Introduction
Colorectal cancer (CRC) is one of the common types
of malignant tumors, as the third most common malignancy and the
second most deadly cancer, CRC accounts for 10% of global cancer
incidence and 9.4% of cancer deaths in 2020 (1). Recently, its incidence has been
continuously increasing, posing a serious threat to lives and
health (2). Therefore, exploring
underlying mechanisms of CRC progression and identifying novel
therapeutic biomarkers for CRC are essential.
As a type of endogenous RNA, circular RNAs
(circRNAs) are generated by the back-splicing of pre-mRNAs, forming
a covalently closed loop structure. circRNAs lack the traditional
5′-end cap structure and 3′-end poly A tail; this makes them more
stable and resistant to exonucleases compared with linear RNA
(3). Accumulating evidence
suggests the key role of circRNAs in cancer development and
progression, with some (such as circRNA_102231 and circRNA CDR1as)
identified as available biomarkers for predicting cancer diagnosis,
treatment and prognosis (4-6).
Multiple circRNAs with different fragment lengths and sequences can
be derived from the same parental gene, such as different circRNAs
are generated from ubiquitin-associated protein 2 (UBAP2) and three
domain family 33 (7-10), which may exert various biological
function. Circular RNA UBAP2 contributed to malignant properties of
prostate cancer and osteosarcoma, however, it inhibited
proliferation and metastasis of clear cell renal cell carcinoma.
Previous study showed that manganese superoxide dismutase (SOD2)
could modulate colorectal tumorigenesis (11), however, the role of its circRNAs
in CRC remains unreported.
The present study aim to explore the circRNAs
derived from the SOD family using databases and clarify its role in
colorectal cancer.
Materials and methods
Clinical specimens
Paired adjacent normal tissue (distance from cancer
tissue greater than 5 cm) and CRC tissue (n=18) was collected from
Anhui Provincial Cancer Hospital (Hefei, China) from January 2023
to December 2023. Two pathologists histologically confirmed the
final diagnosis of CRC. Only patients with pathologically diagnosed
CRC were included, the exclusion criterion was a history of
malignant tumor treatment in other parts of the body. Table SI summarizes the clinical
characteristics of 18 patients with CRC, with patients age ranging
from 41 to 84 years, and the median age was 58. The surgically
excised specimens were stored at −80°C until used. The Medical
Ethics Committee of the Anhui Provincial Cancer Hospital approved
the study (approval no. 2023081). All participants provided written
informed consent according to the Declaration of Helsinki.
Cell lines
Human normal colon epithelial cells FHC (cat. no.
FH1283, Cellosaurus Accession no. CVCL_3688) were purchased from
Fuheng Biology. CRC cell lines HT-29 (cat. no. C5083, CVCL_0320),
SW480 (cat. no. C5233, CVCL_0546), DLD-1 (cat. no. C5060,
CVCL_0248), HCT116 (cat. no. C5229, CVCL_0291), LOVO (cat. no.
C5178, CVCL_0399), COLO205 (cat. no. C5053, CVCL_0218) and Caco-2
(cat. no. C5224; CVCL_0025) were purchased from Zhejiang Baidi
Biotechnology Co., Ltd. All cells were cultured in high-glucose
Dulbecco's modified Eagle's medium (cat. no. 10566016) supplemented
with 1% penicillin and streptomycin (cat. no. 15140122, both Gibco;
Thermo Fisher Scientific, Inc.) and 10% fetal bovine serum (FBS,
cat. no. 40131ES76, Shanghai Yeasen Biotechnology Co., Ltd.). Cells
were cultured in a humidified environment with 5% CO2 at
37°C. All the cell lines were verified via short tandem repeat
analysis.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Eastep Super Total RNA Extraction kit (cat. no.
LS1040, Promega Corporation) was used to extract total tissues/cell
RNA. RNA was converted to cDNA using GoScript Reverse Transcription
Mix (cat. no. A2800, Promega Corporation) according to the
manufacturer's instructions. The GoTaq qPCR Master Mix (cat. no.
A6002, Promega Corporation) was used to perform RT-qPCR. 18S rRNA
was used as an endogenous control to measure the levels of circRNA
(Table SII) and mRNA.
Thermocycling conditions were as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 30 sec. The specificity of amplification was confirmed via
melting curve analysis. The 2−ΔΔCq method was used to
assess gene expression (12).
Primers are listed in Table
SIII. Primers of circ_0004662 and circ_0078541 were designed
based on the sequence of back splicing site and the primers of
circRNAs in the present study were designed and validated by
Guangzhou Geneseed Biotech. Co., Ltd.
RNase R treatment
Total RNA (5 μg) extracted from CRC cells was
treated in the presence or absence of 5 U/μg RNase R (cat.
no. R0300, Guangzhou Geneseed Biotech. Co., Ltd.) at 37°C for 30
min. Then, RNase R was inactivated at 70°C for 10 min. Finally, RNA
was reverse-transcribed using a random primer.
Actinomycin D treatment
CRC cells were treated with 2 μg/ml
actinomycin D (cat. no. 15021S, Cell Signaling Technology, Inc.) at
37°C for 0, 4, 8 and 12 h. Then, total cell RNA was extracted,
followed by RT-qPCR to measure the stability of circRNAs. The gene
expression at 0 h was considered the baseline.
Construction of plasmid vectors and cell
transfection
To silence circ_0004662, short hairpin RNA (shRNA)
targeting the junction sites of circ_0004662 was designed by
Guangzhou RiboBio Co., Ltd. (Table
SIV). The pLshRNA vector (Shanghai Calm Biotechnology Co.,
Ltd.) was used as the shRNA plasmid; empty vector was used as
sh-negative control (NC). cDNA sequence of human circ_0004662 was
synthesized and cloned into pLC5-ciR vector (Guangzhou Geneseed
Biotech. Co., Ltd.) to generate overexpression plasmids. Sanger
sequencing was performed to confirm all constructs. The 2.5
μg constructed plasmids, 1.88 μg PSPAX2 vector
(Shanghai Calm Biotechnology Co., Ltd.) and 0.625 μg PMD2G
vector (Shanghai Calm Biotechnology Co., Ltd.) were then
co-transfected into 3rd generation 293T cells (Zhejiang Baidi
Biotechnology Co., Ltd.) using Lipofectamine 3000 (cat. no.
L3000008, Thermo Fisher Scientific, Inc.) to package lentivirus
according to manufacturer's instruction. After a 48 h incubation at
37°C, 2 ml viral supernatant was collected and filtered with a 0.22
μm filter. Then viral supernatant was added for 8 h, after
which the medium was replaced. The cells were then subjected to
selection using 2 μg/ml puromycin for 3 days, with 1
μg/ml puromycin used for maintenance. Subsequent experiments
were then started. The sequence used in the present study are as
follows: circ_0004662-siRNA#1 (TATGCTGAGAGATGTTACA);
circ_0004662-siRNA#2 (CGATCGTTATGCTGAGAGA); circ_0004662-siRNA#3
(TCGTTATGCTGAGAGATGT).
Cell proliferation and migration
assay
Wound healing and Transwell assays were performed to
assess migration ability of CRC cells. For wound healing assay,
1×106 transfected CRC cells (DLD-1, SW480, HCT116) were
added to complete DMEM supplemented with 10% FBS in 24-well plates.
A 200-μl sterile tip was used to make a scratch in the
monolayer (confluence ~100% at the start of the assay, followed by
culture in medium containing 2-5% FBS as aforementioned. Cells were
photographed at 0 and 48 h (cat. no. IX2-SLP, Olympus) with
magnification is 100X. ImageJ software (version 1.54 g, National
Institutes of Health) was used to measure relative would healing
area.
For Transwell assay, serum-free medium containing
1×105 transfected CRC cells (DLD-1 and SW480) was added
to upper 24-well Transwell chambers (cat. no. 3422, Corning, Inc.),
a complete DMEM (cat. no. 10566016, Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS was added to lower chamber in
24-well plates. Cells were then incubated in a CO2
incubator at 37°C. After 48 h, cells were fixed with 4%
paraformaldehyde for 10 min at room temperature, and stained with
0.1% crystal violet at room temperature for 10 min. The migrated
cells in the bottom chambers were photographed by invert light
microscope (cat. IX2-SLP, Olympus Corporation) with magnification
200X, and counted by ImageJ software (version 1.54 g, National
Institutes of Health).
Subcellular fractionation
Cytoplasmic and Nuclear RNA Purification kit (cat.
no. 21000, Norgen Biotek Corp.) was used to extract cytoplasmic and
nuclear RNA. Briefly, CRC cells were treated with lysis buffer for
15 min and centrifuged at 4°C, 12,000 × g for 5 min. Nuclear RNA
was isolated from the pellets and supernatant was collected to
extract cytoplasmic RNA according to the manufacturer's
instructions. Subsequently, reverse transcription and PCR were
performed as aforementioned. GAPDH was utilized as the cytoplasmic
reference gene and U6 as the nuclear reference gene.
Fluorescence in situ hybridization
(FISH)
RNA FISH probes to target the splicing site of
circ_0004662 (5′CY3-TGTAACATCTCTCAGCATAACG-3′CY3) were designed and
synthesized by Guangzhou Geneseed Biotech. Co., Ltd. A total of
5×104 DLD-1 cells were seeded on slides at the bottom of
24-well plates. When the cell density reaches 70%, slides were
washed with PBS at room temperature for 5 min, then 4%
paraformaldehyde was used to fix cells at room temperature for 5
min and washed with PBS for 5 min. TritonX-100 (cat. no. P0096,
Beyotime Institute of Biotechnology) was added to incubate slides
at room temperature for 15 min, and wash with PBS three times for 5
min each time. Then anhydrous ethanol was added, and the ethanol
was aspirated after 1 min of action at room temperature.
circ_0004662-specific Cy3 probes were added to cells at 37°C
overnight. The nuclei were counterstained with DAPI (cat. no.
C1002, Beyotime Institute of Biotechnology) at room temperature for
5 min. FISH kit (cat. no. H0101, Guangzhou Geneseed Biotech. Co.,
Ltd.) was used according to the manufacturer's instructions. Probe
signals were imaged under a laser scanning confocal microscope
(cat. no. TCS SP2 AOBS, Leica GmbH; magnification is 400X.
Western blotting
Briefly, cells were lysed using RIPA lysis buffer
containing protease and phosphatase and protease inhibitor
cocktails (cat. nos. P0013B, P1009 and P1050, respectively; all
Beyotime Institute of Biotechnology). Protein concentration was
determined by BCA assay. A One-Step PAGE Gel Fast Preparation kit
(8%; cat. no. E302-1, Vazyme Biotech Co., Ltd.) was used to
separate equal amounts of protein (20 μg/lane). The
separated proteins were transferred to polyvinylidene fluoride
membranes (cat. no. IPVH00010, MilliporeSigma). Non-specific
binding was blocked at room temperature for 1 h using Quick Block
Blocking Buffer (cat. no. P0252, Beyotime Institute of
Biotechnology). Then, the membranes were incubated with antibodies
overnight at 4°C as follows: Flag-tagged recombinant rabbit
monoclonal (cat. no. 30504ES50, Shanghai Yeasen Biotechnology Co.,
Ltd.), recombinant anti-SOD2 (1:1,000, cat. no. ab68155, Abcam),
S100 calcium binding protein A9 (S100A9) polyclonal (1:1,000, cat.
no. 26992-1-AP), phosphoglycerate kinase 1 (PGK1) polyclonal
(1:1,000, cat. no. 17811-1-AP), heterogeneous nuclear
ribonucleoprotein M (hnRNPM) polyclonal (1:1,000, cat. no.
26897-1-AP) and β-tubulin monoclonal (1:1,000, cat. no. 30301ES60,
all Proteintech Group, Inc.). After the membranes were washed with
1X TBST (cat. no. ST673, Beyotime Institute of Biotechnology), they
were incubated with horseradish peroxidase-labeled goat anti-mouse
or anti-rabbit IgG (1:1,000, cat. nos. A0216 and A0208,
respectively; Beyotime Institute of Biotechnology) for 1 h at 25°C.
Finally, ECL substrate kit (cat. no. 36222ES60, Shanghai Yeasen
Biotechnology Co., Ltd.) was used to visualize the membranes. A
chemiluminescence imaging system (Tano4600, Shanghai Tianneng
Technology) was used to visualize strips and to perform
semiquantitative analysis.
circ_ 0004662 pull-down assay and liquid
chromatography-mass spectrometry (LC-MS) analysis
MS2 bacteriophage coat protein (MS2-CP) circRNA
pull-down assay was performed to detect RNA-binding proteins (RBPs)
that bind to circ_0004662. MS2 tagging technique is based on the
natural binding between a stem-loop structure of MS2 and MS2-CP
(13-15). Briefly, overexpression vectors
(Guangzhou Geneseed Biotech. Co., Ltd.) carrying circ_0004662-MS2
were constructed and tagged with green fluorescent protein (GFP);
MS2-CP-Flag was tagged with red fluorescent protein (m-Cherry;
Guangzhou Geneseed Biotech. Co., Ltd.). The vectors were
co-transfected into 293T cells as aforementioned. Cell protein was
isolated by 500 μl lysis buffer as aforementioned. A total
of 450 μl lysate was used for each IP reaction using RNA
Immunoprecipitation Kit (cat. no. P0101, Guangzhou Geneseed
Biotech. Co., Ltd.) according to manufacturer's illustration. A
total 5 μg anti-Flag antibody (cat. no. A00170, Nanjing
Kingsray Biotechnology Co., Ltd.) and 100 μl protein A/G
immunoprecipitation magnetic beads (cat. no. B23201, Selleck) were
used to pull down MS2-CP-MS2-circ_0004662 complex. Lysate extracted
from cells without the MS2 tagging system was used as NC. After
detecting captured lysates via RT-qPCR and western blotting as
aforementioned. The completes were isolated by magnetic grate.
LC-MS was performed to analyze the circ_0004662 pull-down complex
and controls. Briefly, the peptide segments were dissolved in
sample solution [0.1% formic acid (cat. no. 64186, Sigma), 2%
acetonitrile (cat. no. A/0626/17, Fisher), and centrifuged at 4°C,
14,000 × g for 20 min, supernatant was collected and performed mass
spectrometry identification. The liquid phase parameters was as
follows: (a) Column information: 300 μm ×5 mm, Acclaim
PepMap RSLC C18, 5 μm, 100A (cat. no. 160454, Thermo);
Acclaim PepMap 75 μm X150 mm, C18, 3 μm, 100A (cat.
no. 160321, Thermo. (b) Mobile phase information: Mobile phase A
(0.1% formic acid); Mobile phase B: 0.1% formic acid, 80%
acetonitrile; Flow rate: 300 nl/min. (c) Analysis time: 45 min. The
separated peptide segments are directly fed into the mass
spectrometer (Q Exactive, Thermo Fisher Scientific, Inc.) for
online detection, with a resolution: 70,000; AGC target: 3e6;
Maximum IT:40 msec; Scan range: 350 to 1,800 m/z. The raw mass
spectrometry files were processed and converted by MM File
Conversion software to obtain MGF format files, and then the
Uniprot database was searched using MASCOT (http://www.matrixscience.com/).
Prediction and verification of open
reading frames (ORFs)
The human circular RNA database (circRNA DB) was
used to predict ORFs in circ_0004662 (16). To verify the functionality of the
predicted ORF sequence in circ_0004662, circ_0004662 sequence was
cloned into pLC5-ciR [translation verification vector (T)0] to
construct T1 (Guangzhou Geneseed Biotech. Co., Ltd.). Further, a
FLAG coding sequence
(5′-GACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAG-3′)
was inserted upstream of the stop codon (TGA) present in the ORF
sequence to construct T2 vector (17,18). As a result, once the potential ORF
sequence was translated, a tagged protein was generated. Vector
with start codon ATG of the predicted ORF was mutated (T3), and
vectors containing Flag tag with deletion of circular elements were
also constructed (T4; Guangzhou Geneseed Biotech. Co., Ltd.). FLAG
sequence was directly cloned into pLC5-ciR (Guangzhou Geneseed
Biotech. Co., Ltd.) and used as a positive control (T5). Cells were
transfected with the aforementioned plasmids and harvested after 3
days as aforementioned. Western blotting was performed using
anti-FLAG antibody (1:1,000, cat. no. 30504ES50; Shanghai Yeasen
Biotechnology Co., Ltd.) to measure the flagged protein according
to previous description.
RNA-binding protein immunoprecipitation
(RIP)
Briefly, 1×107 cells were treated with IP
lysis buffer (cat. no. P0013, Beyotime Institute of Biotechnology),
protease inhibitor, and RNase inhibitor for 10 min on ice. Cells
were centrifuged at 4°C and 10,000 × g for 10 min. A total of 5
μg IP hnRNPM (1:100), S1009A (1:100), PGK1 (1:50) and IgG
control polyclonal antibody (cat no. 30000-0-AP, Proteintech Group,
Inc.) were each bound to 20 μl magnetic beads (cat. no.
P2108, Beyotime Institute of Biotechnology) for 2 h at 4°C. The
cell lysis supernatant was then incubated with the magnetic bead
suspension at room temperature for 2 h. A magnetic separator was
used to collect the magnetic beads. Finally, pellets were treated
with RIPA lysis buffer or TRIZOL (cat. no. 15596018CN, Thermo
Fisher Scientific, Inc.) for western blotting and RNA
extraction.
Animal experiments
A total of 12 male BALB/c nude mice (age, 5-6 weeks
17-19 g) were purchased from Hangzhou Ziyuan Laboratory Animal
Technology (Zhejiang, China). Mice were housed under specific
pathogen-free conditions with a 12/12-h light/dark cycle and
controlled temperature at 24±2°C, and the relative humidity range
was 50±10%. The mice were allowed ad libitum access to water and
food pellets. DLD-1 cells were transfected with sh-circ_0004662 or
sh-NC and subcutaneously injected into the right dorsum to generate
subcutaneous tumors (5×106/mouse; n=6/group). Mice were
monitored daily and tumor volume was measured every 4 days. After
30 days, mice were anesthetized with 3% isoflurane (maintained with
1.5% isoflurane) and euthanized using cervical dislocation.
Physiological indicators such as respiration, heartbeat and pupil
response were monitored to confirm the death of mice, and
subcutaneous tumors were removed. Tumors were subjected to
histological analysis. Tumor volume was calculated as follows:
Volume (mm3)=width2 × length/2. All
experiments were approved and performed according to the guidelines
approved by the Institutional Animal Care and Use Committee of the
First Hospital Affiliated to the University of Science and
Technology of China [approval no. 2022-N(A)-072].
Immunohistochemical analysis
The mouse xenograft tumors were cut to 5-μm
thick sections, and immunohistochemistry was performed according to
established protocols (19).
Primary antibodies utilized included anti-Ki67 (original usage;
cat. no. MAB-0672; Proteintech Group, Inc.), anti-E-cad, anti-N-cad
(1:5,000, cat. nos. 20874-1-AP; 22018-1-AP, Proteintech Group,
Inc.) and anti-vimentin (1:300; cat. no. ab92547, Abcam). Images
were captured using the Olympus CX41 system with cell Sens Standard
software (Olympus, Tokyo, Japan). IHC score were measured using
ImageJ software (version 1.54 g, National Institutes of Health).
The percentage of positive cells was subdivided into four grades:
0, <5%; 1 for 6-25%; 2 for 26-50%; 3 for 51-75% and 4 for
>75%. Staining intensity was assessed by four degrees: 0,
negative; 1, weak; 2, moderate; and 3, strong. The IHC score is the
cell staining intensity score multiplied by the percentage of
positive cells score.
Statistical analysis
SPSS 24.0 software (IBM Corp.) and GraphPad Prism
8.0 (GraphPad Software, Inc.; Dotmatics) were used for statistical
analysis. Data are presented as the mean ± SD. Kolmogorov-Smirnov
test was used to determine the distribution of each group. Paired
student's t-test was used to measure significance between adjacent
tissues and CRC tissues, while unpaired t-test (two-tailed) was
used to determine significance between two groups. One-way ANOVA
followed by Bonferroni's post hoc test was used to analyze >2
groups. A paired t-test was used to analyze circRNAs between CRC
and adjacent normal tissue. All experiments were repeated at least
three times. P<0.05 was considered to indicate a statistically
significant difference.
Results
circ_0004662 is expressed highly in CRC
samples
circBank database was used to identify circRNAs
derived from SOD family genes (SOD1, SOD2 and SOD3). In total, five
potential circRNAs were generated, including two derived from SOD1
(hsa_circ_0061417 and hsa_circ_0115795) and three from SOD2
(hsa_circ_0004662, hsa_circ_0078541 and hsa_circ_0005472). No
circRNAs were generated from SOD3 (Table SII). However, only two circRNAs
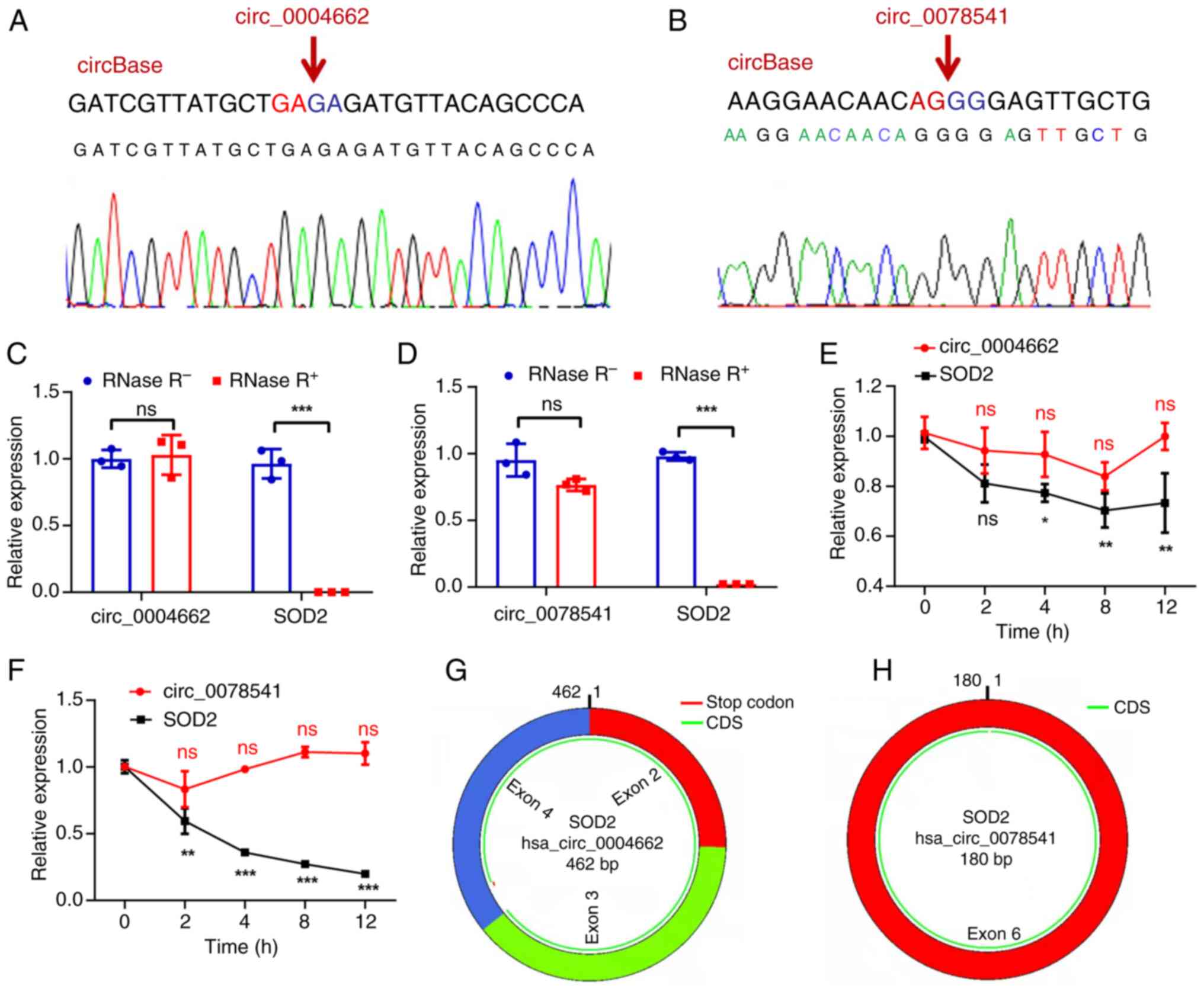
(circ_0004662 and circ_0078541) were successfully validated. Sanger
sequencing was performed to confirm back-splice sites of
circ_0004662 and circ_0078541; this matched the information in
circBase (Fig. 1A and B). These
circRNAs were resistant to RNase R treatment, whereas linear SOD2
were considerably digested with RNase R (Fig. 1C and D). Moreover, compared with
wild type (0 h), treatment with actinomycin D, which can block new
transcription, revealed that circ_0004662 and circ_0078541 were
more stable in comparison to SOD2 mRNA (Fig. 1E and F). This confirmed the
circular structures of circ_0004662 and circ_0078541 (Fig. 1G and H). Further analysis revealed
differential expression of circ_0004662 in cancerous tissues
compared with adjacent normal tissues (Fig. S1A). However, no significant
difference was noted in transcription levels of circ_0078541
(Fig. S1B). Collectively, these
findings underscore the potential importance of circ_0004662 in CRC
pathogenesis.
circ_0004662 promotes the malignant
characteristics of CRC cells both in vitro and in vivo
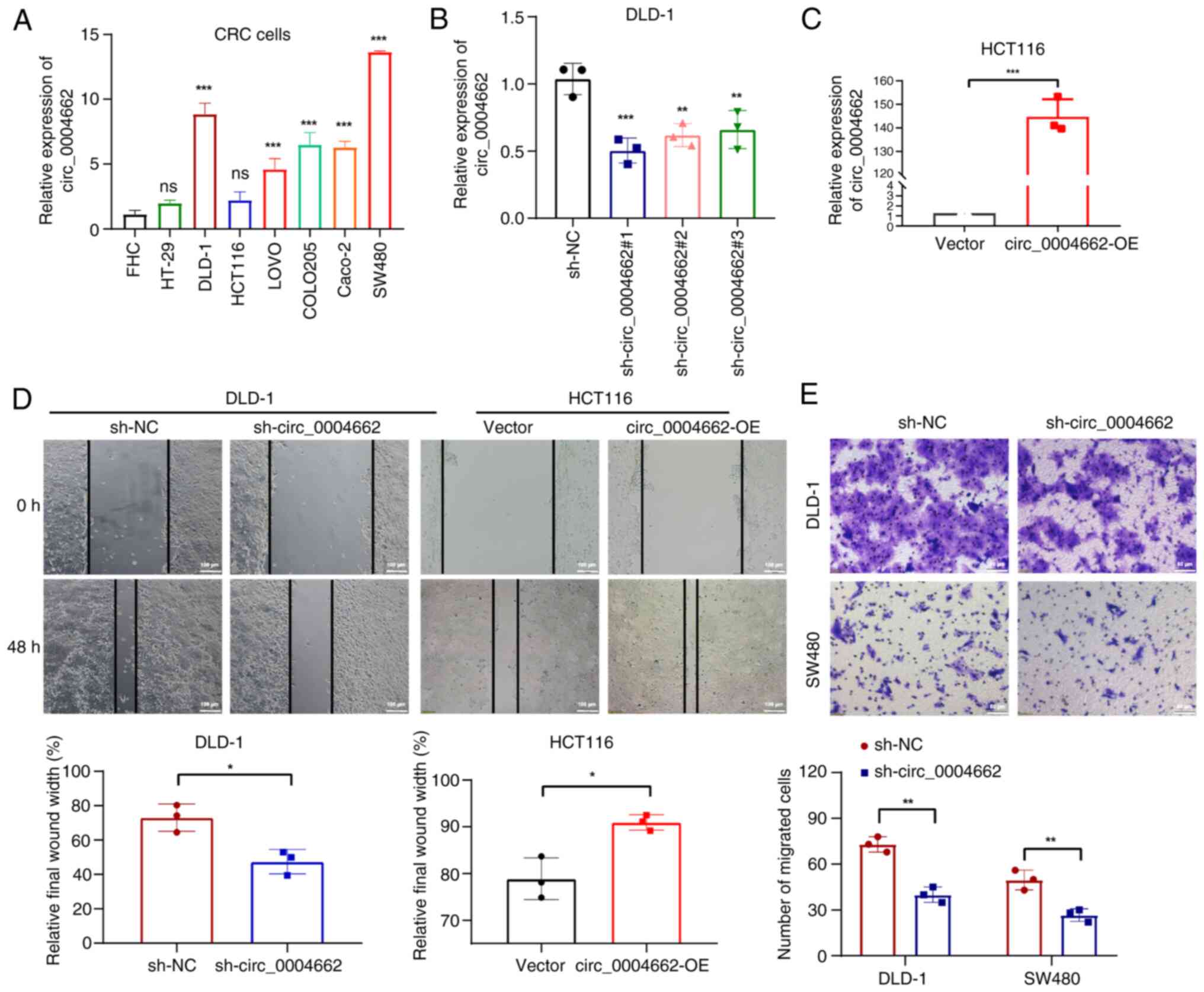
circ_0004662 expression was significantly higher in
most CRC cell lines (DLD-1, LOVO, COLO205, Caco-2, SW480) compared
with normal colon epithelial cells (FHC; Fig. 2A). shRNAs targeting circ_0004662
were transfected into DLD-1 and SW480 cells, whereas overexpression
vector for circ_0004662 was transfected into HCT116 cells.
circ_0004662 expression was significantly decreased in DLD-1 and
SW480 cells (Figs. 2B and
S2A) and circ_0004662 was
significantly upregulated in the HCT116 cells (Fig. 2C).
Functional analysis revealed that circ_0004662
downregulation markedly decrease migratory ability of CRC cells
(Figs. 2D and S2B). By contrast, circ_0004662
overexpression contributed to migration ability of HCT116 cells
(Fig. 2D, right). The Transwell
assay also demonstrated that knockdown of circ_0004662 attenuated
migration capacity in DLD-1 cells (Fig. 2E). circ_0004662 knockdown
attenuated tumor growth in vivo (Fig. 3A-D). Immunohistochemistry showed
that the proportion of Ki-67 in the knockdown group was lower than
that in control group, indicating that knockdown of circ_0004662
decreased the proliferation ability of CRC cells in vivo
(Fig. 3E). E-cadherin (E-cad),
N-cad and vimentin (VIM) are metastasis biomarkers (20,21); E-cad was upregulated, whereas
N-cad were downregulated following knockdown of circ_0004662, while
VIM expression was low in both groups (Fig. 3E).
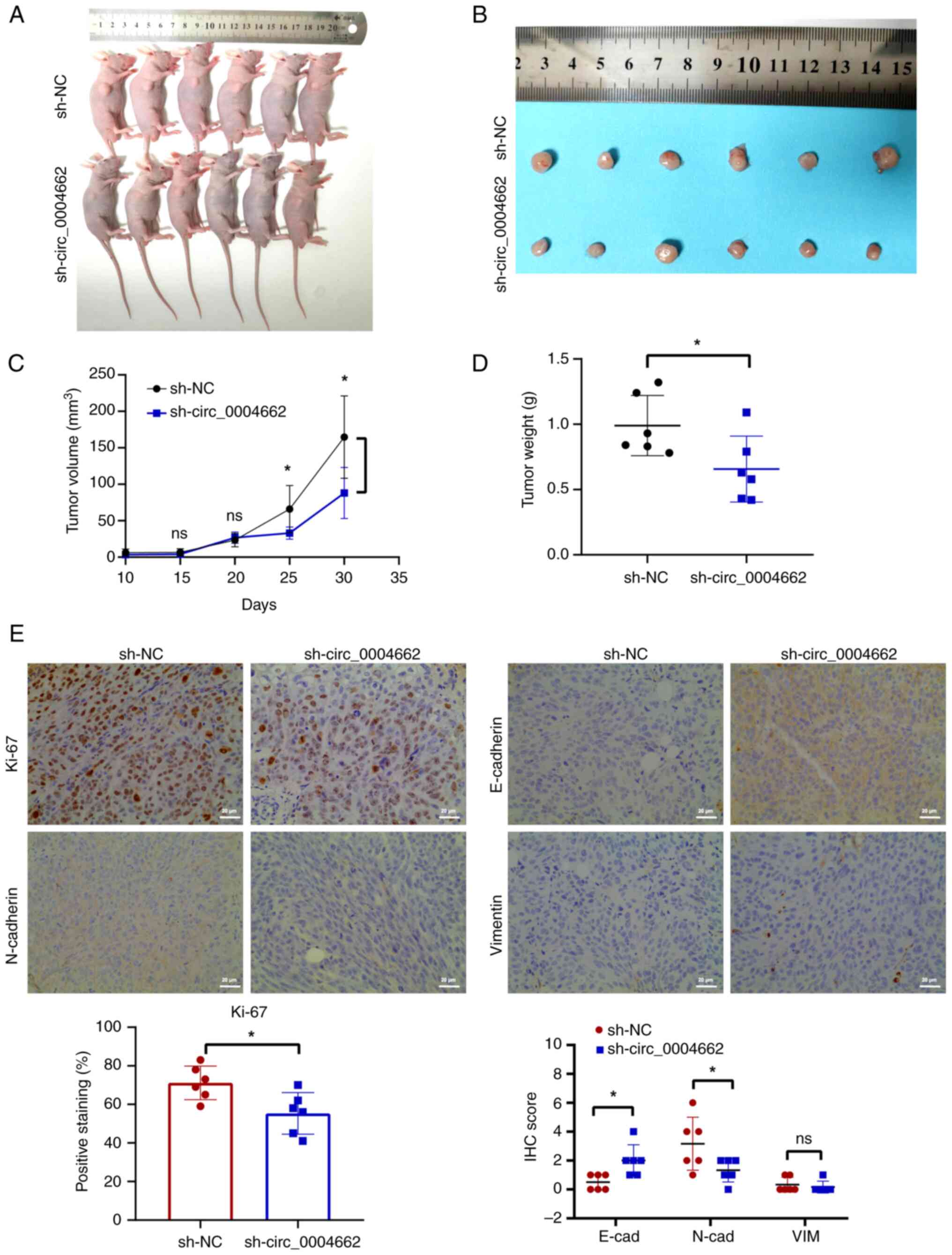 | Figure 3circ_0004662 contributes to
proliferation and migration of colorectal cancer cells in
vivo. (A) Nude mice after subcutaneous tumorigenesis. (B)
Tumors were dissected from nude mice subcutaneously injected with
circ_0004662-knockdown or control DLD-1 cells. (C) Volume and (D)
weight of subcutaneous xenograft tumors. (E) Representative IHC
analysis of Ki-67, E-cad, N-cad and VIM. Scale bar, 20 μm.
*P<0.05 vs. sh-NC group. circ, circular; ns, not
significant; sh, short hairpin; IHC, immunohistochemistry; cad,
cadherin; VIM, vimentin; NC, negative control. |
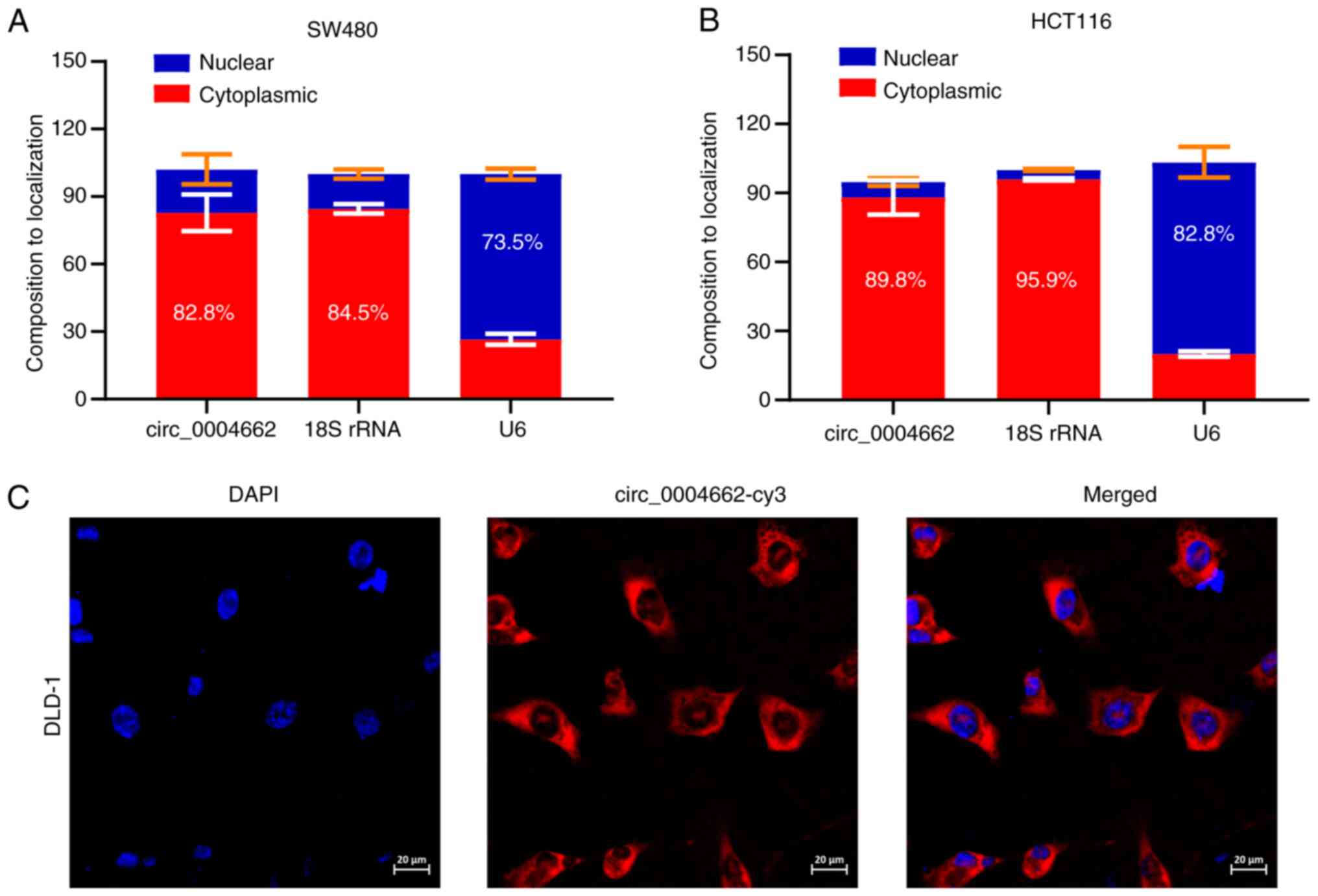
circ_0004662 is localized in the nucleus
and cytoplasm of CRC cells
Because circRNAs exert different functions depending
on cellular localization (22,23), subcellular fractionation analysis
was conducted to investigate localization of circ_0004662.
circ_0004662 was present in the nucleus and cytoplasm of SW480 and
HCT116 cells (Fig. 4A), which was
further confirmed via FISH in DLD-1 cells (Fig. 4B).
circ_ 0004662 is a non-coding RNA in CRC
cells
Using circDB database (reprod.njmu.edu.cn/circrnadb), circ_0004662 was
predicted to contain a potential ORF and a putative internal
ribosome entry site sequence; this suggested that it can encode a
149-amino acid peptide (Fig. 5A).
To investigate whether endogenous circ_0004662 can be translated
into circ_0004662_149aa, Flag-coding sequence was inserted upstream
of the stop codon in the potential ORF sequence (Fig. 5B). Sanger sequencing confirmed the
sequences of the plasmids (Fig.
S3). However, in transfected cells, no FLAG-tagged proteins
were detected at the predicted molecular weight size (Fig. 5C). Furthermore, immunoblotting
using SOD2 antibody failed to identify circ_0004662_149aa at the
expected size in 293T cells (Fig.
5D). Collectively, these findings confirm that circ_0004662 was
a non-coding RNA.
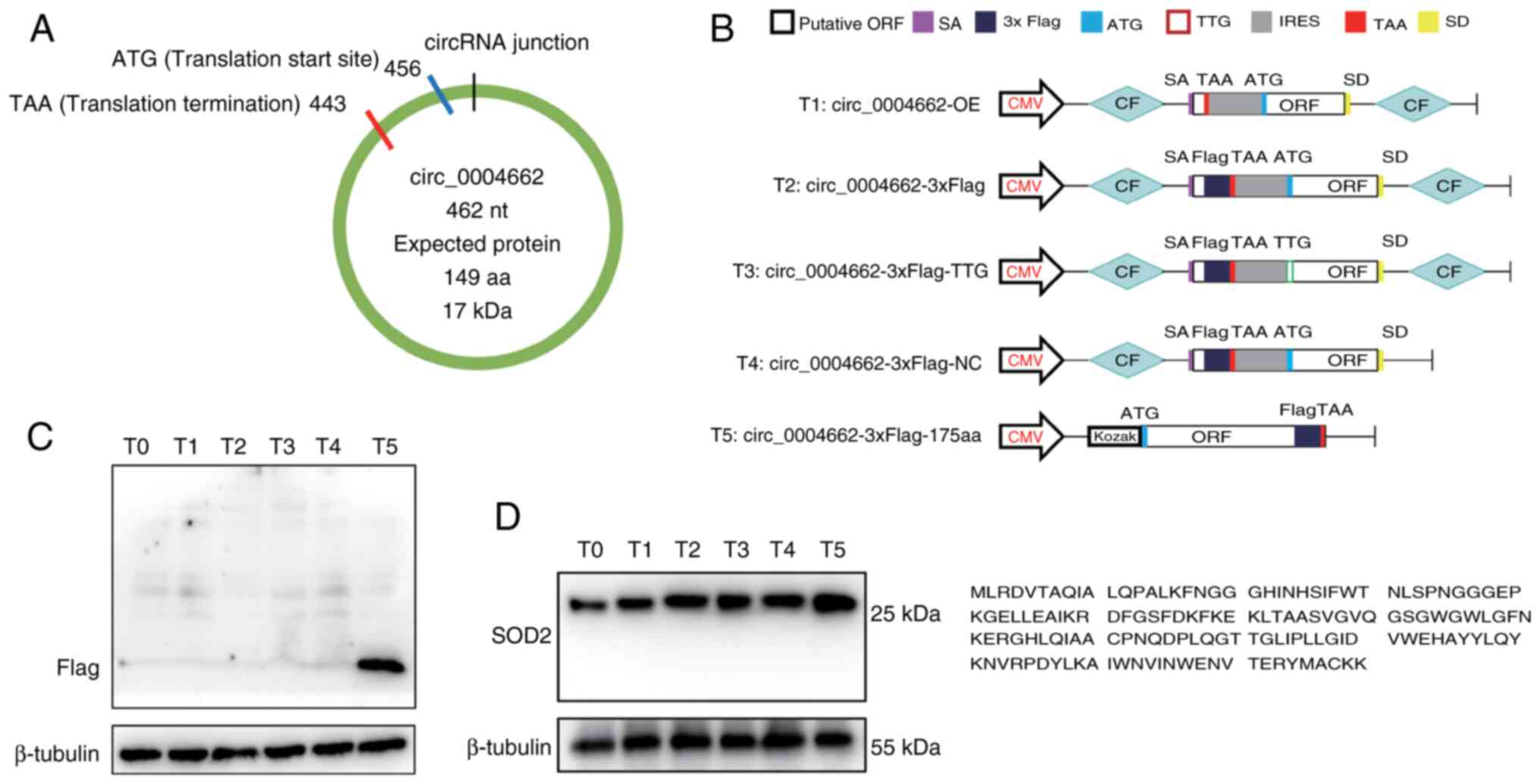 | Figure 5circ_0004662 is a non-coding RNA in
colorectal cancer cells. (A) Putative ORF in circ_0004662. (B)
Construction of Flag-tagged circ_0004662 and the predicted peptide
sequence. Plasmids with start codon mutations and deletion of
downstream flanking sequences could not generate SOD2-149aa.
Positive control was constructed by cloning the linearized
circ_0004662 ORF into a CMV-induced expression vector. (C)
Measurement of flagged protein via western blotting. (D) Detection
of SOD2-149aa using antibody identifying C or N terminal regions of
SOD2. T0, empty vector; T1 circ_0004662-overexpressing vector; T2,
circ_0004662-overexpressing vector containing 3× Flag tag; T3 was
circ_0004662-overexpression vector containing 3× Flag tag, with ATG
mutation; T4 was vectors containing 3× Flag tag, whose circular
elements was deleted, forming a negative control that cannot form a
circular RNA; T5 included the predicted full-length ORF without a
cyclic framework, allowing normal translation and generating
predicted peptides, which was a positive control. circ, circular;
T, translation vector; OE, overexpression; CF, circular frame; ORF,
open reading frame; IRES, internal ribosome entry site; SOD,
superoxide dismutase; aa, amino acid; CMV, CMV promoter; SA, splice
acceptor; SD, splicing donor. |
circ_0004662 binds hnRNPM in CRC
cells
MS2-CP-Flag circRNA pull-down assay was performed to
discover the potential protein partners of circ_0004662 (Fig. 6A). Plasmids expressing
circ_0004662-MS2-GFP and MS2-CP-Flag-mCherry were constructed.
Using the circ_0004662-MS2 tagging system, RIP assay was conducted
after co-transfecting circ_0004662 and MS2-CP-FLAG, resulting in
pull-down of protein complexes between MS2 and MS2-CP by Flag
antibodies (Fig. 6B). Sanger
sequencing confirmed MS2-Flag insertion did not affect circRNA
circularization (Fig. S4A).
RT-qPCR validated overexpression of circ_0004662 in the
circ_0004662-MS2 tagging system (Fig.
6C). Label-free LC-MS analysis revealed that circ_0004662 may
interact with multiple proteins, including several ribosomal
proteins (ribosomal protein L36, ribosomal protein L35A, ribosomal
protein S15A; Table SV), and
genes (subtilisin-like Serine Protease 1, pyruvate kinase,
lipocalin-1, S100 calcium binding Protein A9,) that serve key roles
in cancer progression (Table
SVI). Based on previous literature, PGK1 (24,25), S100A9 (26,27), and hnRNPM (28,29) were selected for further
validation. A RIP assay was conducted in DLD-1 cells using
anti-hnRNPM, anti-PGK1 and anti-S100A9 antibodies, and found
significant enrichment of circ_0004662 after anti-hnRNPM
immunoprecipitation compared with IgG (Fig. 6D); immunoblotting confirmed this
finding (Fig. S4B). hnRNPM can
bind to circRNAs and regulate their generation, thereby affecting
biological functions of cancer cells (29,30). Subsequent hnRNPM knockdown
decreased circ_0004662 expression (Figs. 6E and F and S5) in DLD-1 cells. The enhanced
migration ability induced by circ_0004662 was reversed following
silencing hnRNPM in HCT116 cells (Fig. 6G). In conclusion, these findings
suggested that circ_0004662 promotes CRC progression by interacting
with hnRNPM, highlighting the regulatory role of the
circ_0004662/hnRNPM interaction in CRC cells (Fig. S6).
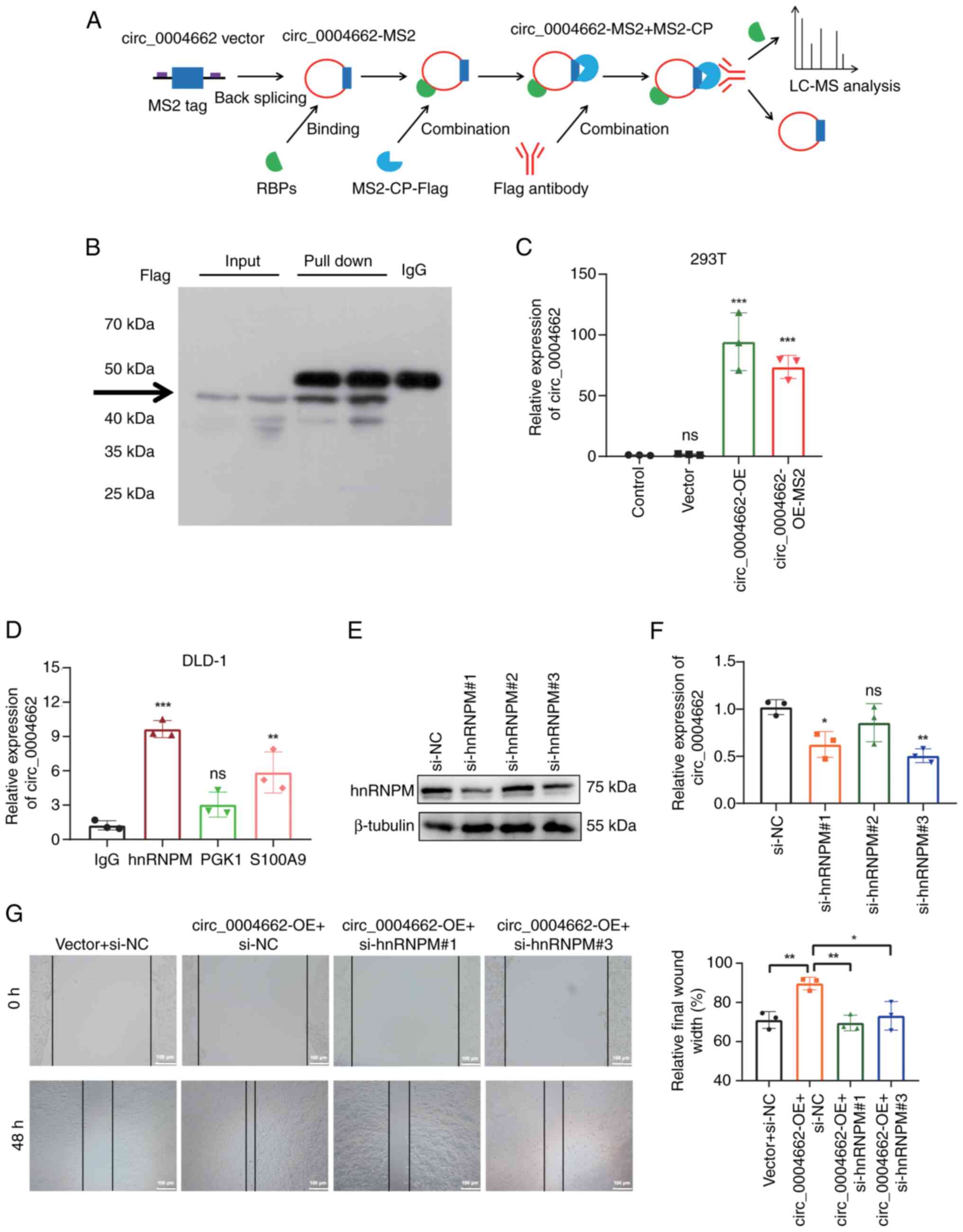 | Figure 6Interaction between circ_0004662 and
hnRNPM in colorectal cancer cells. (A) circ_0004662 pull-down
products using the MS2-tagging system. (B) MS2-CP-Flag was pulled
down by anti-Flag antibodies and subjected to western blotting. (C)
HEK-293T cells were transfected with circ_0004662, control or
MS2-labelled circ_0004662 expression plasmids. Relative expression
of circ_0004662 was measured via PCR. (D) RNA binding protein
immunoprecipitation assay of DLD-1 cells using anti-hnRNPM, PGK-1
and S100A9 antibodies or control IgG. circ_0004662 expression was
measured via RT-qPCR. (E) Interference efficiency of hnRNPM was
measured via western blot. (F) circ_0004662 expression after
knocking down hnRNPM. (G) Effect of hnRNPM interference in HCT116
cells was measured using the wound healing assay.
*P<0.05, **P<0.01,
***P<0.001 vs. vector group or IgG group or NC group.
circ, circular; hnRNPM, heterogeneous nuclear ribonucleoprotein M;
MS2-CP, MS2 bacteriophage coat protein; PGK, phosphoglycerate
kinase; RBP, RNA binding protein; LC-MS, liquid chromatograph mass
spectrometer; OE, overexpression; si, small interfering; NC,
negative control; ns, not significant. |
Discussion
Accumulating evidence indicates that non-coding RNAs
(31,32), particularly circRNAs, serve key
roles in CRC development (33-35). The present study characterized
circRNAs derived from SOD gene family and identified upregulation
of circ_0004662 in CRC cells and tissue. circ_0004662 was present
both in the cytoplasm and nucleus. As a non-coding RNA,
circ_0004662 binds hnRNPM and promotes the migration of CRC cells,
thereby offering potential novel therapeutic targets for
personalized treatment of CRC.
Despite being formed by the same parental genes,
circRNAs exhibit varying roles in cancer progression. For example,
circUBAP2 facilitates malignant characteristics in prostate
(7) and breast cancer (36), hepatocellular carcinoma (19) and osteocarcinoma (8); however, it inhibits proliferation
and metastasis of clear cell renal cell carcinoma cells (10). Furthermore, linear Rho GTPase
activating protein 35 (ARHGAP35) encodes a tumor suppressor, while
circARHGAP35 translates into an oncogenic large protein to promote
cancer progression (37).
However, the specific role of circSODs in regulating biological
characteristics of CRC cells remains unclear. circ_0004662
accelerates osteoarthritis progression via the microRNA
(miR)-424-5p/VEGFA axis (38),
and circ_0004662 drives progression of hepatocellular carcinoma by
serving as a sponge for miR-502-5p and activating the JAK2/STAT3
signaling pathway (39).
Furthermore, the Paired Box 5/circ_0004662/miR-532-3p axis serves
an important role in promoting the proliferation, invasion and
migration of clear cell renal cell carcinoma cells (40). Here, circ_0004662 contributed to
the migration ability of CRC cells in vitro.
E-cadherin, N-cadherin, vimentin, matrix
metalloproteinases, claudins, epithelial cell adhesion molecules
and cytokeratins are common biomarkers for the detection of
epithelial-mesenchymal transition (EMT), which is a vital phenotype
in metastasis (20).
Downregulated E-cad and upregulate N-cad and VIM can promote EMT,
leading to increased invasiveness and metastasis in CRC (21). Here, E-cad was upregulated,
whereas N-cad was downregulated following knockdown in
circ_0004662. circRNA SOD2 promotes EMT in non-small cell lung
cancer advancement via acting as miR-2355-5p competing endogenous
RNA to mediate calmodulin-regulated spectrin associated proteins-2
(41).
Although most circRNAs are non-coding RNAs, many are
translated to peptides and regulate biological characteristics of
cancer cells, affecting tumor progression (42,43). For example, circAXIN1 encodes
AXIN1-295aa and promotes gastric cancer progression by activating
the Wnt/β-catenin signaling pathway (18). A novel protein encoded by
circ_0017272 promotes multiple myeloma progression by regulating
the bone marrow microenvironment and circ_0133744 could encode
proteins promote CRC proliferation and metastasis (44,45). The present study evaluated the
translation potential of circ_0004662 and suggested that it
interacts with multiple ribosomal proteins. However, the predicted
149aa peptide encoded by circ_0004662 was not detected. This
warrants additional investigation considering the complex
translation process.
With advances in high-throughput screening, multiple
RBPs have been implicated in cancer progression (46,47). Interaction between circ_0004662
and RBPs in CRC remains unexplored, the present study noted a
direct interaction between circ_0004662 and hnRNPM. Accumulating
evidence indicates that hnRNPM contributes to cancer cell
metastasis in hepatocellular carcinoma (28,48) and breast cancer (49,50). Notably, as an RBP, hnRNPM can
interact with circRNAs and control their expression via the
alternative splicing of circRNAs (51), which may impair the stability of
target genes. For example, circ_0000921 directly interacts with
hnRNPM to modulate alternative splicing of genes involved in the
process of cell migration, thus regulating gastric cancer
metastasis (29). Further,
combination of hnRNPM with circ_0003764 enhances the ability of
hnRNPM to maintain the stability of IL-6 mRNA and further activates
the STAT3 signaling pathway, promoting progression and sunitinib
resistance in renal cell carcinoma (52). Clinical colon cancer specimens and
mouse carcinogenesis model show that hnRNPM is elevated during the
development of CRC and is associated with poor prognosis (53), and genome-wide transcriptomics and
translatomics analyses have revealed a unique set of
hnRNPM-targeted genes involved in metabolic processes and cancer
neoplasia are selectively translated under hypoxia (53). Further, hnRNPM bind with
carcinoembryonic antigen (CEA), which may participate in the
antiapoptotic role of CEA and mediate the prometastatic properties
of CEA in colon cancer cells, but needs future experiments
(54). In the present study,
hnRNPM knockdown impaired circ_0004662 expression; this indicated
the role of hnRNPM in regulating circ_0004662 in CRC cells.
However, target genes of hnRNPM associated with CRC progression
remain unclear. Therefore, the specific mechanisms of how
circ_0004662 and hnRNPM promotes CRC metastasis should be assessed
in future research.
In conclusion, circ_0004662 promoted CRC cell
migration by interacting with hnRNPM. The present findings may
provide novel insights into the potential strategies for
personalized therapy for patients with CRC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be found
in the iProX database under accession number IPX0009947000 or at
the following URL: https://www.iprox.cn/page/project.html?id=IPX0009947000.
Authors' contributions
YZ conceived the study, conducted experiments and
drafted and revised the manuscript. JW and RQ analyzed data and
wrote the manuscript. LL conceived the study and performed
experiments. YZ and LL confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of the Anhui Provincial Cancer Hospital (approval no.
2023081). All participants in this study gave written informed
consent in accordance with the Declaration of Helsinki. All animal
care and procedures were performed according to guidelines approved
by the Institutional Animal Care and Use Committee of the Anhui
Provincial Cancer Hospital, University of Science and Technology of
China [approval no. 2022-N(A)-072].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by Anhui Provincial Natural
Science Foundation (grant no. 2208085QH259), Youth Fund of Anhui
Cancer Hospital (grant no. 2023YJQN009).
References
|
1
|
Xi Y and Xu P: Global colorectal cancer
burden in 2020 and projections to 2040. Transl Oncol.
14:1011742021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qu R, Ma Y, Zhang Z and Fu W: Increasing
burden of colorectal cancer in China. Lancet Gastroenterol Hepatol.
7:7002022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar
|
|
5
|
Yuan G, Ding W, Sun B, Zhu L, Gao Y and
Chen L: Upregulated circRNA_102231 promotes gastric cancer
progression and its clinical significance. Bioengineered.
12:4936–4945. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li R, Tian X, Jiang J, Qian H, Shen H and
Xu W: CircRNA CDR1as: A novel diagnostic and prognostic biomarker
for gastric cancer. Biomarkers. 28:448–457. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Azhati B, Wang W, Rexiati M, Xing C
and Wang Y: Circular RNA UBAP2 promotes the proliferation of
prostate cancer cells via the miR-1244/MAP3K2 axis. Oncol Lett.
21:4862021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma W, Zhao X, Gao Y, Yao X, Zhang J and Xu
Q: Circular RNA circ_UBAP2 facilitates the progression of
osteosarcoma by regulating microRNA miR-637/high-mobility group box
(HMGB) 2 axis. Bioengineered. 13:4411–4427. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang PF, Wei CY, Huang XY, Peng R, Yang
X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al: Circular RNA
circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress
hepatocellular carcinoma progression. Mol Cancer. 18:1052019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun J, Yin A, Zhang W, Lv J, Liang Y, Li
H, Li Y and Li X: CircUBAP2 inhibits proliferation and metastasis
of clear cell renal cell carcinoma via targeting miR-148a-3p/FOXK2
pathway. Cell Transplant. 29:9636897209257512020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou C, Lyu LH, Miao HK, Bahr T, Zhang QY,
Liang T, Zhou HB, Chen GR and Bai Y: Redox regulation by SOD2
modulates colorectal cancer tumorigenesis through AMPK-mediated
energy metabolism. Mol Carcinog. 59:545–556. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
13
|
Bertrand E, Chartrand P, Schaefer M,
Shenoy SM, Singer RH and Long RM: Localization of ASH1 mRNA
particles in living yeast. Mol Cell. 2:437–445. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou M, Yang Z, Wang D, Chen P and Zhang
Y: The circular RNA circZFR phosphorylates Rb promoting cervical
cancer progression by regulating the SSBP1/CDK2/cyclin E1 complex.
J Exp Clin Cancer Res. 40:482021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng R, Zhang K, Tan S, Gao F, Zhang Y,
Xu W, Wang H, Gu D, Zhu L, Li S, et al: Exosomal circLPAR1
functions in colorectal cancer diagnosis and tumorigenesis through
suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer.
21:492022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Q, Liu J, Deng H, Ma R, Liao JY,
Liang H, Hu J, Li J, Guo Z, Cai J, et al: Targeting
mitochondria-located circRNA SCAR alleviates NASH via reducing mROS
output. Cell. 183:76–93.e22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo
X, Hossain MT, Zhu X, Du K, Hu F, et al: A novel protein
AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin
signaling pathway to promote gastric cancer progression. Mol
Cancer. 20:1582021. View Article : Google Scholar
|
|
19
|
Lyu LH, Zhang CY, Yang WJ, Jin AL, Zhu J,
Wang H, Liu T, Wang BL, Cheng JW, Yang XR and Guo W:
Hsa_circ_0003945 promotes progression of hepatocellular carcinoma
by mediating miR-34c-5p/LGR4/β-catenin axis activity. J Cell Mol
Med. 26:2218–2229. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang M, Sun M and Zhang H: The interaction
between epigenetic changes, EMT, and exosomes in predicting
metastasis of colorectal cancers (CRC). Front Oncol. 12:8798482022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vu T and Datta PK: Regulation of EMT in
colorectal cancer: A culprit in metastasis. Cancers (Basel).
9:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Long H, Zheng Q, Bo X, Xiao X and
Li B: Circular RNA circRHOT1 promotes hepatocellular carcinoma
progression by initiation of NR2F6 expression. Mol Cancer.
18:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma Q, Yang F, Huang B, Pan X, Li W, Yu T,
Wang X, Ran L, Qian K, Li H, et al: CircARID1A binds to IGF2BP3 in
gastric cancer and promotes cancer proliferation by forming a
circARID1A-IGF2BP3-SLC7A5 RNA-protein ternary complex. J Exp Clin
Cancer Res. 41:2512022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He Y, Luo Y, Zhang D, Wang X, Zhang P, Li
H, Ejaz S and Liang S: PGK1-mediated cancer progression and drug
resistance. Am J Cancer Res. 9:2280–2302. 2019.PubMed/NCBI
|
|
25
|
Fu Q and Yu Z: Phosphoglycerate kinase 1
(PGK1) in cancer: A promising target for diagnosis and therapy.
Life Sci. 256:1178632020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Y, Ouyang Y, Li Z, Wang X and Ma J:
S100A8 and S100A9 in cancer. Biochim Biophys Acta Rev Cancer.
1878:1888912023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Markowitz J and Carson WE III: Review of
S100A9 biology and its role in cancer. Biochim Biophys Acta.
1835:100–109. 2013.
|
|
28
|
Qiao L, Xie N, Li Y, Bai Y, Liu N and Wang
J: Downregulation of HNRNPM inhibits cell proliferation and
migration of hepatocellular carcinoma through MAPK/AKT signaling
pathway. Transl Cancer Res. 11:2135–2144. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Li J, Bian X, Wu C, Hua J, Chang
S, Yu T, Li H, Li Y, Hu S, et al: CircURI1 interacts with hnRNPM to
inhibit metastasis by modulating alternative splicing in gastric
cancer. Proc Natl Acad Sci USA. 118:e20128811182021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ho JS, Di Tullio F, Schwarz M, Low D,
Incarnato D, Gay F, Tabaglio T, Zhang J, Wollmann H, Chen L, et al:
HNRNPM controls circRNA biogenesis and splicing fidelity to sustain
cancer cell fitness. Elife. 10:e596542021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sarraf JS, Puty TC, da Silva EM, Allen
TSR, Sarraf YS, de Carvalho LEW, Adami F and de Oliveira EHC:
Noncoding RNAs and colorectal cancer: A general overview. Microrna.
9:336–345. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Z, Liu X, Li Y, Ren P, Zhang C, Wang
L, Du X and Xing B: miR-6716-5p promotes metastasis of colorectal
cancer through downregulating NAT10 expression. Cancer Manag Res.
11:5317–5332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao J and Lu Y: Roles of circRNAs in the
progression of colorectal cancer: Novel strategies for detection
and therapy. Cancer Gene Ther. 31:831–841. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Luo J, Yang W and Ye WC: CircRNAs
in colorectal cancer: Potential biomarkers and therapeutic targets.
Cell Death Dis. 14:3532023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang G, Xu D, Zhang T, Wang G, Qiu L, Gao
X and Miao Y: Biological functions, mechanisms, and clinical
significance of circular RNA in colorectal cancer. Front Oncol.
13:11384812023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang L, Yang X, Zhou F, Sun X and Li S:
Circular RNA UBAP2 facilitates the cisplatin resistance of
triple-negative breast cancer via microRNA-300/anti-silencing
function 1B histone chaperone/PI3K/AKT/mTOR axis. Bioengineered.
13:7197–7208. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T,
Li Y, Lai H, Chen Z, Meng Z, et al: HNRNPL circularizes ARHGAP35 to
produce an oncogenic protein. Adv Sci (Weinh). 8:20017012021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie W, Jiang L, Huang X, You W and Sun W:
Hsa_circ_0004662 accelerates the progression of osteoarthritis via
the microRNA-424-5p/VEGFA axis. Curr Mol Med. 24:217–225. 2024.
View Article : Google Scholar
|
|
39
|
Zhao Z, Song J, Tang B, Fang S, Zhang D,
Zheng L, Wu F, Gao Y, Chen C, Hu X, et al: CircSOD2 induced
epigenetic alteration drives hepatocellular carcinoma progression
through activating JAK2/STAT3 signaling pathway. J Exp Clin Cancer
Res. 39:2592020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yao GS, Fu LM, Dai JS, Chen JW, Liu KZ,
Liang H, Wang Z, Deng Q, Wang JY, Jin MY, et al: Exploring the
oncogenic potential of circSOD2 in clear cell renal cell carcinoma:
A novel positive feedback loop. J Transl Med. 22:5962024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lv C, Hu Y, Zhou X, Zhu Y, Wang J and Zhou
F: CircRNA SOD2 motivates non-small cell lungs cancer advancement
with EMT via acting as microRNA-2355-5p's competing endogenous RNA
to mediate calmodulin regulated spectrin associated proteins-2.
Bioengineered. 13:5756–5768. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Wu C, Du Y, Li Z, Li M, Hou P,
Shen Z, Chu S, Zheng J and Bai J: Expanding uncapped translation
and emerging function of circular RNA in carcinomas and
noncarcinomas. Mol Cancer. 21:132022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang L, Gao H, Li X, Yu F and Li P: The
important regulatory roles of circRNA-encoded proteins or peptides
in cancer pathogenesis (Review). Int J Oncol. 64:192024. View Article : Google Scholar :
|
|
44
|
Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao
S, Hu Y, Yang Y, Du J and Gu C: A novel protein encoded by
circHNRNPU promotes multiple myeloma progression by regulating the
bone marrow microenvironment and alternative splicing. J Exp Clin
Cancer Res. 41:852022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xiong L, Liu HS, Zhou C, Yang X, Huang L,
Jie HQ, Zeng ZW, Zheng XB, Li WX, Liu ZZ, et al: A novel protein
encoded by circINSIG1 reprograms cholesterol metabolism by
promoting the ubiquitin-dependent degradation of INSIG1 in
colorectal cancer. Mol Cancer. 22:722023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang B, Wang YW and Zhang K: Interactions
between circRNA and protein in breast cancer. Gene. 895:1480192024.
View Article : Google Scholar
|
|
47
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu GQ, Wang Y, Wang B, Liu WR, Dong SS,
Chen EB, Cai JL, Wan JL, Du JX, Song LN, et al: Targeting HNRNPM
inhibits cancer stemness and enhances antitumor immunity in
Wnt-activated hepatocellular carcinoma. Cell Mol Gastroenterol
Hepatol. 13:1413–1447. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang WH, Ding MJ, Cui GZ, Yang M and Dai
DL: Heterogeneous nuclear ribonucleoprotein M promotes the
progression of breast cancer by regulating the axin/β-catenin
signaling pathway. Biomed Pharmacother. 105:848–855. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun H, Liu T, Zhu D, Dong X, Liu F, Liang
X, Chen C, Shao B, Wang M and Wang Y: HnRNPM and CD44s expression
affects tumor aggressiveness and predicts poor prognosis in breast
cancer with axillary lymph node metastases. Genes Chromosomes
Cancer. 56:598–607. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gu X, Li X, Jin Y, Zhang Z, Li M, Liu D
and Wei F: CDR1as regulated by hnRNPM maintains stemness of
periodontal ligament stem cells via miR-7/KLF4. J Cell Mol Med.
25:4501–4515. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shou Y, Yue C, Wang Q, Liu J, Xu J, Miao
Q, Liu D, Yang H, Liu Y and Zhang X: circPTPN12 promotes the
progression and sunitinib resistance of renal cancer via
hnRNPM/IL-6/STAT3 pathway. Cell Death Dis. 14:2322023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen TM, Lai MC, Li YH, Chan YL, Wu CH,
Wang YM, Chien CW, Huang SY, Sun HS and Tsai SJ: hnRNPM induces
translation switch under hypoxia to promote colon cancer
development. EBioMedicine. 41:299–309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Laguinge L, Bajenova O, Bowden E, Sayyah
J, Thomas P and Juhl H: Surface expression and CEA binding of hnRNP
M4 protein in HT29 colon cancer cells. Anticancer Res. 25:23–31.
2005.PubMed/NCBI
|