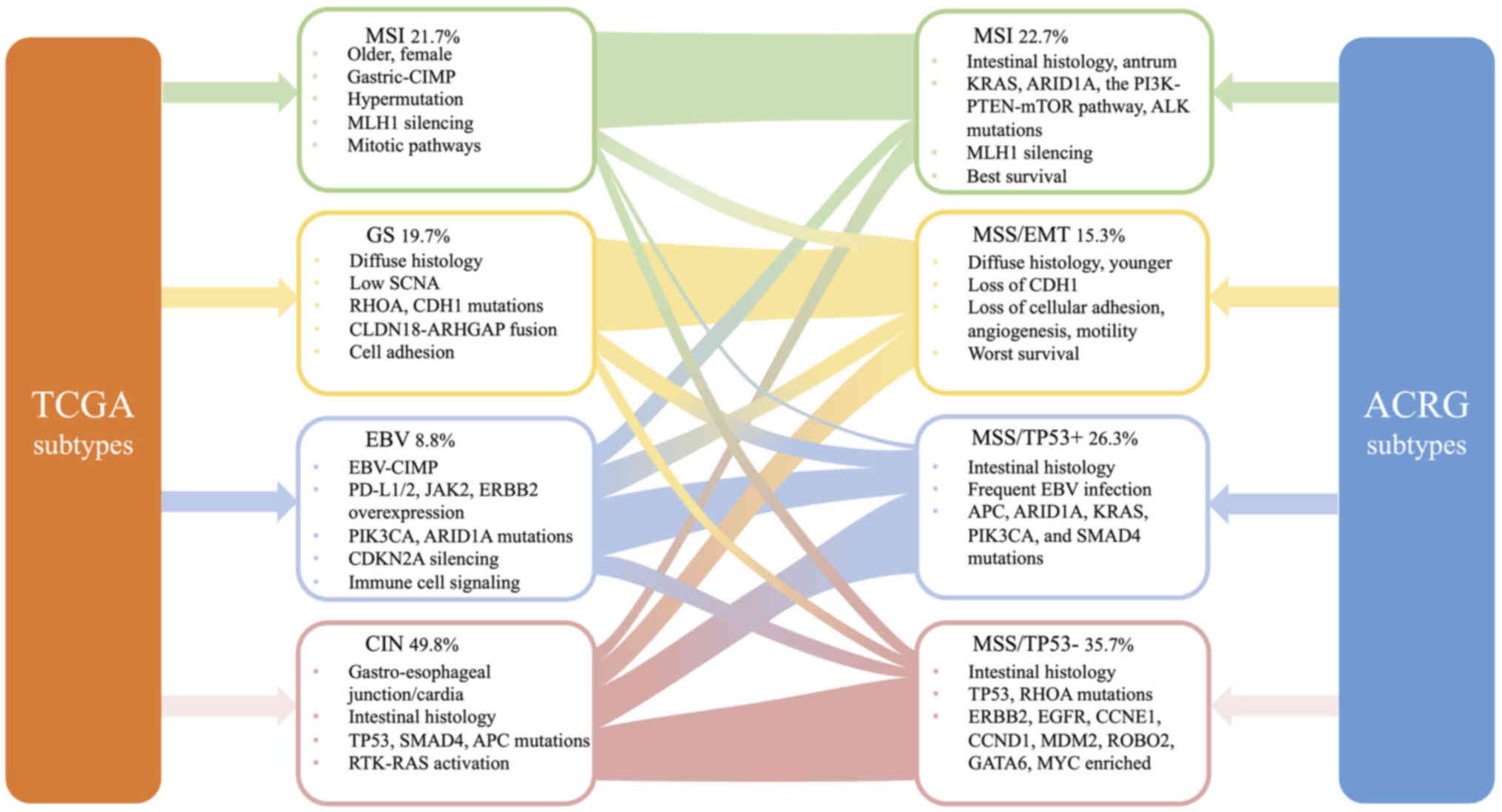
The TCGA database categorizes gastric adenocarcinoma
into four subtypes based on the following features: Epstein-Barr
virus (EBV; 8.8%), microsatellite instability (MSI; 21.7%),
genomically stable (GS; 19.7%) and chromosomal instability (CIN;
49.8%) (16). EBV-positive tumors
exhibit recurrent mutations in phosphatidylinositol 3-kinase
(PIK3CA) and AT-rich interactive domain-containing protein 1A
(ARID1A), as well as extreme DNA hypermethylation and
overexpression of programmed death-ligand 1/2 (PD-L1/2) (41). Amplifications of Janus-activated
kinase 2 (JAK2) and Erb-B2 receptor tyrosine kinase 2 (ERBB2)
(42) were also observed. MSI
tumors, characterized by mismatch repair deficiency, were more
common in elderly (median age 72 years) and female (56%) patients
than in male (early) patients. MSI-high GC also had a high rate of
PD-L1 expression. The GS subtype had low somatic copy number
alterations (SCNAs), was rich in diffuse histological variations,
and had mutations in Ras homolog family member A (RHOA) and
E-cadherin (CDH1). CIN tumors were more common in the
gastroesophageal junction/cardia and exhibited noticeable
non-diploidy and focal amplification of receptor tyrosine
kinase-Ras (RTK/Ras). There was also a high frequency of TP53
mutations in CIN tumors (Fig.
1).
The rapid development of genomics and
transcriptomics has been propelled by the widespread adoption of
NGS technologies. This has significantly enhanced the understanding
of GC biology, providing novel insights into the complex
interactions among tumor cells, normal cells and their
microenvironment (55). Genomic
subtypes and transcriptomic subtypes are inherently interconnected.
The research on genomic/transcriptomic clustering subtypes
encompasses a total of 16 studies (Table II), with four studies (3,18,19,32) characterizing the subtypes
descriptively. These studies cover various aspects but primarily
focus on EMT (3,18,21,22,32), metabolic characteristics (3,19,20,26,27,56), and immune features (18,20-24,27,40,56). The following is a summary of these
aspects.
In total, five articles discussed the differences in
the EMT pathways among subtypes, with two articles providing
characteristic names for them (3,32).
In 2013, Lei et al (3)
classified gastric adenocarcinoma patients into three main
subtypes, with the mesenchymal subtype exhibiting high activity in
the cancer stem cell pathway. Oh et al (32) suggested that the mesenchymal
phenotype (MP) subtype within the intrinsic subtypes exhibits
clinical and molecular features akin to those of the Genomic
Diffuse (G-DIF) tumors classified by Tan et al (15). The ACRG research also reached a
similar conclusion. Furthermore, the MSS/EMT subtype (ACRG
classification) (29) was
considered a subset of the MP subtype, and both were suggested to
be linked with an unfavorable prognosis (18,21,22). Li et al (18) reported that the stroma-enriched
(StE) subtype shares similar characteristics with Oh et al's
MP subtype (32), including high
genomic stability, prominent EMT features, resistance to standard
chemotherapy and poor prognosis. Ning et al (22) suggested that high hypoxia status
is positively correlated with high m6A methylation in tumors of the
MSS/EMT subtype (ACRG classification) (29). Previous research has indicated
that EMT significantly enhances the movement and spreading of
cancer cells, playing a central role in tumor progression (57-59). Additionally, Oh et al
(32) reported high activation of
the insulin-like growth factor 1 (IGF1)/IGF1 receptor (IGF1R)
pathway in patients in the MP group, and this pathway is considered
a crucial therapeutic target for numerous cancers (60-62). Several studies have identified
immune suppression features in groups with enrichment of EMT
characteristics (18,21,22). Previous studies reported that the
activation of EMT and TGF-related signaling pathways led to
weakened transport of T cells to tumors, reducing the cytotoxicity
of tumor cells (63-65).
Disruption of cellular energy metabolism stands as
one of the core hallmarks of cancer cells (66). Lei et al (3) reported a significant correlation
between the metabolic subtype classified in their study and a
pathway associated with spasmolytic polypeptide-expressing
metaplasia (5), considered an
intermediate step in gastric adenocarcinoma development (67). Bornschein et al (56) classified gastroesophageal junction
(GEJ) adenocarcinoma into three groups based on differentially
expressed genes and revealed enrichment of fatty acid metabolism
pathways in the stroma-enhanced and poor prognosis subgroups (Group
1). Previous research has suggested that adipose tissue can create
a proinflammatory microenvironment in obese patients, contributing
to stromal activation associated with more aggressive tumor
behavior and an unfavorable prognosis (68-70). The presence of Barrett's esophagus
was strongly correlated with a Group 1 designation. Group 2 was
characterized by metabolic pathways, which are typically active in
the intestinal and hepatobiliary systems. A total of four studies
(19,20,26,27) conducted analyses on the TCGA
dataset, classifying subtypes based on metabolism-related genes.
Zhu et al (19) classified
the TCGA samples into four subtypes based on the express of
glycolysis-related genes and cholesterol-related genes and
identified abnormal amplification of MYC and TP53 in the
cholesterol subtype. Upregulated MYC expression is linked to a more
invasive phenotype in GC cell lines. MYC amplification represents a
common mechanism of MYC mutation in cancer (71). MYC amplification has been reported
in plasma samples from patients with GC (72). The glycolysis subgroup had
increased expression of cytotoxic T lymphocyte-associated antigen-4
(CTLA-4) and PDCD1. Li et al (20) reported that the C2 subtype, which
has a high TP53 mutation rate, was enriched in bile acid
metabolism. Tao et al (27) reported that cluster 2, which
presented activation of numerous metabolic pathways, also exhibited
activation of nucleotide processing and repair-related pathways and
a higher tumor mutation burden (TMB). This was associated with an
improved prognosis for patients with GC (26,27).
Tumor tissues and cells undergo varying degrees of
metabolic dysregulation and immune dysfunction (73). Li et al (18) classified patients with GC not only
into the StE subgroup but also into the immune-deprived (ImD) and
immune-enriched (ImE) subtypes. The ImD and ImE subtypes share
several common features with Oh et al's epithelial phenotype
(EP) (32), such as high genomic
instability, elevated DNA damage repair activity and sensitivity to
standard chemotherapy. However, Li et al's classification
(18) could capture the
intratumoral heterogeneity within the EP, including significantly
different tumor immune microenvironments, somatic cell mutations
and SCNA patterns, responses to chemotherapy and immunotherapy and
clinical outcomes. Both ImD and ImE subtypes had high TMB, but
their levels of immune infiltration vary significantly. The primary
reason might be that ImD exhibits a high frequency of SCNAs that
suppress antitumor immune responses (74). Similarly, Wu et al
(21) used the TCGA dataset to
classify major classes based on immune-related genes: C1 and C2.
The C1 subtype exhibited immune quiescence, EMT and angiogenesis
pathway activity. By contrast, the C2 subtype primarily exhibited
enrichment of MYC targets, oxidative phosphorylation and the E2F
target pathway, along with a higher TMB. TMB could serve as an
indicator for predicting immunotherapy response, and patients with
high TMB had improved clinical outcomes (75-77). Several studies have reported that
high TMB is associated with a favorable prognosis in patients with
GC (24,26,27,78,79). Interestingly, cluster 1,
classified by Tao et al (27), had a poor prognosis despite the
enrichment of immune cells, conflicting with previous research
results (18,21,23,40,56). Tao et al (27) suggested that this difference might
be due to the innate immune and memory cell status of immune cells
in cluster 1, as well as the presence of various immune cells with
immunosuppressive effects.
Additionally, numerous classification studies have
focused simultaneously on the correlation between their own
molecular classification and the traditional Lauren classification
(3,17,18,21,29,32,36), as well as the classical TCGA
classification (17,18,20,24,27,29,36). It was found that the diffuse
subtype according to Lauren classification was mostly associated
with EMT features (18,21,29), genomic stability (17,18,36) and late-stage GC (18,21). The intestinal subtype is often
found in patients with MSI (18,29), TP53 mutations (3,17,18,36), or high TMB (18,21,36).
Proteomic technologies, predominantly reliant on
liquid chromatography coupled to tandem mass spectrometry, are
gaining traction in cancer research. They are utilized to identify
and quantify proteins and post-translational modifications that
undergo modulation in cancer. These technologies also help
elucidate their associations with copy number variations,
epigenetic silencing and alterations in microRNA (miRNA) expression
(80). In recent years, several
proteomic studies on various cancers have rediscovered numerous of
the same subtypes identified through gene expression and proposed
new disease classifications (81-85).
Four studies classified GC molecularly based on
proteomic subtyping, all of which were conducted on Chinese cohorts
(Table III) (34). Based on gene products
significantly differentially expressed between tumor and normal
tissues, Ge et al (34)
classified 84 patients with diffuse-type GC (DGC) from the Beijing
Cancer Hospital into PX1-3 subtypes. PX1 and PX2 subtypes exhibited
dysregulation throughout the cell cycle, and the PX2 subtype also
displayed promotion of EMT. The PX3 subtype was enriched in
immune-related proteins and drug-target proteins such as TMEM173
(STING), CD276, CD40, FCGR1A, ARG1, SIRPA, NT5E and IDO1 (86-88). DNA mutations in the PI3K-AKT,
CXCR4, and focal adhesion pathways were enriched in the PX3
subtype. According to the prognostic analysis of advanced GC,
patients with PX1 GC had the best prognosis, while patients with
PX3 GC had the worst prognosis, which might be related to the
abnormal enrichment of immune-regulating proteins. Subsequently,
this institution conducted a proteomics analysis of phosphorylated
proteins (35), which classified
83 patients with DGC into the Ph1-3 subtypes. The Ph1 subtype had
the best prognosis and was enriched in TILs and stromal cells.
Elevated levels of intratumoral and stromal TILs have been
confirmed to correlate with improved prognosis in various cancers
(89-92). The Ph1 subtype also showed
upregulation of rRNA processing and RNA polymerase II promoter
activity. The Ph2 subtype mainly presented upregulation of DNA
metabolism and repair processes with loss of essential functions of
the stomach, including gastric acid secretion. The Ph3 subtype
presented upregulation of chromosome segregation with loss of
cell-to-cell interactions and communication. Additionally, compared
with the previous proteomic subtypes, the Ph1 subgroup included
some patients assigned to the PX2 and PX3 groups, but these
patients had improved overall survival (OS) than those in the
original PX2 and PX3 groups, indicating that subtyping based on
phosphorylated proteomic data may be more accurate (35).
The incidence of adenocarcinoma of the
esophagogastric junction (AEG) has been increasing annually
(98,99), and the prognosis has been poor
(100). Li et al
(37) classified 103 AEG tumor
samples based on proteomic clustering. The S-I subtype was more
abundant in Siewert type II patients, while the S-III subtype was
more common in Siewert type III patients. The S-III subgroup had
the best prognosis, followed by the S-II subgroup, and the S-I
subgroup had the worst prognosis. The most common leptin receptor
(LEPR) and significant co-occurrence of the CSMD1 and ANKRD36C
genes were found in the S-I subtype. LEPR is a receptor for leptin,
a protein hormone mainly secreted by adipose tissue (101). LEPR genotypes have been found to
be associated with the risk of various cancers, including
esophageal squamous cell carcinoma (102), breast cancer (103) and GC (104). RYR2 and TTN mutations found in
the S-III subtype were mutually exclusive, and the FAT4 and PRKDC
genes exhibited a significant co-mutation relationship. The RYR2
gene plays a crucial role in steroid metabolism and could reduce
the risk of breast cancer (105). In the S-II subtype, specific
cooccurring mutations in the MUC4 and CPED1 genes were identified,
with NCKAP1 mutations being the most common. In addition, in the
S-I and S-III subtypes, there was a significant correlation between
CDK1/2 and their phosphorylated substrates. Moreover, CSNK2A1 was
found to be significantly correlated with the phosphorylation of
Occludin S408 in the S-II subtype, and CSNK2A1 may be a target for
the S-II subtype. Moreover, CSNK2A1 has been shown to participate
in tumorigenesis by phosphorylating various proteins, including
SIRTs (106,107). Li et al (37) found and validated that the
characteristic protein of the S-II subtype (FBXO44) could promote
tumor progression and metastasis in vitro and in
vivo. Recent research has indicated that FBXO44 serves as a
crucial inhibitor of DNA replication-coupled repeat elements in
human cancer (108).
Metabolomics involves examining metabolites present
in biological fluids, cells and tissues, and is widely utilized for
the identification of biomarkers (111). Wang et al (39) conducted spatial metabolomic
studies on 362 patients with GC and identified three tumor (T1-3)-
and three stroma (S1-3)-specific subtypes with distinct tissue
metabolism patterns. The tumor-specific T1 subtype exhibited
positive correlations with CD3, CD8, FOXP3, MIB1 and HER2
expression, while displaying negative correlations with MMR status.
The T1 subtype exhibited 45 significantly upregulated metabolic
pathways, including 13 associated with carbohydrate metabolism and
10 associated with amino acid metabolism. Notably, nucleotide
metabolism, as well as ascorbic acid and citric acid metabolism,
was upregulated only in T1. In a recent study on psoriasis, the
upregulation of ascorbic acid and citric acid metabolism was shown
to enhance the immunosuppression of Tregs (112). Conversely, the tumor-specific
subtype T2 exhibited downregulation HER2, MIB1, CD3 and FOXP3 but a
high rate of MMR status. Additionally, 17 notably upregulated
metabolic pathways were identified, comprising 7 associated with
carbohydrate metabolism and 4 associated with amino acid
metabolism. Additionally, this subtype is associated with an
unfavorable prognosis. The tumor-specific subtype T3 was shown to
be associated with biotin metabolism and cytoplasmic DNA sensing
pathways. The cGAS-STING pathway was identified as a vital
DNA-sensing mechanism in innate immunity and viral defense. The
cGAS-STING signaling pathway also functions in promoting tumor
metastasis, and chronic activation of this pathway can
paradoxically induce immunosuppressive tumor microenvironments
(113). Subtype similarities
were observed between T1 and S3, T2 and S2, and T3 and S1.
Integrated single-cell genomics, epigenomics,
transcriptomics, proteomics, and/or metabolomics analyses are
reshaping our understanding of cellular biology in health and
disease (114). Molecular
classification based on multi-omics data has been conducted for
various cancers (115-117). Hu et al (28) subtyped GC samples into CS1 and CS2
subtypes based on data for mRNAs, long non-coding RNAs, miRNAs and
DNA methylation CpG sites associated with prognosis. The main
pathways enriched in the CS1 subtype, which has a favorable
prognosis, were involved in the activation of extracellular-related
biological processes, including EMT, cell adhesion tissue, response
to growth factors and cell-matrix adhesion pathways. SMOC2, which
promotes EMT, was significantly upregulated in the CS1 subtype
(118). The CS2 subtype, which
has an unfavorable prognosis, was primarily enriched in pathways
related to the cell cycle, including the G1/S-specific
transcription, G2M checkpoint, E2F targets, DNA replication and
repair biological processes. Patients in the CS2 subgroup exhibited
activated programmed death-1 (PD-1) signaling, which is associated
with most EBV and MSI subtypes found in CS2 patients.
GC is one of the most prevalent malignant tumors of
the digestive system, exhibiting notable heterogeneity and complex
molecular features (122).
Historically, targeted therapies focused on single genes have shown
promise in prolonging survival and improving quality of life
compared with treatment based on pathological or morphological
classifications. Claudin 18.2, due to its unique biological
behaviour-being almost exclusively expressed in the gastric mucosa
and appearing on the tumour cell surface during malignant
transformation-has emerged as a promising target for GC therapy
(123). In several international
multicentre phase II/III clinical trials, zolbetuximab (an
anti-claudin 18.2 monoclonal antibody) demonstrated the ability to
improve OS and progression-free survival in previously untreated
patients with GC with high levels of claudin 18.2 when used in
combination with chemotherapy (124-127). A recent systematic review has
detailed the biological behaviour of claudin 18.2 and the clinical
efficacy of its targeted therapies (123). However, these approaches have
limitations, including limited therapeutic efficacy and severe side
effects. With the advent of the era of precision medicine in GC,
personalized treatments involving multigene clustering are
gradually demonstrating their advantages, as they effectively
address the suboptimal outcomes associated with the high
heterogeneity of GC (128).
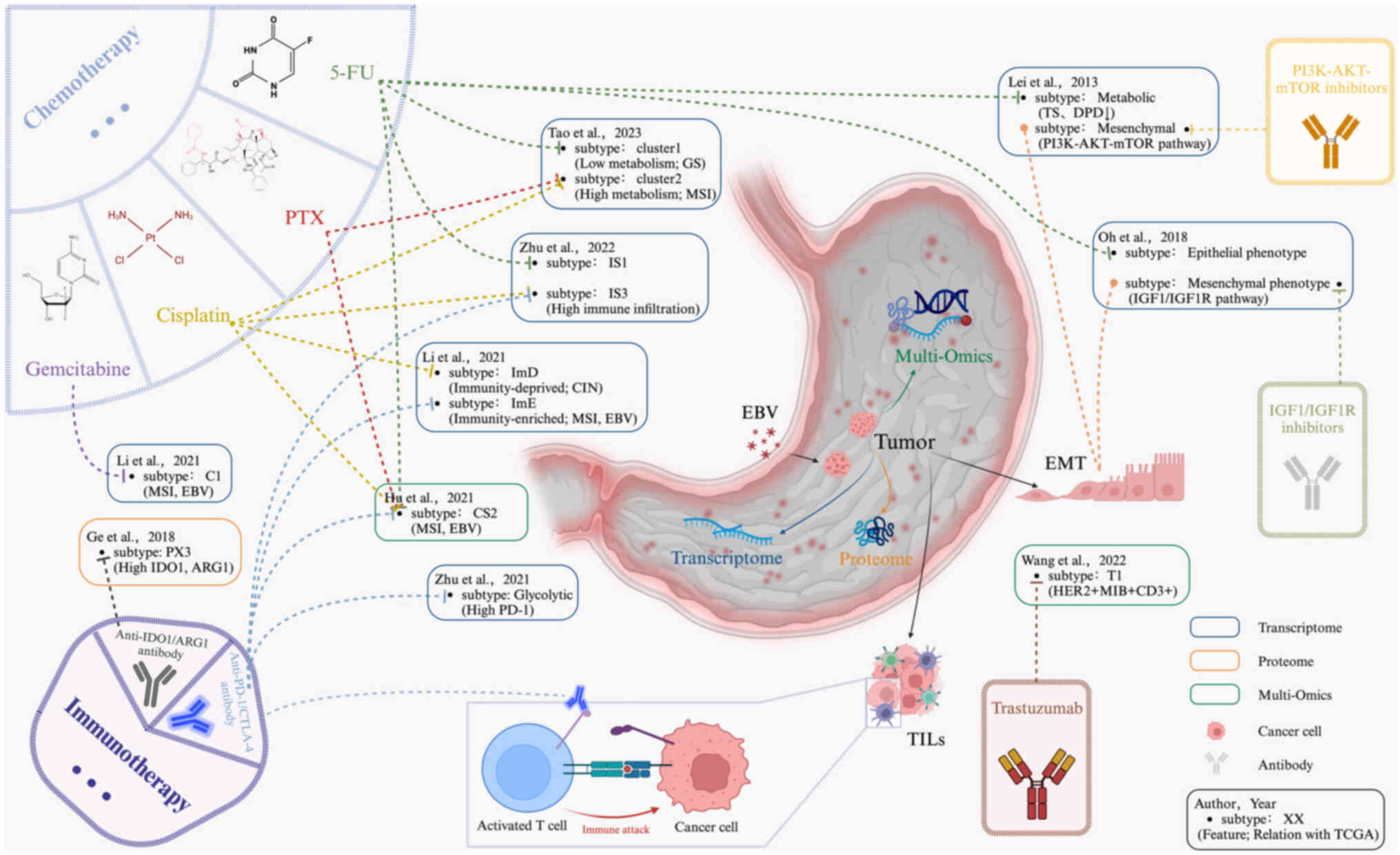
Among 27 studies on GC molecular subtyping, 10 described suitable
treatment options based on their own molecular subtyping results
(Fig. 2).
Adjuvant chemotherapy, primarily based on
fluoropyrimidine, includes single-agent treatment with S1 (a
combination of tegafur, gimeracil and oteracil) or combination
therapy with capecitabine and oxaliplatin or S1 and docetaxel
(129,130). Adjuvant chemotherapy has shown
favorable survival benefits in East Asian countries (2). Lei et al (3) reported that the metabolic subtype of
GC was more sensitive to 5-fluorouracil (5-FU) than were the other
two subtypes, possibly due to the significantly reduced expression
of dihydropyrimidine dehydrogenase and thymidylate synthase (TS)
(131,132). Additionally, sensitivity to 5-FU
in a specific molecular subtype was also identified in three other
studies (23,27,28). A prospective study suggested that
MSI (dMMR) patients with GC may not benefit from 5-FU adjuvant
chemotherapy (133). Li et
al (18) investigated the
response rates to four different chemotherapies (cisplatin,
capecitabine, oxaliplatin and doxorubicin) among GC subtypes. They
found that almost all drugs followed the same sensitivity pattern:
ImE > StE > ImD. Cisplatin had the highest efficacy in
treating ImD (ImD: 73%; StE: 46%; ImE: 67%). This may be due to the
high prevalence of homologous recombination defects in ImD, which
might increase the sensitivity to cisplatin chemotherapy (134,135), a phenomenon extensively studied
in BRCA1/2-negative triple-negative breast cancer (136-138). Cisplatin was also effective in
Tao et al's (27) cluster
1 and Zhu et al's (23)
subtype IS3. A previous study revealed that patients with CIN GC
with a high level of fractional allelic loss were more likely to
benefit from neoadjuvant chemotherapy based on cisplatin (139). Shi et al (38) discovered that patients with
elevated SMARCC1 activity in IGC and reduced NFKB1 activity in DGC
may derive benefits from chemotherapy. Notably, NFKB1 has been
linked to chemotherapy resistance in breast cancer (140,141). SMARCC1 is the core subunit of
the switching or sucrose non-fermentable (SWI/SNF) complex
(142). Multiple pieces of
evidence suggest that SWI/SNF complex alterations can serve as
biomarkers for the efficacy of cancer immunotherapy (143,144). Abnormal expression of the
SWI/SNF complex was identified as an independent adverse prognostic
factor in patients with GS GC (based on TCGA classification).
Detecting abnormalities in the SWI/SNF complex might help identify
patients likely to benefit from novel treatment approaches
(145). A clinical study
revealed that SMARCC1-positive patients benefit from gemcitabine
treatment after recurrence, as SMARCC1 can regulate cancer cell
resistance to gemcitabine (146). Regarding gemcitabine, Li et
al (20) reported that the C1
subtype was more sensitive than the C2 subtype was.
Additionally, in patients with HER2 (also known as
ERBB2) overexpression or amplification, trastuzumab should be added
to first-line cytotoxic chemotherapy (39,128). Immunotherapy has proven to be an
effective treatment for various cancers (150). PD-1 and CTLA-4, belonging to the
immunoglobulin-related receptor family, play diverse roles in
regulating T-cell immune responses (151). Subtypes enriched in immune
cells, such as Li et al's (18) ImE subtype, Zhu et al's
(19) glycolysis subtype, and Zhu
et al's (23) IS3 subtype,
are more likely to benefit from immunotherapy than other subtypes.
The MSI and EBV subtypes (based on TCGA classification) have been
largely confirmed to be sensitive to immunotherapy (18,28). MSI subtype GC, identified in
several studies, might not benefit from chemotherapy, possibly
because of elevated TS levels (152,153). Patients with MSI subtype GC
exhibit increased reactivity to immunotherapy due to elevated PD-L1
expression (153-156). Patients with MSI subtype GC with
high mutation rates in the PAM pathway often exhibit lower TIL
numbers and primary resistance to immune checkpoint inhibitors,
suggesting that immunotherapy could be used as a stratification
approach in patients with advanced MSI-H GC (48,157). Similarly, EBV-positive GC might
also respond to immune checkpoint therapy (158), but its efficacy awaits
verification. High CTLA-4 levels and lower TILs might have impacted
the effectiveness of anti-PD-1 monoclonal antibodies in patients
with EBV GC (156,159). Ge et al (34) reported that the PX3 subtype of
DGC, which exhibits high expression of IDO1 and ARG1, might benefit
from IDO1 and ARG1 inhibitors. Various IDO1 and ARG1 inhibitors
have been evaluated in clinical trials (160,161).
GC, characterized by strong heterogeneity, is a
complex malignancy of the digestive system with unique
epidemiological, histological and molecular differences (13). Cluster-based subtyping has great
value in GC research. Unlike traditional molecular classification
methods, cluster classification methods allow for the subdivision
of GC into subgroups with distinct molecular characteristics, tumor
biological features and clinical presentations. This approach forms
the basis for personalized treatment, optimized clinical trial
designs and improved patient management, driving medical
advancements.
Although cluster-based subtyping of GC provides
crucial information for clinical diagnosis and treatment, there are
still limitations: i) Most research findings are derived from
bioinformatic analyses in basic research, and unlike conventional
pathological classifications or single-gene classifications, they
have not been widely used in clinical settings. This may be the
focus of the next step in GC genomic subtype research; ii) the
current genomic subtyping system for GC, although diverse, has not
been validated in large cohorts (n>1,000), and simultaneous
comparisons in large cohorts to explore the most adaptive genomic
subtype are needed for precise application; iii) with the
development of GC, molecular changes are dynamic, therefore
identifying stable molecules to establish a consistent
classification system is crucial; and iv) GC often exhibits both
inter-tumor and intratumor heterogeneity, thus further exploration
of the role of spatial genomics, single-cell technologies and other
new techniques in GC cluster classification is needed.
In conclusion, GC should not be treated as a single
disease. Cluster-based molecular classifications could aid in GC
research, allowing us to delve deeper into the biological
characteristics of different subtypes of GC, laying the foundation
for personalized treatment and precision medicine. Additionally,
utilizing new information from clustering subtypes will help in the
design of more accurate and targeted clinical trials, enhancing the
effectiveness and credibility of related research. This approach is
expected to accelerate the discovery of new treatment methods,
providing more effective treatment options for patients with GC and
propelling medical research toward more rational and individualized
treatment.
Not applicable.
YM and ZJ wrote and revised the manuscript. ZJ, LY
and ZL conceived and designed the review. YM, ZJ, YZ, LP and RX
performed the literature review. LP and LY revised the manuscript.
ZL and LY acquired funding. Data authentication is not applicable.
All authors have read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the National Key R&D
Program of China (grant no. 2021YFA0910100), the National Natural
Science Foundation of China (grant nos. 82374544, 82204828,
92259302 and 82074245), the Healthy Zhejiang One Million People
Cohort (grant no. K-20230085), the Program of Zhejiang Provincial
TCM Sci-tech Plan (grant nos. GZY-ZJ-KJ-230003 and
GZY-ZJ-KJ-23048), the Medical Science and Technology Project of
Zhejiang (grant nos. 2022KY114, 2018KY305 and 2021KY582) and the
Natural Science Foundation of Zhejiang (grant nos. R24H290003 and
HDMY22H160008).
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lei Z, Tan IB, Das K, Deng N, Zouridis H,
Pattison S, Chua C, Feng Z, Guan YK, Ooi CH, et al: Identification
of molecular subtypes of gastric cancer with different responses to
PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology.
145:554–565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board: The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar :
|
|
5
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and So-called intestinal-type
carcinoma. An attempt at a Histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakamura K, Sugano H and Takagi K:
Carcinoma of the stomach in incipient phase: Its histogenesis and
histological appearances. Gan. 59:251–258. 1968.PubMed/NCBI
|
|
7
|
Korfer J, Lordick F and Hacker UT:
Molecular targets for gastric cancer treatment and future
perspectives from a clinical and translational point of view.
Cancers (Basel). 13:52162021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis GD, Figari I, Fendly B, Wong WL,
Carter P, Gorman C and Shepard HM: Differential responses of human
tumor cell lines to anti-p185HER2 monoclonal antibodies. Cancer
Immunol Immunother. 37:255–263. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park JB, Rhim JS, Park SC, Kimm SW and
Kraus MH: Amplification, overexpression, and rearrangement of the
erbB-2 protooncogene in primary human stomach carcinomas. Cancer
Res. 49:6605–6609. 1989.PubMed/NCBI
|
|
10
|
Maeda K, Chung YS, Ogawa Y, Ko T, Ogawa M,
Onoda N, Kato Y, Arimoto Y, Nitta A and Sowa M: Expression of
vascular endothelial cell growth factor as a predictor of
recurrence in gastric carcinoma. Gan To Kagaku Ryoho. 22:699–701.
1995.In Japanese. PubMed/NCBI
|
|
11
|
Maeda K, Chung YS, Ogawa Y, Takatsuka S,
Kang SM, Ogawa M, Sawada T and Sowa M: Prognostic value of vascular
endothelial growth factor expression in gastric carcinoma. Cancer.
77:858–863. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guan WL, He Y and Xu RH: Gastric cancer
treatment: Recent progress and future perspectives. J Hematol
Oncol. 16:572023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alsina M, Arrazubi V, Diez M and Tabernero
J: Current developments in gastric cancer: From molecular profiling
to treatment strategy. Nat Rev Gastroenterol Hepatol. 20:155–170.
2023. View Article : Google Scholar
|
|
14
|
Shah MA, Khanin R, Tang L, Janjigian YY,
Klimstra DS, Gerdes H and Kelsen DP: Molecular classification of
gastric cancer: A new paradigm. Clin Cancer Res. 17:2693–2701.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan IB, Ivanova T, Lim KH, Ong CW, Deng N,
Lee J, Tan SH, Wu J, Lee MH, Ooi CH, et al: Intrinsic subtypes of
gastric cancer, based on gene expression pattern, predict survival
and respond differently to chemotherapy. Gastroenterology.
141:476–485. e1–e11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Cheng WY, Shi H, Huang S, Chen H,
Liu D, Xu W, Yu J and Wang J: Classifying gastric cancer using
FLORA reveals clinically relevant molecular subtypes and highlights
LINC01614 as a biomarker for patient prognosis. Oncogene.
40:2898–2909. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L and Wang X: Identification of gastric
cancer subtypes based on pathway clustering. NPJ Precis Oncol.
5:462021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu Z, Qin J, Dong C, Yang J, Yang M, Tian
J and Zhong X: Identification of four gastric cancer subtypes based
on genetic analysis of cholesterogenic and glycolytic pathways.
Bioengineered. 12:4780–4793. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li L and Ma J: Molecular characterization
of metabolic subtypes of gastric cancer based on metabolism-related
lncRNA. Sci Rep. 11:214912021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu D, Feng M, Shen H, Shen X, Hu J, Liu J,
Yang Y, Li Y, Yang M, Wang W, et al: Prediction of two molecular
subtypes of gastric cancer based on immune signature. Front Genet.
12:7934942021. View Article : Google Scholar
|
|
22
|
Ning ZK, Hu CG, Liu J, Tian HK, Yu ZL,
Zhou HN, Li H and Zong Z: The hypoxic landscape stratifies gastric
cancer into 3 subtypes with distinct M6a methylation and tumor
microenvironment infiltration characteristics. Front Immunol.
13:8600412022. View Article : Google Scholar
|
|
23
|
Zhu Y, Zhao Y, Cao Z, Chen Z and Pan W:
Identification of three immune subtypes characterized by distinct
tumor immune microenvironment and therapeutic response in stomach
adenocarcinoma. Gene. 818:1461772022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He F, Ding H, Zhou Y, Wang Y, Xie J, Yang
S and Zhu Y: Depiction of Aging-Based molecular phenotypes with
diverse clinical prognosis and immunological features in gastric
cancer. Front Med (Lausanne). 8:7927402021. View Article : Google Scholar
|
|
25
|
Li B, Zhang F, Niu Q, Liu J, Yu Y, Wang P,
Zhang S, Zhang H and Wang Z: A molecular classification of gastric
cancer associated with distinct clinical outcomes and validated by
an XGBoost-based prediction model. Mol Ther Nucleic Acids.
31:224–240. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen T, Zhao L, Chen J, Jin G, Huang Q,
Zhu M, Dai R, Yuan Z, Chen J, Tang M, et al: Identification of
three metabolic subtypes in gastric cancer and the construction of
a metabolic pathway-based risk model that predicts the overall
survival of GC patients. Front Genet. 14:10948382023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tao G, Wen X, Wang X and Zhou Q: Bulk and
single-cell transcriptome profiling reveal the metabolic
heterogeneity in gastric cancer. Sci Rep. 13:87872023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu X, Wang Z, Wang Q, Chen K, Han Q, Bai
S, Du J and Chen W: Molecular classification reveals the diverse
genetic and prognostic features of gastric cancer: A multi-omics
consensus ensemble clustering. Biomed Pharmacother. 144:1122222021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Loh M, Liem N, Vaithilingam A, Lim PL,
Sapari NS, Elahi E, Mok ZY, Cheng CL, Yan B, Pang B, et al: DNA
methylation subgroups and the CpG island methylator phenotype in
gastric cancer: A comprehensive profiling approach. BMC
Gastroenterol. 14:552014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Bai W and Zhang X: Identifying
heterogeneous subtypes of gastric cancer and subtype-specific
subpaths of microRNA-target pathways. Mol Med Rep. 17:3583–3590.
2018.
|
|
32
|
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE,
Park KC, Lee SH, Park JL, Park YY, Lee HS, et al: Clinical and
genomic landscape of gastric cancer with a mesenchymal phenotype.
Nat Commun. 9:17772018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mun DG, Bhin J, Kim S, Kim H, Jung JH,
Jung Y, Jang YE, Park JM, Kim H, Jung Y, et al: Proteogenomic
characterization of human Early-Onset gastric cancer. Cancer Cell.
35:111–124.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ge S, Xia X, Ding C, Zhen B, Zhou Q, Feng
J, Yuan J, Chen R, Li Y, Ge Z, et al: A proteomic landscape of
diffuse-type gastric cancer. Nat Commun. 9:10122018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tong M, Yu C, Shi J, Huang W, Ge S, Liu M,
Song L, Zhan D, Xia X, Liu W, et al: Phosphoproteomics enables
molecular subtyping and nomination of kinase candidates for
individual patients of Diffuse-Type gastric cancer. iScience.
22:44–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Ding Y, Chen Y, Jiang J, Chen Y,
Lu J, Kong M, Mo F, Huang Y, Zhao W, et al: A novel genomic
classification system of gastric cancer via integrating
multidimensional genomic characteristics. Gastric Cancer.
24:1227–1241. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li S, Yuan L, Xu ZY, Xu JL, Chen GP, Guan
X, Pan GZ, Hu C, Dong J, Du YA, et al: Integrative proteomic
characterization of adenocarcinoma of esophagogastric junction. Nat
Commun. 14:7782023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi W, Wang Y, Xu C, Li Y, Ge S, Bai B,
Zhang K, Wang Y, Zheng N, Wang J, et al: Multilevel proteomic
analyses reveal molecular diversity between diffuse-type and
intestinal-type gastric cancer. Nat Commun. 14:8352023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Kunzke T, Prade VM, Shen J, Buck
A, Feuchtinger A, Haffner I, Luber B, Liu DHW, Langer R, et al:
Spatial metabolomics identifies distinct Tumor-Specific subtypes in
gastric cancer patients. Clin Cancer Res. 28:2865–2877. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cao J, Gong J, Li X, Hu Z, Xu Y, Shi H, Li
D, Liu G, Jie Y, Hu B and Chong Y: Unsupervised hierarchical
clustering identifies immune gene subtypes in gastric cancer. Front
Pharmacol. 12:6924542021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Ge J, Wang Y, Xiong F, Guo J,
Jiang X, Zhang L, Deng X, Gong Z, Zhang S, et al: EBV miRNAs BART11
and BART17-3p promote immune escape through the enhancer-mediated
transcription of PD-L1. Nat Commun. 13:8662022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang J, Liu Z, Zeng B, Hu G and Gan R:
Epstein-Barr virus-associated gastric cancer: A distinct subtype.
Cancer Lett. 495:191–199. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hirata T, Yamamoto H, Taniguchi H,
Horiuchi S, Oki M, Adachi Y, Imai K and Shinomura Y:
Characterization of the immune escape phenotype of human gastric
cancers with and without high-frequency microsatellite instability.
J Pathol. 211:516–523. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Corso G, Velho S, Paredes J, Pedrazzani C,
Martins D, Milanezi F, Pascale V, Vindigni C, Pinheiro H, Leite M,
et al: Oncogenic mutations in gastric cancer with microsatellite
instability. Eur J Cancer. 47:443–451. 2011. View Article : Google Scholar
|
|
45
|
Mulkidjan RS, Saitova ES, Preobrazhenskaya
EV, Asadulaeva KA, Bubnov MG, Otradnova EA, Terina DM, Shulga SS,
Martynenko DE, Semina MV, et al: ALK, ROS1, RET and NTRK1-3 Gene
fusions in colorectal and Non-colorectal microsatellite-unstable
cancers. Int J Mol Sci. 24:136102023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chida K, Kawazoe A, Kawazu M, Suzuki T,
Nakamura Y, Nakatsura T, Kuwata T, Ueno T, Kuboki Y, Kotani D, et
al: A low tumor mutational burden and PTEN mutations are predictors
of a negative response to PD-1 blockade in MSI-H/dMMR
gastrointestinal tumors. Clin Cancer Res. 27:3714–3724. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Polom K, Das K, Marrelli D, Roviello G,
Pascale V, Voglino C, Rho H, Tan P and Roviello F: KRAS mutation in
gastric cancer and prognostication associated with microsatellite
instability status. Pathol Oncol Res. 25:333–340. 2019. View Article : Google Scholar
|
|
48
|
Hwang HS, Kim D and Choi J: Distinct
mutational profile and immune microenvironment in
microsatellite-unstable and POLE-mutated tumors. J Immunother
Cancer. 9:e0027972021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Giampieri R, Maccaroni E, Mandolesi A, Del
Prete M, Andrikou K, Faloppi L, Bittoni A, Bianconi M, Scarpelli M,
Bracci R, et al: Mismatch repair deficiency may affect clinical
outcome through immune response activation in metastatic gastric
cancer patients receiving first-line chemotherapy. Gastric Cancer.
20:156–163. 2017. View Article : Google Scholar
|
|
50
|
Kim KJ, Lee KS, Cho HJ, Kim YH, Yang HK,
Kim WH and Kang GH: Prognostic implications of tumor-infiltrating
FoxP3+ regulatory T cells and CD8+ cytotoxic T cells in
microsatellite-unstable gastric cancers. Hum Pathol. 45:285–293.
2014. View Article : Google Scholar
|
|
51
|
Yoshida T, Ogura G, Tanabe M, Hayashi T,
Ohbayashi C, Azuma M, Kunisaki C, Akazawa Y, Ozawa S, Matsumoto S,
et al: Clinicopathological features of PD-L1 protein expression,
EBV positivity, and MSI status in patients with advanced gastric
and esophagogastric junction adenocarcinoma in Japan. Cancer Biol
Ther. 23:191–200. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gu L, Chen M, Guo D, Zhu H, Zhang W, Pan
J, Zhong X, Li X, Qian H, Wang X, et al: PD-L1 and gastric cancer
prognosis: A systematic review and meta-analysis. PLoS One.
12:e01826922017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dislich B, Mertz KD, Gloor B and Langer R:
Interspatial distribution of tumor and immune cells in correlation
with PD-L1 in molecular subtypes of gastric cancers. Cancers
(Basel). 14:17362022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He CY, Qiu MZ, Yang XH, Zhou DL, Ma JJ,
Long YK, Ye ZL, Xu BH, Zhao Q, Jin Y, et al: Classification of
gastric cancer by EBV status combined with molecular profiling
predicts patient prognosis. Clin Transl Med. 10:353–362. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hawkins RD, Hon GC and Ren B:
Next-generation genomics: An integrative approach. Nat Rev Genet.
11:476–486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bornschein J, Wernisch L, Secrier M,
Miremadi A, Perner J, MacRae S, O'Donovan M, Newton R, Menon S,
Bower L, et al: Transcriptomic profiling reveals three molecular
phenotypes of adenocarcinoma at the gastroesophageal junction. Int
J Cancer. 145:3389–3401. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pollak M: Insulin and insulin-like growth
factor signalling in neoplasia. Nat Rev Cancer. 8:915–928. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pollak M: The insulin and insulin-like
growth factor receptor family in neoplasia: An update. Nat Rev
Cancer. 12:159–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Werner H, Meisel-Sharon S and Bruchim I:
Oncogenic fusion proteins adopt the insulin-like growth factor
signaling pathway. Mol Cancer. 17:282018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tauriello DVF, Palomo-Ponce S, Stork D,
Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M,
Ibiza S, Cañellas A, Hernando-Momblona X, et al: TGFβ drives immune
evasion in genetically reconstituted colon cancer metastasis.
Nature. 554:538–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mariathasan S, Turley SJ, Nickles D,
Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita
JL, Cubas R, et al: TGFbeta attenuates tumour response to PD-L1
blockade by contributing to exclusion of T cells. Nature.
554:544–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xie F, Zhou X, Li H, Su P, Liu S, Li R,
Zou J, Wei X, Pan C, Zhang Z, et al: USP8 promotes cancer
progression and extracellular vesicle-mediated CD8+ T cell
exhaustion by deubiquitinating the TGF-β receptor TβRII. EMBO J.
41:e1087912022. View Article : Google Scholar
|
|
66
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Goldenring JR and Nomura S:
Differentiation of the gastric mucosa III. Animal models of oxyntic
atrophy and metaplasia. Am J Physiol Gastrointest Liver Physiol.
291:G999–G1004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ma Y, Zhu J, Chen S, Li T, Ma J, Guo S, Hu
J, Yue T, Zhang J, Wang P, et al: Activated gastric
cancer-associated fibroblasts contribute to the malignant phenotype
and 5-FU resistance via paracrine action in gastric cancer. Cancer
Cell Int. 18:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Howe LR, Subbaramaiah K, Hudis CA and
Dannenberg AJ: Molecular pathways: Adipose inflammation as a
mediator of obesity-associated cancer. Clin Cancer Res.
19:6074–6083. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Manousopoulou A, Hayden A, Mellone M,
Garay-Baquero DJ, White CH, Noble F, Lopez M, Thomas GJ, Underwood
TJ and Garbis SD: Quantitative proteomic profiling of primary
cancer-associated fibroblasts in oesophageal adenocarcinoma. Br J
Cancer. 118:1200–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Calcagno DQ, Leal MF, Assumpcao PP, Smith
MA and Burbano RR: MYC and gastric adenocarcinoma carcinogenesis.
World J Gastroenterol. 14:5962–5968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Park KU, Lee HE, Park DJ, Jung EJ, Song J,
Kim HH, Choe G, Kim WH and Lee HS: MYC quantitation in cell-free
plasma DNA by real-time PCR for gastric cancer diagnosis. Clin Chem
Lab Med. 47:530–536. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao L, Liu Y, Zhang S, Wei L, Cheng H and
Wang J and Wang J: Impacts and mechanisms of metabolic
reprogramming of tumor microenvironment for immunotherapy in
gastric cancer. Cell Death Dis. 13:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Davoli T, Uno H, Wooten EC and Elledge SJ:
Tumor aneuploidy correlates with markers of immune evasion and with
reduced response to immunotherapy. Science. 355:eaaf83992017.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gibney GT, Weiner LM and Atkins MB:
Predictive biomarkers for checkpoint inhibitor-based immunotherapy.
Lancet Oncol. 17:e542–e551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gandara DR, Paul SM, Kowanetz M,
Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G,
Malboeuf C, et al: Blood-based tumor mutational burden as a
predictor of clinical benefit in non-small-cell lung cancer
patients treated with atezolizumab. Nat Med. 24:1441–1448. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mandal R, Samstein RM, Lee KW, Havel JJ,
Wang H, Krishna C, Sabio EY, Makarov V, Kuo F, Blecua P, et al:
Genetic diversity of tumors with mismatch repair deficiency
influences anti-PD-1 immunotherapy response. Science. 364:485–491.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Cai L, Li L, Ren D, Song X, Mao B, Han B
and Zhang H: Prognostic impact of gene copy number instability and
tumor mutation burden in patients with resectable gastric cancer.
Cancer Commun (Lond). 40:63–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang D, Wang N, Li X, Chen X, Shen B, Zhu
D, Zhu L, Xu Y, Yu Y and Shu Y: Tumor mutation burden as a
biomarker in resected gastric cancer via its association with
immune infiltration and hypoxia. Gastric Cancer. 24:823–834. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mani DR, Krug K, Zhang B, Satpathy S,
Clauser KR, Ding L, Ellis M, Gillette MA and Carr SA: Cancer
proteogenomics: Current impact and future prospects. Nat Rev
Cancer. 22:298–313. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mertins P, Mani DR, Ruggles KV, Gillette
MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al:
Proteogenomics connects somatic mutations to signalling in breast
cancer. Nature. 534:55–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pozniak Y, Balint-Lahat N, Rudolph JD,
Lindskog C, Katzir R, Avivi C, Pontén F, Ruppin E, Barshack I and
Geiger T: System-wide clinical proteomics of breast cancer reveals
global remodeling of tissue homeostasis. Cell Syst. 2:172–184.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vasaikar S, Huang C, Wang X, Petyuk VA,
Savage SR, Wen B, Dou Y, Zhang Y, Shi Z, Arshad OA, et al:
Proteogenomic analysis of human colon cancer reveals new
therapeutic opportunities. Cell. 177:1035–1049.e19. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jayavelu AK, Wolf S, Buettner F, Alexe G,
Häupl B, Comoglio F, Schneider C, Doebele C, Fuhrmann DC, Wagner S,
et al: The proteogenomic subtypes of acute myeloid leukemia. Cancer
Cell. 40:301–317.e12. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Tong Y, Sun M, Chen L, Wang Y, Li Y, Li L,
Zhang X, Cai Y, Qie J, Pang Y, et al: Proteogenomic insights into
the biology and treatment of pancreatic ductal adenocarcinoma. J
Hematol Oncol. 15:1682022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang C, Chong X, Jiang F, Gao J, Chen Y,
Jia K, Fan M, Liu X, An J, Li J, et al: Plasma extracellular
vesicle derived protein profile predicting and monitoring
immunotherapeutic outcomes of gastric cancer. J Extracell Vesicles.
11:e122092022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Melero I, Berman DM, Aznar MA, Korman AJ,
Perez Gracia JL and Haanen J: Evolving synergistic combinations of
targeted immunotherapies to combat cancer. Nat Rev Cancer.
15:457–472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang
S and Li Y: Targeting the IDO1 pathway in cancer: From bench to
bedside. J Hematol Oncol. 11:1002018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Barnes TA and Amir E: HYPE or HOPE: The
prognostic value of infiltrating immune cells in cancer. Br J
Cancer. 117:451–460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kim RS, Song N, Gavin PG, Salgado R,
Bandos H, Kos Z, Floris G, Eynden GGGMVD, Badve S, Demaria S, et
al: Stromal tumor-infiltrating lymphocytes in NRG Oncology/NSABP
B-31 adjuvant trial for Early-Stage HER2-Positive breast cancer. J
Natl Cancer Inst. 111:867–871. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Paijens ST, Vledder A, de Bruyn M and
Nijman HW: Tumor-infiltrating lymphocytes in the immunotherapy era.
Cell Mol Immunol. 18:842–859. 2021. View Article : Google Scholar :
|
|
92
|
Liu D, Heij LR, Czigany Z, Dahl E, Lang
SA, Ulmer TF, Luedde T, Neumann UP and Bednarsch J: The role of
tumor-infiltrating lymphocytes in cholangiocarcinoma. J Exp Clin
Cancer Res. 41:1272022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lascarez-Lagunas LI, Nadarajan S,
Martinez-Garcia M, Quinn JN, Todisco E, Thakkar T, Berson E, Eaford
D, Crawley O, Montoya A, et al: ATM/ATR kinases link the
synaptonemal complex and DNA double-strand break repair pathway
choice. Curr Biol. 32:4719–4726.e4. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Goel S, DeCristo MJ, Watt AC, BrinJones H,
Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, et
al: CDK4/6 inhibition triggers anti-tumour immunity. Nature.
548:471–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pritzl CJ, Luera D, Knudson KM, Quaney MJ,
Calcutt MJ, Daniels MA and Teixeiro E: IKK2/NFkB signaling controls
lung resident CD8+ T cell memory during influenza
infection. Nat Commun. 14:43312023. View Article : Google Scholar
|
|
96
|
Chang CP, Su YC, Lee PH and Lei HY:
Targeting NFKB by autophagy to polarize hepatoma-associated
macrophage differentiation. Autophagy. 9:619–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zinatizadeh MR, Schock B, Chalbatani GM,
Zarandi PK, Jalali SA and Miri SR: The Nuclear Factor Kappa B
(NF-kB) signaling in cancer development and immune diseases. Genes
Dis. 8:287–297. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chevallay M, Bollschweiler E, Chandramohan
SM, Schmidt T, Koch O, Demanzoni G, Mönig S and Allum W: Cancer of
the gastroesophageal junction: A diagnosis, classification, and
management review. Ann N Y Acad Sci. 1434:132–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Vial M, Grande L and Pera M: Epidemiology
of adenocarcinoma of the esophagus, gastric cardia, and upper
gastric third. Recent Results Cancer Res. 182:1–17. 2010.PubMed/NCBI
|
|
100
|
Gao Y, Xin L, Lin H, Yao B, Zhang T, Zhou
AJ, Huang S, Wang JH, Feng YD, Yao SH, et al: Machine
learning-based automated sponge cytology for screening of
oesophageal squamous cell carcinoma and adenocarcinoma of the
oesophagogastric junction: A nationwide, multicohort, prospective
study. Lancet Gastroenterol Hepatol. 8:432–445. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Menghi F, Orzan FN, Eoli M, Farinotti M,
Maderna E, Pisati F, Bianchessi D, Valletta L, Lodrini S, Galli G,
et al: DNA microarray analysis identifies CKS2 and LEPR as
potential markers of meningioma recurrence. Oncologist.
16:1440–1450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Qiu H, Lin X, Tang W, Liu C, Chen Y, Ding
H, Kang M and Chen S: Investigation of TCF7L2, LEP and LEPR
polymorphisms with esophageal squamous cell carcinomas. Oncotarget.
8:109107–109119. 2017. View Article : Google Scholar
|
|
103
|
Liu CR, Li Q, Hou C, Li H, Shuai P, Zhao
M, Zhong XR, Xu ZP and Li JY: Changes in body mass index, leptin,
and leptin receptor polymorphisms and breast cancer risk. DNA Cell
Biol. 37:182–188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yu H, Pan R, Qi Y, Zheng Z, Li J, Li H,
Ying J, Xu M and Duan S: LEPR hypomethylation is significantly
associated with gastric cancer in males. Exp Mol Pathol.
116:1044932020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wei Y, Wang X, Zhang Z, Zhao C, Chang Y,
Bian Z and Zhao X: Impact of NR5A2 and RYR2 3'UTR polymorphisms on
the risk of breast cancer in a Chinese Han population. Breast
Cancer Res Treat. 183:1–8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH,
Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, et al: Expression of
DBC1 and SIRT1 is associated with poor prognosis of gastric
carcinoma. Clin Cancer Res. 15:4453–4459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Bae JS, Park SH, Jamiyandorj U, Kim KM,
Noh SJ, Kim JR, Park HJ, Kwon KS, Jung SH, Park HS, et al:
CK2alpha/CSNK2A1 Phosphorylates SIRT6 and is involved in the
progression of breast carcinoma and predicts shorter survival of
diagnosed patients. Am J Pathol. 186:3297–3315. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shen JZ, Qiu Z, Wu Q, Finlay D, Garcia G,
Sun D, Rantala J, Barshop W, Hope JL, Gimple RC, et al: FBXO44
promotes DNA replication-coupled repetitive element silencing in
cancer cells. Cell. 184:352–369.e23. 2021. View Article : Google Scholar :
|
|
109
|
Mattei AL, Bailly N and Meissner A: DNA
methylation: A historical perspective. Trends Genet. 38:676–707.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Papanicolau-Sengos A and Aldape K: DNA
methylation profiling: An emerging paradigm for cancer diagnosis.
Annu Rev Pathol. 17:295–321. 2022. View Article : Google Scholar
|
|
111
|
Johnson CH, Ivanisevic J and Siuzdak G:
Metabolomics: Beyond biomarkers and towards mechanisms. Nat Rev Mol
Cell Biol. 17:451–459. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Peng L, Chen L, Wan J, Liu W, Lou S and
Shen Z: Single-cell transcriptomic landscape of immunometabolism
reveals intervention candidates of ascorbate and aldarate
metabolism, fatty-acid degradation and PUFA metabolism of T-cell
subsets in healthy controls, psoriasis and psoriatic arthritis.
Front Immunol. 14:11798772023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kwon J and Bakhoum SF: The cytosolic
DNA-Sensing cGAS-STING pathway in cancer. Cancer Discov. 10:26–39.
2020. View Article : Google Scholar :
|
|
114
|
Vandereyken K, Sifrim A, Thienpont B and
Voet T: Methods and applications for single-cell and spatial
multi-omics. Nat Rev Genet. 24:494–515. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Liu Z, Zhao Y, Kong P, Liu Y, Huang J, Xu
E, Wei W, Li G, Cheng X, Xue L, et al: Integrated multi-omics
profiling yields a clinically relevant molecular classification for
esophageal squamous cell carcinoma. Cancer Cell. 41:181–195.e9.
2023. View Article : Google Scholar
|
|
116
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar :
|
|
117
|
Xu Y, She Y, Li Y, Li H, Jia Z, Jiang G,
Liang L and Duan L: Multi-omics analysis at epigenomics and
transcriptomics levels reveals prognostic subtypes of lung squamous
cell carcinoma. Biomed Pharmacother. 125:1098592020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Feng D, Gao P, Henley N, Dubuissez M, Chen
N, Laurin LP, Royal V, Pichette V and Gerarduzzi C: SMOC2 promotes
an epithelial-mesenchymal transition and a pro-metastatic phenotype
in epithelial cells of renal cell carcinoma origin. Cell Death Dis.
13:6392022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Mullen J, Kato S, Sicklick JK and Kurzrock
R: Targeting ARID1A mutations in cancer. Cancer Treat Rev.
100:1022872021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Hansford S, Kaurah P, Li-Chang H, Woo M,
Senz J, Pinheiro H, Schrader KA, Schaeffer DF, Shumansky K,
Zogopoulos G, et al: Hereditary diffuse gastric cancer syndrome:
CDH1 mutations and beyond. JAMA Oncol. 1:23–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Cochran BJ, Ong KL, Manandhar B and Rye
KA: APOA1: A protein with multiple therapeutic functions. Curr
Atheroscler Rep. 23:112021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chia NY and Tan P: Molecular
classification of gastric cancer. Ann Oncol. 27:763–769. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Nakayama I, Qi C, Chen Y, Nakamura Y, Shen
L and Shitara K: Claudin 18.2 as a novel therapeutic target. Nat
Rev Clin Oncol. 21:354–369. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Shitara K, Lordick F, Bang YJ, Enzinger P,
Ilson D, Shah MA, Van Cutsem E, Xu RH, Aprile G, Xu J, et al:
Zolbetuximab plus mFOLFOX6 in patients with CLDN18.2-positive,
HER2-negative, untreated, locally advanced unresectable or
metastatic gastric or gastro-oesophageal junction adenocarcinoma
(SPOTLIGHT): A multicentre, randomised, double-blind, phase 3
trial. Lancet. 401:1655–1668. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Shah MA, Shitara K, Ajani JA, Bang YJ,
Enzinger P, Ilson D, Lordick F, Van Cutsem E, Gallego Plazas J,
Huang J, et al: Zolbetuximab plus CAPOX in CLDN18.2-positive
gastric or gastroesophageal junction adenocarcinoma: The
randomized, phase 3 GLOW trial. Nat Med. 29:2133–2141. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sahin U, Tureci O, Manikhas G, Lordick F,
Rusyn A, Vynnychenko I, Dudov A, Bazin I, Bondarenko I, Melichar B,
et al: FAST: A randomised phase II study of zolbetuximab (IMAB362)
plus EOX versus EOX alone for first-line treatment of advanced
CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma.
Ann Oncol. 32:609–619. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Klempner SJ, Lee KW, Shitara K, Metges JP,
Lonardi S, Ilson DH, Fazio N, Kim TY, Bai LY, Moran D, et al:
ILUSTRO: Phase II multicohort trial of zolbetuximab in patients
with advanced or metastatic claudin 18.2-Positive gastric or
gastroesophageal junction adenocarcinoma. Clin Cancer Res.
29:3882–3891. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:264–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bang YJ, Kim YW, Yang HK, Chung HC, Park
YK, Lee KH, Lee KW, Kim YH, Noh SI, Cho JY, et al: Adjuvant
capecitabine and oxaliplatin for gastric cancer after D2
gastrectomy (CLASSIC): A phase 3 open-label, randomised controlled
trial. Lancet. 379:315–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
De Vita F, Giuliani F, Galizia G, Belli C,
Aurilio G, Santabarbara G, Ciardiello F, Catalano G and Orditura M:
Neo-adjuvant and adjuvant chemotherapy of gastric cancer. Ann
Oncol. 18(Suppl 6): vi120–vi123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Salonga D, Danenberg KD, Johnson M,
Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman
L, Diasio RB and Danenberg PV: Colorectal tumors responding to
5-fluorouracil have low gene expression levels of dihydropyrimidine
dehydrogenase, thymidylate synthase, and thymidine phosphorylase.
Clin Cancer Res. 6:1322–1327. 2000.PubMed/NCBI
|
|
132
|
White C, Scott RJ, Paul C, Ziolkowski A,
Mossman D, Fox SB, Michael M and Ackland S: Dihydropyrimidine
dehydrogenase deficiency and implementation of upfront DPYD
genotyping. Clin Pharmacol Ther. 112:791–802. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhao F, Li E, Shen G, Dong Q, Ren D, Wang
M, Zhao Y, Liu Z, Ma J, Xie Q, et al: Correlation between mismatch
repair and survival of patients with gastric cancer after
5-FU-based adjuvant chemotherapy. J Gastroenterol. 58:622–632.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Hoppe MM, Sundar R, Tan DSP and
Jeyasekharan AD: Biomarkers for homologous recombination deficiency
in cancer. J Natl Cancer Inst. 110:704–713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Golan T, O'Kane GM, Denroche RE,
Raitses-Gurevich M, Grant RC, Holter S, Wang Y, Zhang A, Jang GH,
Stossel C, et al: Genomic features and classification of homologous
recombination deficient pancreatic ductal adenocarcinoma.
Gastroenterology. 160:2119–2132.e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Rodler E, Sharma P, Barlow WE, Gralow JR,
Puhalla SL, Anders CK, Goldstein L, Tripathy D, Brown-Glaberman UA,
Huynh TT, et al: Cisplatin with veliparib or placebo in metastatic
triple-negative breast cancer and BRCA mutation-associated breast
cancer (S1416): A randomised, double-blind, placebo-controlled,
phase 2 trial. Lancet Oncol. 24:162–174. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Telli ML, Timms KM, Reid J, Hennessy B,
Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, et
al: Homologous recombination deficiency (HRD) score predicts
response to platinum-containing neoadjuvant chemotherapy in
patients with Triple-Negative breast cancer. Clin Cancer Res.
22:3764–3773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhu Y, Hu Y, Tang C, Guan X and Zhang W:
Platinum-based systematic therapy in triple-negative breast cancer.
Biochim Biophys Acta Rev Cancer. 1877:1886782022. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Ott K, Vogelsang H, Mueller J, Becker K,
Müller M, Fink U, Siewert JR, Höfler H and Keller G: Chromosomal
instability rather than p53 mutation is associated with response to
neoadjuvant cisplatin-based chemotherapy in gastric carcinoma. Clin
Cancer Res. 9:2307–2315. 2003.PubMed/NCBI
|
|
140
|
Kumar S, Nandi A, Singh S, Regulapati R,
Li N, Tobias JW, Siebel CW, Blanco MA, Klein-Szanto AJ, Lengner C,
et al: Dll1(+) quiescent tumor stem cells drive chemoresistance in
breast cancer through NF-kappaB survival pathway. Nat Commun.
12:4322021. View Article : Google Scholar
|
|
141
|
Kuo WY, Hwu L, Wu CY, Lee JS, Chang CW and
Liu RS: STAT3/NF-κB-regulated lentiviral TK/GCV suicide gene
therapy for cisplatin-resistant triple-negative breast cancer.
Theranostics. 7:647–663. 2017. View Article : Google Scholar :
|
|
142
|
Liu W, Wang Z, Liu S, Zhang X, Cao X and
Jiang M: RNF138 inhibits late inflammatory gene transcription
through degradation of SMARCC1 of the SWI/SNF complex. Cell Rep.
42:1120972023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Mittal P and Roberts CWM: The SWI/SNF
complex in cancer-biology, biomarkers and therapy. Nat Rev Clin
Oncol. 17:435–448. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Botta GP, Kato S, Patel H, Fanta P, Lee S,
Okamura R and Kurzrock R: SWI/SNF complex alterations as a
biomarker of immunotherapy efficacy in pancreatic cancer. JCI
Insight. 6:e1504532021. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Gluckstein MI, Dintner S, Arndt TT,
Vlasenko D, Schenkirsch G, Agaimy A, Müller G, Märkl B and Grosser
B: Comprehensive immunohistochemical study of the SWI/SNF complex
expression status in gastric cancer reveals an adverse prognosis of
SWI/SNF deficiency in genomically stable gastric carcinomas.
Cancers (Basel). 13:38942021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Iwagami Y, Eguchi H, Nagano H, Akita H,
Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y, et
al: miR-320c regulates gemcitabine-resistance in pancreatic cancer
via SMARCC1. Br J Cancer. 109:502–511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Deng J, Bai X, Feng X, Ni J, Beretov J,
Graham P and Li Y: Inhibition of PI3K/Akt/mTOR signaling pathway
alleviates ovarian cancer chemoresistance through reversing
epithelial-mesenchymal transition and decreasing cancer stem cell
marker expression. BMC Cancer. 19:6182019. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Peng Y, Wang Y, Zhou C, Mei W and Zeng C:
PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we
making headway? Front Oncol. 12:8191282022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yu M, Chen J, Xu Z Yang B, He Q, Luo P,
Yan H and Yang X: Development and safety of PI3K inhibitors in
cancer. Arch Toxicol. 97:635–650. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Rowshanravan B, Halliday N and Sansom DM:
CTLA-4: A moving target in immunotherapy. Blood. 131:58–67. 2018.
View Article : Google Scholar
|
|
152
|
Pereira MA, Dias AR, Ramos MFKP, Cardili
L, Moraes RDR, Zilberstein B, Nahas SC, Mello ES and Ribeiro U Jr:
Gastric cancer with microsatellite instability displays increased
thymidylate synthase expression. J Surg Oncol. 126:116–124. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Puliga E, Corso S, Pietrantonio F and
Giordano S: Microsatellite instability in gastric cancer: Between
lights and shadows. Cancer Treat Rev. 95:1021752021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Liu X, Choi MG, Kim K, Kim KM, Kim ST,
Park SH, Cristescu R, Peter S and Lee J: High PD-L1 expression in
gastric cancer (GC) patients and correlation with molecular
features. Pathol Res Pract. 216:1528812020. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Kim TS, da Silva E, Coit DG and Tang LH:
Intratumoral immune response to gastric cancer varies by molecular
and histologic subtype. Am J Surg Pathol. 43:851–860. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Bai Y, Xie T, Wang Z, Tong S, Zhao X, Zhao
F, Cai J, Wei X, Peng Z and Shen L: Efficacy and predictive
biomarkers of immunotherapy in Epstein-Barr virus-associated
gastric cancer. J Immunother Cancer. 10:e0040802022. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Wang Z, Wang X, Xu Y, Li J, Zhang X, Peng
Z, Hu Y, Zhao X, Dong K, Zhang B, et al: Mutations of PI3K-AKT-mTOR
pathway as predictors for immune cell infiltration and
immunotherapy efficacy in dMMR/MSI-H gastric adenocarcinoma. BMC
Med. 20:1332022. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Panda A, Mehnert JM, Hirshfield KM,
Riedlinger G, Damare S, Saunders T, Kane M, Sokol L, Stein MN,
Poplin E, et al: Immune activation and benefit from avelumab in
EBV-positive gastric cancer. J Natl Cancer Inst. 110:316–320. 2018.
View Article : Google Scholar
|
|
159
|
Saito M and Kono K: Landscape of
EBV-positive gastric cancer. Gastric Cancer. 24:983–989. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Cheong JE and Sun L: Targeting the
IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy-challenges and
opportunities. Trends Pharmacol Sci. 39:307–325. 2018. View Article : Google Scholar
|
|
161
|
Niu F, Yu Y, Li Z, Ren Y, Li Z, Ye Q, Liu
P, Ji C, Qian L and Xiong Y: Arginase: An emerging and promising
therapeutic target for cancer treatment. Biomed Pharmacother.
149:1128402022. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Ichikawa H, Nagahashi M, Shimada Y, Hanyu
T, Ishikawa T, Kameyama H, Kobayashi T, Sakata J, Yabusaki H,
Nakagawa S, et al: Actionable gene-based classification toward
precision medicine in gastric cancer. Genome Med. 9:932017.
View Article : Google Scholar : PubMed/NCBI
|