Breast cancer (BC) is the most commonly diagnosed
malignancy affecting women, as well as the leading cause of
cancer-related mortality among women worldwide (1). According to GLOBOCAN 2020
statistics, the number of new female BC cases globally was
estimated to be 2.3 million (11.7% of all newly diagnosed cancer
cases reported), along with 685,000 (6.9% of all cancer mortality
cases reported) mortality cases in 2020 (2). Researchers from the International
Agency for Research on Cancer predicted an increase in the future
burden of female BC, and the numbers are estimated to increase to 3
million new cases and 1 million deaths in 2040 (3). In Malaysia, female BC is highly
prevalent among women of all ethnic groups, with 1 in every 19
women at risk of developing BC (4). Family history, dietary factors,
lifestyle, sex, old age, hormonal factors and reproductive factors
are among the multitude of risk factors that might predispose an
individual to an increased risk of BC (5).
BC can be broadly categorized into two
histopathological subtypes: Non-invasive BC and invasive BC
(6). In non-invasive BC, cells
are confined to the milk ducts and do not invade fatty and
connective tissues of the breast. Ductal carcinoma in situ
and lobular carcinoma in situ are the two types of
non-invasive BC (7,8). In invasive BC, the cells break
through the duct and lobular wall, invading the tissues of the
breast. Examples of invasive BC include infiltrating lobular
carcinoma and infiltrating ductal carcinoma (IDC) (9). In addition, there are a few types of
less commonly occurring BC, including medullary carcinoma, mucinous
carcinoma and Paget's disease of the nipple, which have been
clinically observed and diagnosed (10). In addition, BC subtypes can also
be characterized according to their distinct and diverse molecular
patterns, which involves profiling the hormone receptor status of
the patient. Hormone receptor-positive subtypes such as the
estrogen receptor-positive (ER+) and progesterone
receptor-positive subtypes are reliant on their respective hormones
for cancer growth and proliferation (11). HER2-positive individuals exhibit
upregulation of the receptor HER2 and/or amplification of the gene
HER2 (12). Triple-negative BC
(TNBC) is characterized by the lack of all three hormone receptors
and is highly heterogenous with poorer prognosis compared with the
other BC subtypes (13). The
heterogeneity in BC molecular subtypes reflects the different
metabolic phenotypes of BC. Disruption of normal cell metabolism
has underlying effects in breast carcinogenesis and tumorigenicity,
which is the rationale behind the heterogeneity and aggressiveness
of BC subtypes (14,15).
The hallmarks of cancer are a concept used to
illustrate the framework in understanding cancer pathology
(16). One distinct feature is
the reprogramming of cellular metabolism. Cancer cells have the
ability to exploit and rewire different metabolic pathways in order
to sustain the increased nutritional and energy requirements for
tumorigenic proliferation and metastasis (14). In a review by Hanahan (17), the author highlighted the
deregulation of cellular metabolism as one of the eight core
hallmarks of cancer. Deregulation of cell metabolism and cell
signaling are caused as a result of metabolic reprogramming,
characterized by increased synthesis of macromolecules and
increased proliferation, giving rise to more aggressive cancer
phenotypes and drug resistance (18). As aforementioned, BC cells have
the ability to exhibit different metabolic phenotypes depending on
their molecular subtypes. Intrinsic factors such as gene
amplifications and mutations, and extrinsic factors such as
hypoxia, oxidative stress and acidosis are contributing factors for
the different metabolic phenotypes observed in BC (19). BC cells have the ability to alter
glucose, lipid and amino acid metabolic pathways, which are usually
regulated by genes to promote uncontrolled cell proliferation and
induce breast carcinogenesis. For example, non-cancerous cells
under normal conditions are able to catabolize glucose to produce
ATP via the mitochondrial oxidative phosphorylation pathway.
However, cancer cells have the preference of utilizing the
glycolysis pathway as an alternative to produce energy and disrupt
tumor microenvironments to promote carcinogenesis and cancer
invasion, known as the Warburg phenomenon (20). It has come to the attention of
researchers that microRNA (miRNA/miR) dysregulation occurs in
various human diseases, including BC, and that miRNAs have the
ability to influence the metabolism of cells by regulating various
metabolic pathways for tumor growth and sustenance (21-23). Therefore, it has been proposed
that dysregulated miRNA expression can be linked to the alteration
of metabolic pathways in BC cells.
miRNAs are small, highly conserved, non-coding RNA
sequences that are 18-25 nucleotides long and have the ability to
exert biological effects by post-transcriptionally regulating mRNAs
(24). miRNAs regulate gene
expression by binding to the 3′ untranslated region (3′-UTR) of the
target mRNA (25). Various
studies have revealed that dysregulation of miRNA levels
contributes to tumor onset, growth and metastasis in individuals
affected by BC (26-28). miRNA-mediated gene expression is
important for normal cellular responses to environmental stresses.
The deregulation of miRNA levels interrupts the normal regulation
of oncogenic and tumor-suppressive target genes, which are
implicated in the pathogenesis of BC tumors (29). Metabolic mechanisms and networks
underlying BC heterogeneity are still poorly understood due to the
inherent complexity of BC tumors. Therefore, further studies are
required to gain a deeper understanding of the metabolic machinery
of BC cells to stimulate future developments in BC diagnosis, BC
prognosis and the identification of personalized treatments for
subtype-specific BC. The present comprehensive review discusses
miRNAs that are involved in BC cellular metabolism, including
mainly glucose, lipid and amino acid metabolism, and also
highlights future perspectives and clinical implications of miRNAs
in BC metabolism.
Glucose is central to energy consumption in
mammalian cells. It can be produced by breaking down complex
molecules, carbohydrates, serving as primary metabolic fuel for
mammals. In addition, glucose can be synthesized from
non-carbohydrate sources, such as proteins and lipids, through
gluconeogenesis, a process which occurs within mitochondria of
liver cells (30). Glucose can
also be synthesized through glycogenolysis where glycogen is broken
down into glucose-1-phosphate and glucose. Glycogenolysis takes
place in hepatocytes and myocytes, and is regulated by enzymes,
phosphorylase kinase and glycogen phosphorylase (31). In addition, glucose metabolism
also involves the process of glycogenesis, where glycogen, the
principal storage form of glucose and primary source of
non-oxidative glucose for skeletal muscle and the liver, is formed
(32).
At the cellular level, glucose is essential in the
production of ATP, which is the main source of energy for use and
storage. ATP is synthesized through the process of cellular
respiration, where glucose is catabolized into acetyl-CoA,
producing high energy electron carriers that are oxidized during
oxidative phosphorylation. ATP is regarded as the 'energy currency'
as it provides readily released energy by breaking the bond of
phosphate groups. It is required in a number of processes,
including intracellular signaling, DNA and RNA synthesis,
purinergic signaling, synaptic signaling, active transport, and
muscle contraction (33).
The regulation of glucose metabolism in BC cells by
miRNAs has gained attention from researchers worldwide. Glycolysis
is part of the glucose metabolic pathway, and it is the first step
in cellular respiration, which entails the oxidation of glucose
molecules in the body (41). A
series of glycolytic enzymes are involved in catabolizing the
glucose molecules, and thus, pyruvates and water molecules are
formed as the end products (42).
Table I summarizes the glycolytic
enzymes or genes targeted by miRNAs and the changes of BC cells
following the expression of the miRNAs.
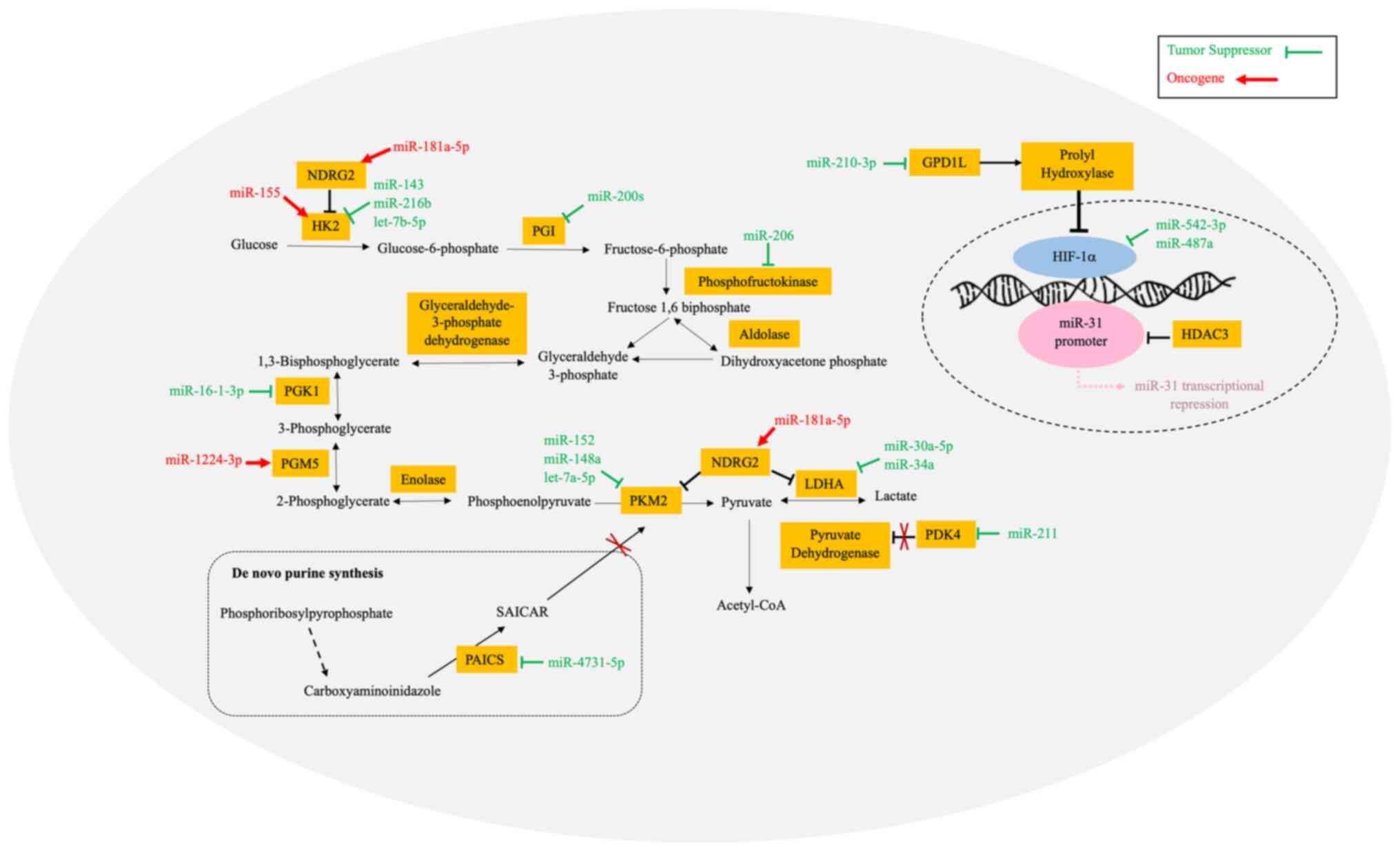
Hexokinase (HK) is the enzyme that catalyzes the
first step of glycolysis, in which glucose molecules are
phosphorylated into glucose-6-phosphate (43). Jiang et al (44) reported that miR-155 upregulated
HK2 via two distinct mechanisms. Firstly, HK2 transcription was
promoted by miR-155 via activation of STAT3. Following upregulation
of miR-155 in BC cells, glucose consumption and lactate production
were found to be increased, and pro-inflammatory cytokines such as
TNF-α, IL-1β and IFN-γ were upregulated. In addition, miR-155
promoted HK2 expression at the post-transcriptional level by
repressing miR-143 by targeting CCAAT/enhancer-binding protein β.
The downregulation of miR-143 increased glucose consumption and
lactate production, and thus, elevated the proliferation and
migration of BC cells, as well as xenograft tumor growth (44). Another study by Liu et al
(45) concluded that miR-216b
inhibited the progression of BC by targeting HK2, which resulted in
mTOR signaling pathway inactivation. The study also showed that
increased levels of miR-216b following transfection of miR-216b
mimics inhibited proliferation, migration and invasion in MCF-7 and
MDA-MB-231 cells. In addition, HK2 silencing led to autophagy of BC
cells, cell cycle arrest and apoptosis of BC cells (45). Furthermore, Li et al
(46) demonstrated that Let-7b-5p
restrained breast tumor growth and metastasis both in vitro
and in vivo by suppressing HK2 expression. A decreased
extracellular acidification rate (ECAR) and increased oxygen
consumption were observed following Let-7b-5p upregulation
(46).
Another glycolytic enzyme, namely pyruvate kinase
(PK), has been extensively studied by numerous researchers in
cancer metabolism (47-49). PK is a metabolic enzyme involved
in the last step of the glycolysis process, catalyzing the
irreversible transphosphorylation between phosphoenolpyruvate and
ADP to produce pyruvate and ATP (50). Four PK isoforms have been
identified: PK isoform L, PK isoform R, PKM1 and PKM2 (51). Among all isoforms, PKM2 has gained
much interest as it can be identified as a cancer biomarker due to
its expression in most human cancers (52). Wen et al (53) reported that the proliferation and
colony formation of MCF-7 and MDA-MB-231 cells were inhibited by
high miR-152 expression via inhibition of β-catenin and PKM2
expression. The authors revealed that, upon expression of
insulin-like growth factor 1 (IGF-1), a binding protein that is
involved in BC progression, β-catenin and PKM2 expression was
induced. IGF-1-induced expression of β-catenin and PKM2 was shown
to enhance the interaction between β-catenin and PKM2, leading to
transcriptional activation of miR-152. Therefore, the results
demonstrated a regulatory circuit among miR-152, β-catenin and PKM2
in BC (53). Xu et al
(54) demonstrated that upon
upregulation of miR-148a and miR-152, the levels of PKM2 were
downregulated. Xu et al (54) also reported that miR-148a and
miR-152 regulated the Warburg effect of BC cells, which was
demonstrated by low glucose consumption and lactate production
levels following the overexpression miR-148a and miR-152 cells. The
authors proposed that the PKM2/NF-кB/miR-148a/miR-152 pathway could
regulate tumor angiogenesis and cancer cell proliferation (54). Similar outcomes were reported by
Yao et al (55) who
revealed that the upregulation of let-7a-5p inhibited aerobic
glycolysis and proliferation of MCF-7 and MDA-MB-231 cells, and
decreased the protein levels of PKM2.
Phosphoglycerate kinase 1 (PGK1) is another pivotal
glycolytic enzyme, which catalyzes the reversible transfer of a
high-energy phosphate group from 1,3-biphosphoglycerate to ADP,
producing 3-phosphoglycerate and ATP (56). Ye et al (57) demonstrated that miR-16-1-3p
inhibited PGK1 expression, resulting in suppression of aerobic
glycolysis by decreasing the glucose uptake, lactate and ATP
production and ECAR, and increasing the oxygen consumption rate in
BC cells. Furthermore, the downregulation of the phosphoglucomutase
(PGM) family member PGM5 in patients with BC has also gained the
attention of researchers. Ran et al (58) reported that miR-1224-3p, an
oncogene that inhibited PGM5, caused an increase in the
proliferation, migration and glycolytic function in BC cells.
Another enzyme, namely lactate dehydrogenase A (LDHA), was found to
be suppressed by miR-30a-5p, thus decreasing glucose uptake,
lactate production, ATP generation and the ECAR of BC cells
(59). Xiao et al
(60) showed that miR-34a
suppressed glycolysis and proliferation of BC cells by
downregulating LDHA.
Another glycolytic enzyme,
6-phosphofructose-2-kinase (PFKFB3), which regulates glycolysis by
controlling the levels of fructose-2,6-bisphosphate (F2,6BP), was
found to be upregulated in BC, while miR-206 was downregulated
(61). Further analysis has shown
that miR-206 overexpression decreased PFKFB3 protein expression,
cell proliferation and migration, as well as F2,6BP and lactate
production of BC cells (61,62).
Phosphoglucose isomerase (PGI) is a key enzyme in
glycolysis, which catalyzes the interconversion of
glucose-6-phosphate and fructose-6-phosphate (63). Ahmad et al (64) demonstrated that PGI was
downregulated following overexpression of miR-200s (miR-200a,
miR-200b and miR-200c), leading to inhibition of wound healing,
colony formation and metastasis in BC cells. Additionally, Guda
et al (65) concluded that
miR-211 is a robust inhibitor of the Warburg effect, and pyruvate
dehydrogenase kinase 4 was silenced by miR-211. Following the
upregulation of miR-211, decreased ECAR and increased OCR were
reported in BC cells. The expression of miR-211 ultimately induced
mitochondrial apoptosis through mitochondrial dysfunction (65).
Although the aforementioned studies demonstrated the
role of miRNAs in regulating the glycolytic enzymes in BC cells,
which could be useful in identifying alternative strategies for BC
treatment, more investigations could be implemented to study other
enzymes, such as aldolase, glyceraldehyde-3-phosphate dehydrogenase
and enolase, and the associated miRNAs in the regulation of BC
progression. Fig. 1 shows the
aerobic glycolysis pathway with all the involved glycolytic
enzymes, as well as the miRNAs that regulate the enzymatic
reactions.
Studies in the past two decades have documented the
transcriptional regulators in aerobic glycolysis of cancers, namely
hypoxia-inducible factor 1 (HIF-1) (66), HK2 (43), histone deacetylase-3 (HDAC3)
(67), phosphoribosyl
aminoimidazole carboxylase and phosphoribosyl aminoimidazole
succinocarboxamide synthase (PAICS) (68). All the aforementioned genes serve
crucial roles in cellular aerobic glycolysis, and miRNAs have been
found to regulate the expression levels of said genes.
HIF-1 is a heterodimeric transcription factor that
consists of two subunits, namely HIF-1α and HIF-1β (69). HIF-1 serves a key role in
reprogramming of cancer metabolism by activating transcription of
genes encoding glucose transporters and glycolytic enzymes, which
take up glucose and convert it to lactate (70). In a study by Du et al
(71), the upregulation of
miR-210-3p, glucose uptake, production of lactate and the ECAR were
promoted in MDA-MB-231 and Hs578T cells. The authors proposed that
miR-210-3p targeted glycerol-3-phosphate dehydrogenase 1 like to
sustain the stability of HIF-1α, and cytoglobin repressed p53
activity (71). Another study by
Jiang et al (72) revealed
that miR-542-3p expression was downregulated in BC tissues and cell
lines. miR-542-3p overexpression inhibited glycolysis and
proliferation, and promoted apoptosis of BC cells by directly
targeting HIF-1α. However, HIF-1α overexpression could reverse the
inhibitory effect of miR-542-3p, resulting in enhanced glycolysis
and cancer cell proliferation, and decreased apoptosis of BC cells
(72). Cao et al (73) highlighted the circular RNA ring
finger protein 20 (circRNF20)/miR-487a/HIF-1α/HK2 axis in BC
progression and the Warburg effect. The authors revealed that
cirRNF20 acted as a sponge of miR-487a, where the decreased
expression of miR-487a led to the upregulation of HIF-1α levels,
which promoted the transcription of HK2, resulting in increased
glycolysis in BC cells (73).
Amino acids are essential nutrients that serve
important roles in the regulation of essential cellular functions
such as protein and nucleotide synthesis needed for cellular
proliferation. Other than being the building blocks for protein
synthesis, amino acids are also essential for the production of
non-essential amino acids to facilitate other metabolic pathways
such as glucose and lipid conversion (75). Amino acids are also important in
the production of nitrogen-containing metabolite precursors that
are used for the synthesis of nucleic acids and neurotransmitters,
as well as activation of important pathways such as nutrient
transport, epigenetic regulation and ferroptosis regulation
(76-78). All these roles that amino acids
serve highlight the extensive effects of amino acid metabolism in
cells (79). The enhanced ability
of cancer cells to acquire and exploit nutrients results in a
greater demand for amino acids to supply and sustain the increased
energy requirements of the tumor (80).
Glutamine, in particular, is a non-essential amino
acid with high versatility and is abundantly available within the
human body (81). Glutamine
serves an integral role in cancer amino acid metabolism as it
serves as the major nitrogen and carbon source for amino acid,
lipid and nucleic acid biosynthesis (82). The heavy reliance on glutamine for
tumor survival and proliferation in cancer cells is known as the
'glutamine addiction' phenotype (83). Glutamine metabolism is closely
linked to various metabolic networks that are essential for cancer
cell survival (84).
Glutaminolysis is the process of conversion whereby glutamine is
catabolized through various metabolic enzymes, namely
phosphate-dependent glutaminase (GLS) and glutamate dehydrogenase
1, to yield glutamate and other tricarboxylic acid (TCA) cycle
metabolites for ATP generation or macromolecule synthesis (85). Oncogenes such as MYC have the
ability to stimulate glutamine consumption and metabolism through
gene activation or miRNA regulation by upregulating GLS (86). BC tissues reportedly exhibit
increased levels of glutamate compared with normal breast tissues,
highlighting the dysregulation of glutamine metabolism and the
importance of glutamine in BC (87). Previous studies have reported that
glutamine and/or glutamate dependence contributes to invasiveness
in other human cancer types, including pancreatic cancer, prostate
cancer and natural killer T-cell lymphoma (88-90). It is crucial to understand that
glutamine requirements in cancer cells are highly heterogenous and
that there are various factors that could collectively influence
the role of glutamine in cancer (91). Altered amino acid and glutamine
metabolic profiles could be unique in different molecular subtypes
and stages of BC, guiding biomarker identification and drug
development for personalized treatment of BC (92).
Aminotransferases, also known as transaminases, are
important metabolic enzymes that catalyzes the interconversion of
amino acids to allow repurposing of relevant amino acid derivatives
essential for cellular function (93). Transaminases such as GLS,
glutamine synthetase and branched-chain amino acid transaminase 1,
and other key metabolic enzymes, such as glutamate dehydrogenase,
have been reported to be deregulated in BC amino acid metabolism
(94-97). Although there are various
scientific works that have investigated the relationship among
amino acid metabolism, its metabolic enzymes and BC (94,98-100), there is no study yet that
focused on the role of miRNAs in the regulation of BC amino acid
metabolism by specifically targeting these metabolic enzymes.
Transporters are membrane bound proteins that serves
mediatory roles in amino acid metabolism by engaging in amino acid
transfer in and out of the cell required for cellular function and
signaling (101). To meet the
increased amino acid demand, cancer cells can modulate the
expression of specific amino acid transporters to suit their
metabolic needs (79). Solute
carrier family 7 member 11 (SLC7A11) is a well-known amino acid
transporter, functioning as an antiporter by exchanging cystine for
glutamate to facilitate glutathione biosynthesis and reduce
reactive oxygen species (ROS)-mediated stress in cells (102). As such, SLC7A11 is also
implicated in ROS homeostasis and ferroptosis regulation in BC
(103). miRNAs such as miR-5096
(102), miR-26b (104) and miR-382-5p (105) have been reported to directly
target SLC7A11 at the SLC7A11 3′-UTR and regulate its expression in
BC cells, thus influencing BC amino acid metabolism.
Circulating miRNAs secreted in extracellular
vesicles (EVs) have also been implicated in cancer by
post-transcriptionally regulating gene expression in cells to
promote cancer formation, malignant transformation, angiogenesis
and metastasis (117-120). A study conducted on BC
patient-derived cancer-associated fibroblasts treated with
MDA-MB-231 EV-encapsulated miR-105 revealed decreased expression of
the MAX interactor 1, dimerization protein (MXI1) gene along with
elevated levels of the MYC protein. These observed changes in MXI1
and MYC expression due to the presence of miR-155 ultimately
enhanced glutamine consumption, glutaminolysis and metabolite
transport in BC in vivo models (80).
One major obstacle in BC therapy are the varied
sensitivities in chemotherapeutic responses observed in different
individuals due to the heterogenous nature of BC (121). Muciño-Olmos et al
(122) utilized multicellular
tumor spheroids (MCTs) from the BC cell line MCF7 to study
miRNA-mRNA interactive pairs that serve a regulatory role in
cellular metabolism in various metabolic phenotypes of MCF7 MCTs
observed during different cell cycle stages. The authors reported
downregulation of miR-663a and miR-1184 along with upregulation of
glutamate-ammonia ligase and phosphoglycerate mutase 1 mRNAs,
respectively, resulting in overall decreased amino acid
biosynthesis in proliferative MCTs. In monoculture cells, miR-320c
and miR-940 were found to inhibit phosphoribosyl pyrophosphate
synthetase 1 and pyrroline-5-carboxylate reductase 1 mRNAs,
respectively, to downregulate amino acid biosynthesis. miR-320c,
miR-19a-3p, miR-454-3p and miR-1226-3p also contributed to the
downregulation of amino acid degradation by regulating their
respective target mRNAs in monoculture cells. As a consequence,
dysregulated expression of all these miRNAs led to altered
metabolic pathways and rapid cancer cell proliferation (122). Table II shows the miRNAs involved in
the regulation of amino acid and glutamine metabolism in BC cell
physiology.
Lipids are a diverse class of hydrophobic organic
biomolecules that include fatty acids (FAs), glycerides,
non-glyceride lipids and lipoproteins, all of which are vital in
maintaining cellular integrity and serving as energy reserves to
fuel cellular activity (123).
FAs are the major components in the structural make up of complex
lipid molecules and can be synthesized de novo from
different carbon sources derived from other metabolic pathways.
Alternatively, FAs can also be acquired exogenously through the
diet (124). Structural
variations seen among complex lipids and FAs often result in
functional differences, which could directly influence cellular
metabolism (125). Various
biological metabolites such as triacylglycerol, diacylglycerol,
monoacylglycerol and acyl-CoAs generated from lipid metabolism are
also important energy sources and modulators of cellular signaling
that governs cellular growth, proliferation, differentiation,
apoptosis, survival and membrane homeostasis (123,126).
Lipid metabolism is tightly regulated by a series of
enzyme-catalyzed reactions and comprises various different
pathways, including but not limited to, FA transport, de
novo synthesis, FA storage, FA mobilization and FA b-oxidation
(127). Cancer cells upregulate
lipid metabolism to support oncogenic development, such as
malignant transformation, cancer development, metastatic
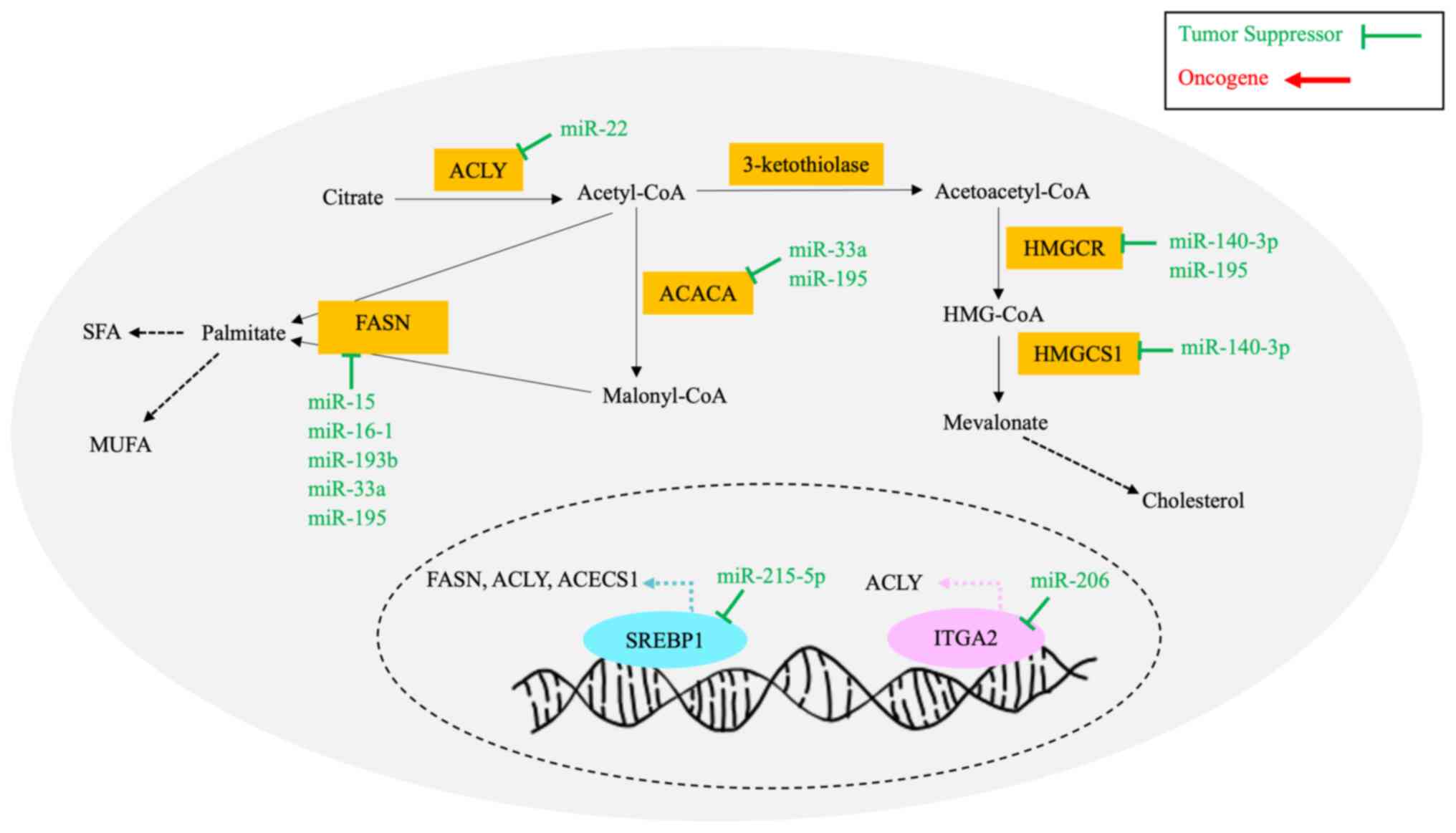
colonization and therapeutic resistance (128). De novo lipogenesis (DNL)
is of particular importance when discussing lipid metabolism in
cancer. TCA cycle-derived citrate acts as a substrate for ATP
citrate lyase (ACLY), providing acetyl-CoA for FA biogenesis.
Palmitate formed from acetyl-CoA and malonyl-CoA undergoes further
processing and elongation to form FA chains, such as saturated FAs
and monosaturated FAs, which can be utilized as building blocks for
cellular membranes and for cellular activity (129). Fig. 2 illustrates the DNL and
cholesterol synthesis pathways that are regulated by a number of
miRNAs, enzymes and genes. Cells usually depend on circulating
exogenous lipids and FAs to fuel normal cellular activity and lipid
storage. However, cancer cells preferentially utilize endogenous
FAs to support oncogenic growth, proliferation and metastasis
(130). The term 'lipogenic
phenotype' has been used to characterize the phenotypic alteration
observed in cancer cells with enhanced DNL and increased endogenous
FA levels regardless of circulating exogenous FA levels (131).
Lipogenic enzymes, including FA synthase (FASN),
ACLY and acetyl-CoA carboxylase (ACACA), are responsible for
cellular lipid metabolism and have been implicated in BC cancer
development and survival, and thus, are recognized as potential
targets for drug discovery in cancer therapeutics (132-134). For instance, FASN, an enzyme
responsible for catalyzing endogenous long-chain FAs, has been
reported to be upregulated in BC cells, and is associated with
tumorigenesis and metastasis, leading to poor BC prognosis
(135). Table III summarizes the targeted
enzymes and genes regulated by miRNAs and the effects in BC lipid
metabolism following miRNA expression in BC cells.
ACLY is a lipogenic enzyme responsible for FA
synthesis regulation. Increased ACLY expression has been associated
with BC development and could be a potential prognostic biomarker
for BC recurrence (138). Liu
et al (139) demonstrated
that overexpression of miR-22 in MCF-7 BC cell lines could
potentially inhibit BC progression and proliferation via
downregulation of the expression of the proto-oncogene ACLY.
Despite miR-22 showing independent tumor-suppressing abilities in
BC, cell tumor formation was enhanced when both miR-22 and ACLY
were overexpressed. Therefore, these results demonstrated the
possibility of miR-22 exerting oncogenic effects when coupled with
ACLY expression (139).
Integrin α-2 (ITGA2) is an integrin gene that
encodes the surface integrin receptor CD49b, and is suspected to be
involved in BC lipid metabolism (140). Adorno-Cruz et al
(140) reported that miR-206
inhibited transcription of ITGA2 by directly binding to the 3′-UTR
of the ITGA2 mRNA. BC cells with miR-206 overexpression exhibited
decreased CD49b levels. Downregulation of the ACLY gene was
observed in ITGA2-knockdown TNBC cell models compared with
controls. ACLY enzyme expression and the cellular concentration of
acetyl-CoA were observed to be reduced following ITGA2 knockdown.
Therefore, miR-206 might have an effect on CD49b signaling pathways
and ACLY expression, which may ultimately affect lipid metabolism
pathways in BC (140).
Previous studies have reported that cancer cells
have a preference in synthesizing FA via DNL for membrane and
energy production to support rapid cell proliferation (141,142). A study conducted by Singh et
al (143) identified FASN,
ACACA and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR)
transcripts as direct gene targets of miR-195 in BC. Overexpression
of miR-195 decreased cellular levels of cholesterol and
triglycerides by reducing the gene expression of FASN, ACACA and
HMGCR transcripts in MCF-7 BC cell lines. These results confirmed
that miR-195 directly targeted key genes involved in the regulation
of DNL and cholesterol biosynthesis, and dysregulated miR-195
expression might have implications in BC tumorigenesis and
progression (143).
Cholesterol is a vital component in lipid rafts that
regulates various cell signaling pathways such as cell binding and
cholesterol biosynthesis that are implicated in cancer cell
migration and metastasis (144,145). There are two key enzymes
involved in the cholesterol synthesis pathway: HMGCR and
hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1) (146). The tumor suppressor miRNA
miR-140-3p-1 was found to be downregulated during TNBC progression
in a study by Bhardwaj et al (147). Further analysis also revealed
that miR-140-3p-1 directly bound to HMGCR and HMGCS1 gene
transcripts at the 3′-UTR to repress transcript activity,
ultimately impacting enzyme and cholesterol levels, which could
contribute to BC development or progression (147).
Sterol regulatory element-binding proteins (SREBPs)
are a family of transcription factors that are involved in cellular
lipid metabolism by activating genes that encode integral lipogenic
enzymes needed for the synthesis of FAs and choles terol (148). The tumor suppressor miRNA
miR-215-5p was reported to negatively regulate SREBP1 expression by
directly binding to the 3′-UTR of the SREBP1 mRNA in BC cells by Wu
et al (149). SREBP1 was
highly expressed in MCF-7 and MDA-MB-231 cells, and patients with
BC compared with controls following the downregulation of
miR-215-5p levels. Furthermore, SREBP1 expression stimulated lipid
metabolism by increasing the expression of lipogenic enzymes such
as FASN, ACLY and acetyl Co-A synthetase 1 to promote cell
migration, invasion and EMT in BC. On the other hand, miR-215-5p
exerted its antitumor effect by downregulating SREBP1 expression
and inhibiting lipid metabolism in BC cells. The results of the
study highlighted the role of the miR-215-5p/SREBP1 axis in the
regulation of BC lipid metabolism, resulting in attenuation of EMT
in BC cells (149).
Early-onset BC was reported to be among the cancers
with the highest mortality rate and disease burden in the year 2019
(150). BC has relatively higher
survival rate compared with other cancer types when detected early;
however, extensive genetic heterogeneity between primary and
disseminated BC due to genomic evolution occurring during BC
development could result in poor prognostic outcomes and strong
limitations in diagnosing and treating patients with BC (151). Early BC detection and diagnosis
are important for effective therapeutic management to improve the
overall survival and decrease the mortality rate of patients with
BC. Although mammograms and ultrasounds are commonly used for
initial diagnosis of the tumor, the existing probability of
acquiring false-positive or false-negative results cannot be
overlooked (152). Furthermore,
expensive and invasive procedures such as tissue biopsies are often
performed to confirm tumor malignancy, and such procedures are
often accompanied by physical discomfort, contributing to the
psychological and financial burden of the patient (153). Therefore, researchers have
shifted their focus to less invasive and more accessible approaches
for early BC detection and risk prediction in the form of human
biofluids, including blood, saliva and urine (154-157).
The concept of metabolic reprogramming observed in
malignant cells has been gaining widespread interest due to
advances in cancer metabolomics (172). 'Omics'-based approaches,
including metabolomics, proteomics, transcriptomics and genomics,
have a wide range of applications in cancer research, including but
not limited to improving the understanding of underlying mechanisms
that lead to BC pathology, and also aid in the identification of
potential diagnostic and prognostic markers, and novel drug targets
for BC therapy (173).
Metabolomics is an emerging, high-throughput technique used in
cancer research with the purpose of measuring and detecting changes
in metabolite levels present in a metabolome or a given biological
sample during malignant cell proliferation and transformation
(174). Metabolite detection is
often performed using techniques such as liquid chromatography-mass
spectrometry, gas chromatography-mass spectrometry and nuclear
magnetic resonance (175). The
metabolome of an organism is largely defined by its genome, and
thus, alterations in the respective genome of an individual due to
diseases such as cancer often show a reflective result in the
cellular function and metabolomic profile of said individual
(176).
Metabolites such as glutamine have been observed to
be upregulated due to the enhanced expression of the amino acid
transporter SLC1A5 in more aggressive forms of BC, with varying
glutamine and β-alanine profiles observed between ER+
and ER− patients with BC (177). Furthermore, 20 different
metabolites involved in arginine, proline, glycerophospholipid,
phenylalanine, tyrosine and tryptophan metabolism pathways have
been observed to be deregulated in patients with IDC compared with
healthy patients (178). Thus,
dysregulated metabolite profiles are capable of differentiating
between healthy individuals and patients with breast cancer
(178). Metabolomic profiling
has also been used as an effective diagnostic tool for cancer
biomarker detection and stratification of cancer subtypes in lung,
colorectal and cervical cancer (179-181), further accentuating the idea
that metabolic profiles have the potential of being utilized as
promising biosignatures in BC. The multifaceted nature of BC tumors
means they can be challenging to treat due to the diverse clinical
presentations and tumor responses to anticancer therapy. In BC
therapeutics, metabolomic studies could help with the
identification of metabolic pathways responsible for mechanisms
involved in cancer drug resistance and anticancer drug responses.
As reported by Granit et al (182), the lipid metabolism metabolite
valproic acid has been found to enhance the anticancer effect of
cisplatin in order to counter cisplatin resistance in TNBC
cells.
Despite their advantages, '-omics' based approaches
do not come without its limitations. Xiao et al (183) highlighted the limitations of
using transcriptomic and genomic data in metabolic research. As
metabolic regulatory networks in cancer cells are complex, solely
relying on either transcriptomic or genomic data to characterize
the complexity of cancer is often proven to be insufficient or
unreliable (183). The inherent
complexity of BC tumors among different individuals due to inter-
and intra-tumor heterogeneity poses challenges in genotype and
phenotype mapping. Therefore, by merging metabolomic-,
transcriptomic- and genomic-based approaches, this research gap
could be minimized to improve BC characterization and BC subtype
refinement for further applications in precision medicine and to
enhance diagnostic and prognostic accuracy in BC (184). Furthermore, operating analytical
equipment and data analysis can be complicated, and thus, advanced
bioinformatics knowledge and skills might be required for more
accurate interpretation and analysis of data. Furthermore, study
limitations, including selection of internal controls, selection of
mass spectrometers and analysis equipment, and sample sizes, need
to be optimized to obtain reliable and valid results (183,184).
Molecular heterogeneity of BC tumors still poses a
challenge to overcome in BC treatment prediction and patient
prognosis. miRNAs can regulate metabolism in tumors either directly
or indirectly by modulating genes and/or enzymes involved in these
pathways. With the knowledge that dysregulated miRNA profiles
reflect changes in metabolism, unique metabolic profiles can be
generated to assist with understanding the pathology behind BC
subtypes (23). There have been
numerous studies that have analyzed the involvement of miRNAs in BC
metabolism in different BC molecular subtypes (109,115,137). For example, the studies by
Msheik et al (109) and
Bacci et al (115) both
looked into the role of miRNAs affecting amino acid transporters
and amino acid metabolism in ER+ BC. miR-216
upregulation coupled with decreased expression of its target,
SLC7A5, was shown to be associated with improved overall survival
in patients with ER+ BC (109). Additionally, altered amino acid
metabolism due to miR-23b-3p overexpression resulted in endocrine
therapy resistance in ER+ BC, offering targetable
pathways to predict and combat endocrine therapy resistance in
ER+ BC (115). The
results of these studies showed that miRNAs could be potential
prognostic and predictive biomarkers by targeting the amino acid
metabolic dependencies observed in ER+ BC to possibly
monitor patient prognosis and predict treatment responses in
patients with ER+ BC (109,115). TNBC is a BC subtype with poor
patient prognosis and has been clinically challenging to treat due
to its non-hormone-dependent nature (13). A study on miR-193b showed that
miR-193b directly targeted the FASN enzyme, which is needed for
TNBC cell survival (137). The
metformin-induced upregulation of miR-193b resulted in decreased
FASN levels, followed by TNBC cell death (137). The findings of the study
provided an insight into the therapeutic effect of miRNAs on
targetable metabolic pathways in aggressive BC subtypes such as
TNBC (137). HER2-positive BC is
another BC subtype with poor patient prognosis and response to
treatment (185). To the best of
our knowledge, there have been no studies that focused on the role
of miRNAs in HER2-positive BC metabolism to date. The combination
of metabolic footprinting and miRNA profiling has potential in BC
subtype stratification refinement and also provides opportunities
for identifying novel drug targets for subtype-specific BC
treatment approaches (186).
However, no studies could be found comparing metabolic profiles
between BC subtypes and investigating the differential role of
miRNAs according to these metabolic profiles thus far.
Research towards understanding the roles miRNAs
serve in BC metabolism is still in its early stages, with more that
remains to be investigated. The aforementioned studies have shown
evidence that miRNAs hold indisputable influence over BC
metabolism, and thus, such insights into the cellular function of
BC pathology is information worth seeking. The combination of
metabolomics, transcriptomics and genomics could be a valuable
diagnostic and prognostic tool in BC when properly validated and
implemented in clinical settings. Further studies on cancer
metabolism integrating '-omics' based approaches and multiomic data
could create reformative and promising avenues for future BC
diagnostics, prognostics and therapeutics, which will ultimately
improve the precision of BC treatment and reduce the global disease
burden of BC.
BC is a highly heterogenous malignancy that affect
millions of women worldwide regardless of their age and ethnicity,
placing a substantial disease burden on the global population. In
the advent of scientific technological advancement, miRNAs have
garnered considerable attention amongst researchers as promising
non-invasive biomarkers for early diagnosis and prognosis of BC,
and as emerging therapeutic targets for individualized BC treatment
in a subtype-specific manner. The present review highlights the
pivotal role miRNAs serve in regulating and reprogramming cellular
metabolism in BC. miRNAs have been shown to be implicated in major
metabolism pathways, including glucose, amino acid, glutamine and
lipid metabolism pathways, by inhibiting or promoting gene
expression and metabolic enzyme expression in BC through their
oncogenic and/or tumor-suppressive abilities. The complex interplay
between miRNAs and metabolic cell signaling pathways contributes to
BC tumorigenesis and oncogenic development, conferring more
'aggressive' BC phenotypes such as increased invasiveness and
metastasis, high risk of relapse and therapeutic resistance, which
could be clinically challenging to treat. Advances in understanding
pathophysiological mechanisms of miRNAs in BC metabolic
dysregulation by implementing '-omics'-based approaches in
scientific research can instigate future breakthroughs in cancer
therapeutics and provide potential developments in precision
medicine for subtype-specific BC treatment to further improve
clinical outcomes and patient survival in BC.
Not applicable.
WXL conceived the idea for the manuscript. WXL and
BSY drafted the manuscript, and designed the tables and figures.
The manuscript was revised by YKC, RM, GCT and MIAW. Data
authentication is not applicable. All authors have read and
approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the Ministry of Science,
Technology and Innovation of Malaysia under the Technology
Development Fund 1 [grant no. TeD (1)/2023/5450832
(TDF06221585)].
|
1
|
International Agency for Research on
Cancer: Global Cancer Observatory. Cancer Today. Accessed on
September 22, 2024https://gco.iarc.fr/today/online-analysis-multi-bars.
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S
and Soerjomataram I: Current and future burden of breast cancer:
Global statistics for 2020 and 2040. Breast. 66:15–23. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee MS, 'Azmiyaty Amar Ma' Ruf C, Nadhirah
Izhar DP, Nafisah Ishak S, Wan Jamaluddin WS, Ya'acob SNM and
Kamaluddin MN: Awareness on breast cancer screening in Malaysia: A
cross sectional study. Biomedicine (Taipei). 9:182019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Momenimovahed Z and Salehiniya H:
Epidemiological characteristics of and risk factors for breast
cancer in the world. Breast Cancer (Dove Med Press). 11:151–164.
2019.PubMed/NCBI
|
|
6
|
Malhotra GK, Zhao X, Band H and Band V:
Histological, molecular and functional subtypes of breast cancers.
Cancer Biol Ther. 10:955–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watkins EJ: Overview of breast cancer.
JAAPA. 32:13–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Posner MC and Wolmark N: Non-invasive
breast carcinoma. Breast Cancer Res Treat. 21:155–164. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Corben AD: Pathology of invasive breast
disease. Surg Clin North Am. 93:363–392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma GN, Dave R, Sanadya J, Sharma P and
Sharma KK: Various types and management of breast cancer: An
overview. J Adv Pharm Technol Res. 1:109–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yip CH and Rhodes A: Estrogen and
progesterone receptors in breast cancer. Future Oncol.
10:2293–2301. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iqbal N and Iqbal N: Human epidermal
growth factor receptor 2 (HER2) in cancers: Overexpression and
therapeutic implications. Mol Biol Int. 2014:8527482014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Derakhshan F and Reis-Filho JS:
Pathogenesis of triple-negative breast cancer. Annu Rev Pathol.
17:181–204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan J and Le A: The heterogeneity of
breast cancer metabolism. Adv Exp Med Biol. 1311:89–101. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahn S, Woo JW, Lee K and Park SY: HER2
status in breast cancer: Changes in guidelines and complicating
factors for interpretation. J Pathol Transl Med. 54:34–44. 2020.
View Article : Google Scholar :
|
|
16
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Serrano-Carbajal EA, Espinal-Enríquez J
and Hernández-Lemus E: Targeting metabolic deregulation landscapes
in breast cancer subtypes. Front Oncol. 10:972020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Zhang S and Wang X: The metabolic
mechanisms of breast cancer metastasis. Front Oncol. 10:6024162021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan B, Manley J, Lee J and Singh SR: The
emerging roles of microRNAs in cancer metabolism. Cancer Lett.
356:301–308. 2015. View Article : Google Scholar
|
|
21
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muñoz JP, Pérez-Moreno P, Pérez Y and
Calaf GM: The role of MicroRNAs in breast cancer and the challenges
of their clinical application. Diagnostics (Basel). 13:30722023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suriya Muthukumaran N, Velusamy P, Akino
Mercy CS, Langford D, Natarajaseenivasan K and Shanmughapriya S:
MicroRNAs as regulators of cancer cell energy metabolism. J Pers
Med. 12:13292022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar
|
|
25
|
Diener C, Keller A and Meese E: The
miRNA-target interactions: An underestimated intricacy. Nucleic
Acids Res. 52:1544–1557. 2024. View Article : Google Scholar
|
|
26
|
Liu L, He J, Wei X, Wan G, Lao Y, Xu W, Li
Z, Hu H, Hu Z, Luo X, et al: MicroRNA-20a-mediated loss of
autophagy contributes to breast tumorigenesis by promoting genomic
damage and instability. Oncogene. 36:5874–5884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: MiR-23a promotes TGF-β1-induced EMT and
tumor metastasis in breast cancer cells by directly targeting CDH1
and activating Wnt/β-catenin signaling. Oncotarget. 8:69538–69550.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao F and Tian J: FOXK1, regulated by
miR-365-3p, promotes cell growth and EMT indicates unfavorable
prognosis in breast cancer. Onco Targets Ther. 13:623–634. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakrani MN, Wineland RH and Anjum F:
Physiology, glucose metabolism. StatPearls [Internet]. StatPearls
Publishing; Treasure Island, FL: 2023
|
|
31
|
Paredes-Flores MA and Mohiuddin SS:
Biochemistry, glycogenolysis. StatPearls [Internet]. StatPearls
Publishing; Treasure Island, FL: 2022
|
|
32
|
Patino SC and Orrick JA: Biochemistry,
glycogenesis. StatPearls [Internet]. StatPearls Publishing;
Treasure Island, FL: 2023
|
|
33
|
Dunn J and Grider MH: Physiology,
adenosine triphosphate. StatPearls [Internet]. StatPearls
Publishing; Treasure Island, FL: 2023
|
|
34
|
Pavlova NN, Zhu J and Thompson CB: The
hallmarks of cancer metabolism: Still emerging. Cell Metab.
34:355–377. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pascale RM, Calvisi DF, Simile MM, Feo CF
and Feo F: The Warburg effect 97 years after its discovery. Cancers
(Basel). 12:28192020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu L, Chen X, Wang L and Chen S: The sweet
trap in tumors: Aerobic glycolysis and potential targets for
therapy. Oncotarget. 7:38908–38926. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nong S, Han X, Xiang Y, Qian Y, Wei Y,
Zhang T, Tian K, Shen K, Yang J and Ma X: Metabolic reprogramming
in cancer: Mechanisms and therapeutics. MedComm (2020). 4:e2182023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chaudhry R and Varacallo M: Biochemistry,
glycolysis. StatPearls [Internet]. StatPearls Publishing; Treasure
Island, FL: 2023
|
|
42
|
Lenzen S: A fresh view of glycolysis and
glucokinase regulation: History and current status. J Biol Chem.
289:12189–12194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roberts DJ and Miyamoto S: Hexokinase II
integrates energy metabolism and cellular protection: Akting on
mitochondria and TORCing to autophagy. Cell Death Differ.
22:248–257. 2015. View Article : Google Scholar :
|
|
44
|
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH,
Liang S, Li B, Li Y, Li D, Wang ED and Liu MF: A novel
miR-155/miR-143 cascade controls glycolysis by regulating
hexokinase 2 in breast cancer cells. EMBO J. 31:1985–1998. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu T, Ye P, Ye Y and Han B: MicroRNA-216b
targets HK2 to potentiate autophagy and apoptosis of breast cancer
cells via the mTOR signaling pathway. Int J Biol Sci. 17:2970–2983.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li L, Zhang X, Lin Y, Ren X, Xie T, Lin J,
Wu S and Ye Q: Let-7b-5p inhibits breast cancer cell growth and
metastasis via repression of hexokinase 2-mediated aerobic
glycolysis. Cell Death Discov. 9:1142023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li L, Peng G, Liu X, Zhang Y, Han H and
Liu ZR: Pyruvate kinase M2 coordinates metabolism switch between
glycolysis and glutaminolysis in cancer cells. iScience.
23:1016842020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hsu MC and Hung WC: Pyruvate kinase M2
fuels multiple aspects of cancer cells: From cellular metabolism,
transcriptional regulation to extracellular signaling. Mol Cancer.
17:352018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Park B, Kim JY, Riffey OF, Dowker-Key P,
Bruckbauer A, McLoughlin J, Bettaieb A and Donohoe DR: Pyruvate
kinase M1 regulates butyrate metabolism in cancerous colonocytes.
Sci Rep. 12:87712022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Schormann N, Hayden KL, Lee P, Banerjee S
and Chattopadhyay D: An overview of structure, function, and
regulation of pyruvate kinases. Protein Sci. 28:1771–1784. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Amin S, Yang P and Li Z: Pyruvate kinase
M2: A multifarious enzyme in non-canonical localization to promote
cancer progression. Biochim Biophys Acta Rev Cancer. 1871:331–341.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Israelsen WJ and Vander Heiden MG:
Pyruvate kinase: Function, regulation and role in cancer. Semin
Cell Dev Biol. 43:43–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wen YY, Liu WT, Sun HR, Ge X, Shi ZM, Wang
M, Li W, Zhang JY, Liu LZ and Jiang BH: IGF-1-mediated
PKM2/β-catenin/miR-152 regulatory circuit in breast cancer. Sci
Rep. 7:158972017. View Article : Google Scholar
|
|
54
|
Xu Q, Liu LZ, Yin Y, He J, Li Q, Qian X,
You Y, Lu Z, Peiper SC, Shu Y and Jiang BH: Regulatory circuit of
PKM2/NF-κB/miR-148a/152-modulated tumor angiogenesis and cancer
progression. Oncogene. 34:5482–5493. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yao A, Xiang Y, Si YR, Fan LJ, Li JP, Li
H, Guo W, He HX, Liang XJ, Tan Y, et al: PKM2 promotes glucose
metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback
loop in breast cancer cells. J Cell Biochem. 120:6542–6554. 2019.
View Article : Google Scholar
|
|
56
|
Chen Y, Cen L, Guo R, Huang S and Chen D:
Roles and mechanisms of phosphoglycerate kinase 1 in cancer. Bull
Cancer. 109:1298–1307. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ye T, Liang Y, Zhang D and Zhang X:
MicroRNA-16-1-3p represses breast tumor growth and metastasis by
inhibiting PGK1-mediated warburg effect. Front Cell Dev Biol.
8:6151542020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ran F, Zhang Y, Shi Y, Liu J, Li H, Ding L
and Ye Q: miR-1224-3p promotes breast cancer cell proliferation and
migration through PGM5-mediated aerobic glycolysis. J Oncol.
2021:55297702021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li L, Kang L, Zhao W, Feng Y, Liu W, Wang
T, Mai H, Huang J, Chen S, Liang Y, et al: miR-30a-5p suppresses
breast tumor growth and metastasis through inhibition of
LDHA-mediated Warburg effect. Cancer Lett. 400:89–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xiao X, Huang X, Ye F, Chen B, Song C, Wen
J, Zhang Z, Zheng G, Tang H and Xie X: The miR-34a-LDHA axis
regulates glucose metabolism and tumor growth in breast cancer. Sci
Rep. 6:217352016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ge X, Lyu P, Cao Z, Li J, Guo G, Xia W and
Gu Y: Overexpression of miR-206 suppresses glycolysis,
proliferation and migration in breast cancer cells via PFKFB3
targeting. Biochem Biophys Res Commun. 463:1115–1121. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Telang S, Yalcin A, Clem AL, Bucala R,
Lane AN, Eaton JW and Chesney J: Ras transformation requires
metabolic control by 6-phosphofructo-2-kinase. Oncogene.
25:7225–7234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kim JW and Dang CV: Multifaceted roles of
glycolytic enzymes. Trends Biochem Sci. 30:142–150. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ahmad A, Aboukameel A, Kong D, Wang Z,
Sethi S, Chen W, Sarkar FH and Raz A: Phosphoglucose
isomerase/autocrine motility factor mediates epithelial-mesenchymal
transition regulated by miR-200 in breast cancer cells. Cancer Res.
71:3400–3409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guda MR, Asuthkar S, Labak CM, Tsung AJ,
Alexandrov I, Mackenzie MJ, Prasad DV and Velpula KK: Targeting
PDK4 inhibits breast cancer metabolism. Am J Cancer Res.
8:1725–1738. 2018.PubMed/NCBI
|
|
66
|
Lu H, Forbes RA and Verma A:
Hypoxia-inducible factor 1 activation by aerobic glycolysis
implicates the Warburg effect in carcinogenesis. J Biol Chem.
277:23111–23115. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhai Z, Mu T, Zhao L, Li Y, Zhu D and Pan
Y: MiR-181a-5p facilitates proliferation, invasion, and glycolysis
of breast cancer through NDRG2-mediated activation of PTEN/AKT
pathway. Bioengineered. 13:83–95. 2022. View Article : Google Scholar :
|
|
68
|
Lang L, Tao J, Yang C and Li W: Tumor
suppressive role of microRNA-4731-5p in breast cancer through
reduction of PAICS-induced FAK phosphorylation. Cell Death Discov.
8:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ziello JE, Jovin IS and Huang Y:
Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its
potential for therapeutic intervention in malignancy and ischemia.
Yale J Biol Med. 80:51–60. 2007.PubMed/NCBI
|
|
70
|
Semenza GL: HIF-1: Upstream and downstream
of cancer metabolism. Curr Opin Genet Dev. 20:51–56. 2010.
View Article : Google Scholar :
|
|
71
|
Du Y, Wei N, Ma R, Jiang SH and Song D: A
miR-210-3p regulon that controls the Warburg effect by modulating
HIF-1α and p53 activity in triple-negative breast cancer. Cell
Death Dis. 11:7312020. View Article : Google Scholar
|
|
72
|
Jiang Y, Zhang M, Yu D, Hou G, Wu J and Li
F: CircRBM33 downregulation inhibits hypoxia-induced glycolysis and
promotes apoptosis of breast cancer cells via a
microRNA-542-3p/HIF-1α axis. Cell Death Discov. 8:1262022.
View Article : Google Scholar
|
|
73
|
Cao L, Wang M, Dong Y, Xu B, Chen J, Ding
Y, Qiu S, Li L, Karamfilova Zaharieva E, Zhou X and Xu Y: Circular
RNA circRNF20 promotes breast cancer tumorigenesis and Warburg
effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 11:1452020.
View Article : Google Scholar
|
|
74
|
Zhao Y, He J, Yang L, Luo Q and Liu Z:
Histone deacetylase-3 modification of MicroRNA-31 promotes cell
proliferation and aerobic glycolysis in breast cancer and is
predictive of poor prognosis. J Breast Cancer. 21:112–123. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kurmi K and Haigis MC: Nitrogen metabolism
in cancer and immunity. Trends Cell Biol. 30:408–424. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant
GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et
al: Metabolism. Lysosomal amino acid transporter SLC38A9 signals
arginine sufficiency to mTORC1. Science. 347:188–194. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yeon A, You S, Kim M, Gupta A, Park MH,
Weisenberger DJ, Liang G and Kim J: Rewiring of cisplatin-resistant
bladder cancer cells through epigenetic regulation of genes
involved in amino acid metabolism. Theranostics. 8:4520–4534. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gao M, Monian P, Quadri N, Ramasamy R and
Jiang X: Glutaminolysis and transferrin regulate ferroptosis. Mol
Cell. 59:298–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wei Z, Liu X, Cheng C, Yu W and Yi P:
Metabolism of amino acids in cancer. Front Cell Dev Biol.
8:6038372021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu
J, Liu X, Chen CH, Fadare O, Pizzo DP, et al: Cancer-cell-secreted
exosomal miR-105 promotes tumour growth through the MYC-dependent
metabolic reprogramming of stromal cells. Nat Cell Biol.
20:597–609. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Cruzat V, Macedo Rogero M, Noel Keane K,
Curi R and Newsholme P: Glutamine: Metabolism and immune function,
supplementation and clinical translation. Nutrients. 10:15642018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Choi YK and Park KG: Targeting glutamine
metabolism for cancer treatment. Biomol Ther (Seoul). 26:19–28.
2018. View Article : Google Scholar
|
|
83
|
Wise DR and Thompson CB: Glutamine
addiction: A new therapeutic target in cancer. Trends Biochem Sci.
35:427–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jin J, Byun JK, Choi YK and Park KG:
Targeting glutamine metabolism as a therapeutic strategy for
cancer. Exp Mol Med. 55:706–715. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jin L, Alesi GN and Kang S: Glutaminolysis
as a target for cancer therapy. Oncogene. 35:3619–3625. 2016.
View Article : Google Scholar
|
|
86
|
Haikala HM, Marques E, Turunen M and
Klefström J: Myc requires RhoA/SRF to reprogram glutamine
metabolism. Small GTPases. 9:274–282. 2018. View Article : Google Scholar :
|
|
87
|
Budczies J, Pfitzner BM, Györffy B, Winzer
KJ, Radke C, Dietel M, Fiehn O and Denkert C: Glutamate enrichment
as new diagnostic opportunity in breast cancer. Int J Cancer.
136:1619–1628. 2015. View Article : Google Scholar
|
|
88
|
Herner A, Sauliunaite D, Michalski CW,
Erkan M, De Oliveira T, Abiatari I, Kong B, Esposito I, Friess H
and Kleeff J: Glutamate increases pancreatic cancer cell invasion
and migration via AMPA receptor activation and Kras-MAPK signaling.
Int J Cancer. 129:2349–2359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mukha A, Kahya U, Linge A, Chen O, Löck S,
Lukiyanchuk V, Richter S, Alves TC, Peitzsch M, Telychko V, et al:
GLS-driven glutamine catabolism contributes to prostate cancer
radiosensitivity by regulating the redox state, stemness and
ATG5-mediated autophagy. Theranostics. 11:7844–7868. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Xiong J, Wang N, Zhong HJ, Cui BW, Cheng
S, Sun R, Chen JY, Xu PP, Cai G, Wang L, et al: SLC1A1 mediated
glutamine addiction and contributed to natural killer T-cell
lymphoma progression with immunotherapeutic potential.
EBioMedicine. 72:1036142021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cluntun AA, Lukey MJ, Cerione RA and
Locasale JW: Glutamine metabolism in cancer: Understanding the
heterogeneity. Trends Cancer. 3:169–180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
El Ansari R, McIntyre A, Craze ML, Ellis
IO, Rakha EA and Green AR: Altered glutamine metabolism in breast
cancer; subtype dependencies and alternative adaptations.
Histopathology. 72:183–190. 2018. View Article : Google Scholar
|
|
93
|
Lieu EL, Nguyen T, Rhyne S and Kim J:
Amino acids in cancer. Exp Mol Med. 52:15–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kung HN, Marks JR and Chi JT: Glutamine
synthetase is a genetic determinant of cell type-specific glutamine
independence in breast epithelia. PLoS Genet. 7:e10022292011.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lampa M, Arlt H, He T, Ospina B, Reeves J,
Zhang B, Murtie J, Deng G, Barberis C, Hoffmann D, et al:
Glutaminase is essential for the growth of triple-negative breast
cancer cells with a deregulated glutamine metabolism pathway and
its suppression synergizes with mTOR inhibition. PLoS One.
12:e01850922017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Thewes V, Simon R, Hlevnjak M, Schlotter
M, Schroeter P, Schmidt K, Wu Y, Anzeneder T, Wang W, Windisch P,
et al: The branched-chain amino acid transaminase 1 sustains growth
of antiestrogen-resistant and ERα-negative breast cancer. Oncogene.
36:4124–4134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Craze ML, El-Ansari R, Aleskandarany MA,
Cheng KW, Alfarsi L, Masisi B, Diez-Rodriguez M, Nolan CC, Ellis
IO, Rakha EA and Green AR: Glutamate dehydrogenase (GLUD1)
expression in breast cancer. Breast Cancer Res Treat. 174:79–91.
2019. View Article : Google Scholar
|
|
98
|
Cao Y, Lin SH, Wang Y, Chin YE, Kang L and
Mi J: Glutamic pyruvate transaminase GPT2 promotes tumorigenesis of
breast cancer cells by activating sonic hedgehog signaling.
Theranostics. 7:3021–3033. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhang L and Han J: Branched-chain amino
acid transaminase 1 (BCAT1) promotes the growth of breast cancer
cells through improving mTOR-mediated mitochondrial biogenesis and
function. Biochem Biophys Res Commun. 486:224–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Masisi BK, El Ansari R, Alfarsi L, Craze
ML, Jewa N, Oldfield A, Cheung H, Toss M, Rakha EA and Green AR:
The biological and clinical significance of glutaminase in luminal
breast cancer. Cancers (Basel). 13:39632021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kandasamy P, Gyimesi G, Kanai Y and
Hediger MA: Amino acid transporters revisited: New views in health
and disease. Trends Biochem Sci. 43:752–789. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yadav P, Sharma P, Sundaram S, Venkatraman
G, Bera AK and Karunagaran D: SLC7A11/xCT is a target of miR-5096
and its restoration partially rescues miR-5096-mediated ferroptosis
and anti-tumor effects in human breast cancer cells. Cancer Lett.
522:211–224. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Liu Y, Hu Y, Jiang Y, Bu J and Gu X:
Targeting ferroptosis, the achilles' heel of breast cancer: A
review. Front Pharmacol. 13:10361402022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX,
Cao DX, He M, Chen GQ, He JR and Zhao Q: MicroRNA-26b is
underexpressed in human breast cancer and induces cell apoptosis by
targeting SLC7A11. FEBS Lett. 585:1363–1367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Sun D, Li YC and Zhang XY: Lidocaine
promoted ferroptosis by targeting miR-382-5p/SLC7A11 axis in
ovarian and breast cancer. Front Pharmacol. 12:6812232021.
View Article : Google Scholar
|
|
106
|
Wang J, Yang K, Cao J and Li L: Knockdown
of circular RNA septin 9 inhibits the malignant progression of
breast cancer by reducing the expression of solute carrier family 1
member 5 in a microRNA-149-5p-dependent manner. Bioengineered.
12:10624–10637. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
van Geldermalsen M, Wang Q, Nagarajah R,
Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N,
et al: ASCT2/SLC1A5 controls glutamine uptake and tumour growth in
triple-negative basal-like breast cancer. Oncogene. 35:3201–3108.
2016. View Article : Google Scholar :
|
|
108
|
Kinslow CJ, Tang A, Chaudhary KR and Cheng
SK: Prevalence of estrogen receptor alpha (ESR1) somatic mutations
in breast cancer. JNCI Cancer Spectr. 6:pkac0602022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Msheik ZS, Nassar FJ, Chamandi G, Itani
AR, Gadaleta E, Chalala C, Alwan N and Nasr RR: miR-126 decreases
proliferation and mammosphere formation of MCF-7 and predicts
prognosis of ER+ breast cancer. Diagnostics (Basel). 12:7452022.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yanagida O, Kanai Y, Chairoungdua A, Kim
DK, Segawa H, Nii T, Cha SH, Matsuo H, Fukushima J, Fukasawa Y, et
al: Human L-type amino acid transporter 1 (LAT1): Characterization
of function and expression in tumor cell lines. Biochim Biophys
Acta. 1514:291–302. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Saito Y and Soga T: Amino acid
transporters as emerging therapeutic targets in cancer. Cancer Sci.
112:2958–2965. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Li Y, Wang W, Wu X, Ling S, Ma Y and Huang
P: SLC7A5 serves as a prognostic factor of breast cancer and
promotes cell proliferation through activating AKT/mTORC1 signaling
pathway. Ann Transl Med. 9:8922021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kurozumi S, Kaira K, Matsumoto H, Kurosumi
M, Yokobori T, Kanai Y, Sekine C, Honda C, Katayama A, Furuya M, et
al: Association of L-type amino acid transporter 1 (LAT1) with the
immune system and prognosis in invasive breast cancer. Sci Rep.
12:27422022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Törnroos R, Tina E and Göthlin Eremo A:
SLC7A5 is linked to increased expression of genes related to
proliferation and hypoxia in estrogen-receptor-positive breast
cancer. Oncol Rep. 47:172022. View Article : Google Scholar
|
|
115
|
Bacci M, Lorito N, Ippolito L, Ramazzotti
M, Luti S, Romagnoli S, Parri M, Bianchini F, Cappellesso F, Virga
F, et al: Reprogramming of amino acid transporters to support
aspartate and glutamate dependency sustains endocrine resistance in
breast cancer. Cell Rep. 28:104–118.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Delgir S, Ilkhani K, Safi A, Rahmati Y,
Montazari V, Zaynali-Khasraghi Z, Seif F, Bastami M and Alivand MR:
The expression of miR-513c and miR-3163 was downregulated in tumor
tissues compared with normal adjacent tissue of patients with
breast cancer. BMC Med Genomics. 14:1802021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O'Connor STF, Li S, Chin R, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Figueira I, Godinho-Pereira J, Galego S,
Maia J, Haskó J, Molnár K, Malhó R, Costa-Silva B, Wilhelm I,
Krizbai IA and Brito MA: MicroRNAs and extracellular vesicles as
distinctive biomarkers of precocious and advanced stages of breast
cancer brain metastases development. Int J Mol Sci. 22:52142021.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Lu C, Zhao Y, Wang J, Shi W, Dong F, Xin
Y, Zhao X and Liu C: Breast cancer cell-derived extracellular
vesicles transfer miR-182-5p and promote breast carcinogenesis via
the CMTM7/EGFR/AKT axis. Mol Med. 27:782021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Yang M, Zhang Y, Li M, Liu X and Darvishi
M: The various role of microRNAs in breast cancer angiogenesis,
with a special focus on novel miRNA-based delivery strategies.
Cancer Cell Int. 23:242023. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Guo L, Kong D, Liu J, Zhan L, Luo L, Zheng
W, Zheng Q, Chen C and Sun S: Breast cancer heterogeneity and its
implication in personalized precision therapy. Exp Hematol Oncol.
12:32023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Muciño-Olmos EA, Vázquez-Jiménez A,
López-Esparza DE, Maldonado V, Valverde M and Resendis-Antonio O:
MicroRNAs regulate metabolic phenotypes during multicellular tumor
spheroids progression. Front Oncol. 10:5823962020. View Article : Google Scholar
|
|
123
|
Fu Y, Zou T, Shen X, Nelson PJ, Li J, Wu
C, Yang J, Zheng Y, Bruns C, Zhao Y, et al: Lipid metabolism in
cancer progression and therapeutic strategies. MedComm (2020).
2:27–59. 2020. View Article : Google Scholar
|
|
124
|
Gyamfi D, Ofori Awuah E and Owusu S:
Chapter 2-lipid metabolism: An overview. Patel VB: The Molecular
Nutrition of Fats. Academic Press; Cambridge, MA, USA: pp. 17–32.
2019
|
|
125
|
Burdge GC and Calder PC: Introduction to
fatty acids and lipids. World Rev Nutr Diet. 112:1–16. 2015.
View Article : Google Scholar
|
|
126
|
Zechner R, Zimmermann R, Eichmann TO,
Kohlwein SD, Haemmerle G, Lass A and Madeo F: FAT SIGNALS-lipases
and lipolysis in lipid metabolism and signaling. Cell Metab.
15:279–291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Monaco ME: Fatty acid metabolism in breast
cancer subtypes. Oncotarget. 8:29487–29500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Park JK, Coffey NJ, Limoges A and Le A:
The Heterogeneity of lipid metabolism in cancer. Adv Exp Med Biol.
1063:33–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Vasseur S and Guillaumond F: Lipids in
cancer: A global view of the contribution of lipid pathways to
metastatic formation and treatment resistance. Oncogenesis.
11:462022. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Koundouros N and Poulogiannis G:
Reprogramming of fatty acid metabolism in cancer. Br J Cancer.
122:4–22. 2020. View Article : Google Scholar :
|
|
131
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Alo' PL, Visca P, Marci A, Mangoni A,
Botti C and Di Tondo U: Expression of fatty acid synthase (FAS) as
a predictor of recurrence in stage I breast carcinoma patients.
Cancer. 77:474–482. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chajès V, Cambot M, Moreau K, Lenoir GM
and Joulin V: Acetyl-CoA carboxylase alpha is essential to breast
cancer cell survival. Cancer Res. 66:5287–5294. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Mashima T, Seimiya H and Tsuruo T: De novo
fatty-acid synthesis and related pathways as molecular targets for
cancer therapy. Br J Cancer. 100:1369–1372. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xu S, Chen T, Dong L, Li T, Xue H, Gao B,
Ding X, Wang H and Li H: Fatty acid synthase promotes breast cancer
metastasis by mediating changes in fatty acid metabolism. Oncol
Lett. 21:272021.
|
|
136
|
Wang J, Zhang X, Shi J, Cao P, Wan M,
Zhang Q, Wang Y, Kridel SJ, Liu W, Xu J, et al: Fatty acid synthase
is a primary target of MiR-15a and MiR-16-1 in breast cancer.
Oncotarget. 7:78566–78576. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wahdan-Alaswad RS, Cochrane DR, Spoelstra
NS, Howe EN, Edgerton SM, Anderson SM, Thor AD and Richer JK:
Metformin-induced killing of triple-negative breast cancer cells is
mediated by reduction in fatty acid synthase via miRNA-193b. Horm
Cancer. 5:374–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Chen Y, Li K, Gong D, Zhang J, Li Q, Zhao
G and Lin P: ACLY: A biomarker of recurrence in breast cancer.
Pathol Res Pract. 216:1530762020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Liu H, Huang X and Ye T: MiR-22
down-regulates the proto-oncogene ATP citrate lyase to inhibit the
growth and metastasis of breast cancer. Am J Transl Res.
10:659–669. 2018.PubMed/NCBI
|
|
140
|
Adorno-Cruz V, Hoffmann AD, Liu X,
Dashzeveg NK, Taftaf R, Wray B, Keri RA and Liu H: ITGA2 promotes
expression of ACLY and CCND1 in enhancing breast cancer stemness
and metastasis. Genes Dis. 8:493–508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Daniëls VW, Smans K, Royaux I, Chypre M,
Swinnen JV and Zaidi N: Cancer cells differentially activate and
thrive on de novo lipid synthesis pathways in a low-lipid
environment. PLoS One. 9:e1069132014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Simeone P, Tacconi S, Longo S, Lanuti P,
Bravaccini S, Pirini F, Ravaioli S, Dini L and Giudetti AM:
Expanding roles of De Novo lipogenesis in breast cancer. Int J
Environ Res Public Health. 18:35752021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1.
Sci Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Yang Z, Qin W, Chen Y, Yuan B, Song X,
Wang B, Shen F, Fu J and Wang H: Cholesterol inhibits
hepatocellular carcinoma invasion and metastasis by promoting CD44
localization in lipid rafts. Cancer Lett. 429:66–77. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yang YF, Jan YH, Liu YP, Yang CJ, Su CY,
Chang YC, Lai TC, Chiou J, Tsai HY, Lu J, et al: Squalene synthase
induces tumor necrosis factor receptor 1 enrichment in lipid rafts
to promote lung cancer metastasis. Am J Respir Crit Care Med.
190:675–687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Vona R, Iessi E and Matarrese P: Role of
cholesterol and lipid rafts in cancer signaling: A promising
therapeutic opportunity? Front Cell Dev Biol. 9:6229082021.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Bhardwaj A, Singh H, Trinidad CM,
Albarracin CT, Hunt KK and Bedrosian I: The isomiR-140-3p-regulated
mevalonic acid pathway as a potential target for prevention of
triple negative breast cancer. Breast Cancer Res. 20:1502018.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
DeBose-Boyd RA and Ye J: SREBPs in lipid
metabolism, insulin signaling, and beyond. Trends Biochem Sci.
43:358–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wu CL, Xu LL, Peng J and Zhang DH: Al-MPS
obstructs EMT in breast cancer by inhibiting lipid metabolism via
miR-215-5p/SREBP1. Endocrinology. 163:bqac0402022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhao J, Xu L, Sun J, Song M, Wang L, Yuan
S, Zhu Y, Wan Z, Larsson S, Tsilidis K, et al: Global trends in
incidence, death, burden and risk factors of early-onset cancer
from 1990 to 2019. BMJ Oncol. 2:e0000492023. View Article : Google Scholar
|
|
151
|
Ellsworth RE, Blackburn HL, Shriver CD,
Soon-Shiong P and Ellsworth DL: Molecular heterogeneity in breast
cancer: State of the science and implications for patient care.
Semin Cell Dev Biol. 64:65–72. 2017. View Article : Google Scholar
|
|
152
|
Ho TQH, Bissell MCS, Kerlikowske K,
Hubbard RA, Sprague BL, Lee CI, Tice JA, Tosteson ANA and
Miglioretti DL: Cumulative probability of false-positive results
after 10 years of screening with digital breast tomosynthesis vs
digital mammography. JAMA Netw Open. 5:e2224402022. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
El Hachem Z, Zoghbi M and Hallit S:
Psychosocial consequences of false-positive results in screening
mammography. J Family Med Prim Care. 8:419–425. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Park S, Ahn S, Kim JY, Kim J, Han HJ,
Hwang D, Park J, Park HS, Park S, Kim GM, et al: Blood test for
breast cancer screening through the detection of tumor-associated
circulating transcripts. Int J Mol Sci. 23:91402022. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Gilson Sena IF, Fernandes LL, Lorandi LL,
Santana TV, Cintra L, Lima IF, Iwai LK, Kramer JM, Birbrair A and
Heller D: Identification of early biomarkers in saliva in
genetically engineered mouse model C(3)1-TAg of breast cancer. Sci
Rep. 12:115442022. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Giró Benet J, Seo M, Khine M, Gumà Padró
J, Pardo Martnez A and Kurdahi F: Breast cancer detection by
analyzing the volatile organic compound (VOC) signature in human
urine. Sci Rep. 12:148732022. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Zhang L, Xiao H, Karlan S, Zhou H, Gross
J, Elashoff D, Akin D, Yan X, Chia D, Karlan B and Wong DT:
Discovery and preclinical validation of salivary transcriptomic and
proteomic biomarkers for the non-invasive detection of breast
cancer. PLoS One. 5:e155732010. View Article : Google Scholar
|
|
158
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Garrido-Palacios A, Rojas Carvajal AM,
Núñez-Negrillo AM, Cortés-Martín J, Sánchez-García JC and
Aguilar-Cordero MJ: MicroRNA dysregulation in early breast cancer
diagnosis: A systematic review and meta-analysis. Int J Mol Sci.
24:82702023. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Kashyap D and Kaur H: Cell-free miRNAs as
non-invasive biomarkers in breast cancer: Significance in early
diagnosis and metastasis prediction. Life Sci. 246:1174172020.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Papadaki C, Stoupis G, Tsalikis L,
Monastirioti A, Papadaki M, Maliotis N, Stratigos M, Mastrostamatis
G, Mavroudis D and Agelaki S: Circulating miRNAs as a marker of
metastatic disease and prognostic factor in metastatic breast
cancer. Oncotarget. 10:966–981. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Chen X, Wang YW, Zhu WJ, Li Y, Liu L, Yin
G and Gao P: A 4-microRNA signature predicts lymph node metastasis
and prognosis in breast cancer. Hum Pathol. 76:122–132. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Gong C, Tan W, Chen K, You N, Zhu S, Liang
G, Xie X, Li Q, Zeng Y, Ouyang N, et al: Prognostic value of a
BCSC-associated MicroRNA signature in hormone receptor-positive
HER2-negative breast cancer. EBioMedicine. 11:199–209. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Fu Z, Wang L, Li S, Chen F, Au-Yeung KK
and Shi C: MicroRNA as an important target for anticancer drug
development. Front Pharmacol. 12:7363232021. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Chakrabortty A, Patton DJ, Smith BF and
Agarwal P: miRNAs: Potential as biomarkers and therapeutic targets
for cancer. Genes (Basel). 14:13752023. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai
CY, Hou MF, Lee JN, Wu DC, Wang SC and Tsai EM: miR-125a-5p is a
prognostic biomarker that targets HDAC4 to suppress breast
tumorigenesis. Oncotarget. 6:494–509. 2015. View Article : Google Scholar :
|
|
167
|
Søkilde R, Persson H, Ehinger A, Pirona
AC, Fernö M, Hegardt C, Larsson C, Loman N, Malmberg M, Rydén L, et
al: Refinement of breast cancer molecular classification by miRNA
expression profiles. BMC Genomics. 20:5032019. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C,
Wang X, Luo Z, Wang J, Liu S, et al: microRNA-21 promotes breast
cancer proliferation and metastasis by targeting LZTFL1. BMC
Cancer. 19:7382019. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Arisan ED, Rencuzogullari O,
Cieza-Borrella C, Miralles Arenas F, Dwek M, Lange S and
Uysal-Onganer P: MiR-21 is required for the epithelial-mesenchymal
transition in MDA-MB-231 breast cancer cells. Int J Mol Sci.
22:15572021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Wang J, Wang Q, Guan Y, Sun Y, Wang X,
Lively K, Wang Y, Luo M, Kim JA, Murphy E, et al: Breast cancer
cell-derived microRNA-155 suppresses tumor progression via
enhancing immune cell recruitment and antitumor function. J Clin
Invest. 132:e1572482022. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Xu W, Song C, Wang X, Li Y, Bai X, Liang
X, Wu J and Liu J: Downregulation of miR-155-5p enhances the
anti-tumor effect of cetuximab on triple-negative breast cancer
cells via inducing cell apoptosis and pyroptosis. Aging (Albany
NY). 13:228–240. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Schmidt DR, Patel R, Kirsch DG, Lewis CA,
Vander Heiden MG and Locasale JW: Metabolomics in cancer research
and emerging applications in clinical oncology. CA Cancer J Clin.
71:333–358. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Rossi C, Cicalini I, Cufaro MC, Consalvo
A, Upadhyaya P, Sala G, Antonucci I, Del Boccio P, Stuppia L and De
Laurenzi V: Breast cancer in the era of integrating 'Omics'
approaches. Oncogenesis. 11:172022. View Article : Google Scholar
|
|
174
|
Danzi F, Pacchiana R, Mafficini A, Scupoli
MT, Scarpa A, Donadelli M and Fiore A: To metabolomics and beyond:
A technological portfolio to investigate cancer metabolism. Signal
Transduct Target Ther. 8:1372023. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Fan S, Shahid M, Jin P, Asher A and Kim J:
Identification of metabolic alterations in breast cancer using mass
spectrometry-based metabolomic analysis. Metabolites. 10:1702020.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Subramani R, Poudel S, Smith KD, Estrada A
and Lakshmanaswamy R: Metabolomics of breast cancer: A review.
Metabolites. 12:6432022. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Budczies J, Brockmöller SF, Müller BM,
Barupal DK, Richter-Ehrenstein C, Kleine-Tebbe A, Griffin JL,
Orešič M, Dietel M, Denkert C and Fiehn O: Comparative metabolomics
of estrogen receptor positive and estrogen receptor negative breast
cancer: Alterations in glutamine and beta-alanine metabolism. J
Proteomics. 94:279–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Amiri-Dashatan N, Yekta RF, Koushki M,
Arefi Oskouie A, Esfahani H, Taheri S and Kazemian E: Metabolomic
study of serum in patients with invasive ductal breast carcinoma
with LC-MS/MS approach. Int J Biol Markers. 37:349–359. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Shestakova KM, Moskaleva NE, Boldin AA,
Rezvanov PM, Shestopalov AV, Rumyantsev SA, Zlatnik EY, Novikova
IA, Sagakyants AB, Timofeeva SV, et al: Targeted metabolomic
profiling as a tool for diagnostics of patients with non-small-cell
lung cancer. Sci Rep. 13:110722023. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Gold A, Choueiry F, Jin N, Mo X and Zhu J:
The application of metabolomics in recent colorectal cancer
studies: A state-of-the-art review. Cancers (Basel). 14:7252022.
View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Nam M, Seo SS, Jung S, Jang SY, Lee J,
Kwon M, Khan I, Ryu DH, Kim MK and Hwang GS: Comparable plasma
lipid changes in patients with high-grade cervical intraepithelial
neoplasia and patients with cervical cancer. J Proteome Res.
20:740–750. 2021. View Article : Google Scholar
|
|
182
|
Granit A, Mishra K, Barasch D,
Peretz-Yablonsky T, Eyal S and Kakhlon O: Metabolomic profiling of
triple negative breast cancer cells suggests that valproic acid can
enhance the anticancer effect of cisplatin. Front Cell Dev Biol.
10:10147982022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Xiao Y, Ma D, Yang YS, Yang F, Ding JH,
Gong Y, Jiang L, Ge LP, Wu SY, Yu Q, et al: Comprehensive
metabolomics expands precision medicine for triple-negative breast
cancer. Cell Res. 32:477–490. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Iyer A, Hamers AAJ and Pillai AB:
CyTOF® for the masses. Front Immunol. 13:8158282022.
View Article : Google Scholar
|
|
185
|
Fogazzi V, Kapahnke M, Cataldo A,
Plantamura I, Tagliabue E, Di Cosimo S, Cosentino G and Iorio MV:
The role of MicroRNAs in HER2-positive breast cancer: Where we are
and future prospective. Cancers (Basel). 14:53262022. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Cappelletti V, Iorio E, Miodini P,
Silvestri M, Dugo M and Daidone MG: Metabolic footprints and
molecular subtypes in breast cancer. Dis Markers. 2017:76878512017.
View Article : Google Scholar
|