Bladder cancer (BCa) is the most frequently
occurring neoplasm in the urinary tract, originating from the
urothelium. It is a widespread malignancy that carries notable
morbidity and mortality rates. Annually, there are >500,000 new
cases and more than 200,000 deaths reported worldwide This disease
primarily affects individuals aged ≥55 years (1,2).
In general, compared with muscle-invasive BCa (MIBC), non-MIBC
(NMIBC) exhibits more favorable prognosis and a decreased
propensity for extravesical spread. Low grade NMIBC carries a
higher risk of recurrence. Specifically, the 5-year recurrence
rates range from 31 to 78%, while the rates of progression are
between 1 and 45% (3). A total of
60-70% of patients with NMIBC encounter frequent recurrences
following transurethral resection of bladder tumor. The high
recurrence rate can be attributed to incomplete piecemeal resection
and subsequent tumor re-implantation (4). In addition, patients diagnosed with
MIBC at stage T2-T4 display a poor treatment response and
prognosis. This is primarily due to the tumor susceptibility to
rapid growth and distant metastasis, which leads to an unfavorable
clinical outcome and a decreased lifespan of ~15 months (5). Currently, cystoscopy is the
recommended standard procedure for diagnosing and continuously
monitoring treatment outcomes of BCa (6). This is key for patients at a high
risk of recurrence or progression, who require regular surveillance
(3,7).
Regular follow-up cystoscopy is crucial for the
early detection of any new or recurrent tumors, particularly when
monitoring patients with NMIBC who have recurrence rates up to
74.3% (8). Furthermore, high-risk
patients requires more frequent cystoscopy, usually scheduled every
3-4 months within the first 2 years of follow-up (9). However, discomfort and potential
complications, invasiveness, cost, accessibility and the frequency
of follow-up examinations all contribute to the complexity of
implementing and maintaining regular surveillance (10-12). While cystoscopy combined with
biopsy serves an indispensable role in disease monitoring and
recurrence prevention, the development of emerging liquid
biopsy-based biomarkers has decreased reliance on cystoscopy. These
non-invasive approaches may revolutionize management options for
BCa, leading to improved patient outcomes and decreased healthcare
costs (13). Efforts have been
made to develop non-invasive diagnostic tools for urinary tract
cancer by utilizing urine (14)
as urine comes into direct contact with urothelial tumor cells and
reflects the local tumor microenvironment. Analysis of urine
composition, including cell-specific characteristics,
protein/antigen biomarkers and genetic mutations, holds great
promise for revolutionizing the diagnosis and monitoring of bladder
conditions. Urine biomarkers with high sensitivity and negative
predictive value (NPV) may effectively exclude the presence of BCa
(15-17), decreasing the need for additional
diagnostic tests performed in the operating theatre.
As an adjunct measure for detecting BCa, urine
cytology serves a key role in the objective assessment of cell
characteristics, including morphology, nuclear features and
cellularity. Urine cytology enables the classification of lesions
into cyto-diagnostic categories, including non-diagnostic cases,
neoplastic high-grade urothelial carcinoma (UC), atypical
urothelial cells, those suspicious for high-grade UC, high-grade
UC, and other malignancies. This classification provides valuable
insight into the response of tumors to therapeutic interventions,
such as chemotherapy or radiation (18,19). Currently, urine cytology
demonstrates high specificity (80-95%) in detecting high-grade UC.
However, its sensitivity is inadequate, especially for detecting
low-grade UC (overall sensitivity, 4-31%; well differentiated grade
1, 12%; moderately differentiated, grade 2, 26%) (20,21). Recent research has focused on
evaluating the effectiveness of urine cytology as a screening
method for monitoring low-risk NMIBC (22,23). Among urine cytology tests
conducted, only a small percentage (2.9%) of patients in all risk
groups consistently show positive results consistent with
cystoscopy and pathological findings (22,23). Non-invasive urine cytology and
urography present >30% variation in sensitivity and specificity,
depending on the location and grading of the UC. High-grade tumors,
including carcinoma in situ, can show a positivity rate up
to 84%. By contrast, low-grade tumors exhibit a considerably lower
sensitivity, with a positivity rate of 16% (24,25).
The present review discusses the utilization of
urine-based liquid biopsy technology to address concerns regarding
the high costs and low patient compliance associated with follow-up
procedures. It systematically summarizes urinary biomarkers,
including protein assays, characterization of nucleic acids (such
as DNA and RNA) and emerging potential biomarkers, as well as
innovative diagnostic tool (Table
I). The present reviewed aimed to evaluate advancements in
urinary biomarkers, with an emphasis on performance variability and
the ongoing challenges in the diagnosis, prognosis and monitoring
of patients with BCa. Additionally, the present review compares the
efficacy of urine and blood biomarkers in the detection of
urological tumors, especially in BCa.
At present, several protein-based tests, such as
NMP22 (MDxHealth), bladder tumor antigen (BTA) (Polymedco Inc.
Cortlandt Manor, NY, USA), UroVysion (Abbott Laboratories) and
ImmunoCyt (Diagnocure Inc., Québec, Canada), have been approved by
the United States FDA and are commercially available for the
diagnosis and surveillance of BCa (26). NMP22, a key component of the
nuclear mitotic apparatus protein, serves a vital role in cellular
processes, including DNA replication, RNA transcription and gene
expression. Understanding of the NMP22 regulatory network is key
for the proper functioning and regulation of cells; disruption in
these mechanisms can have notable implications for cancer
proliferation. The concentration of NMP22 in malignant transitional
cells is 80-fold higher in BCa compared with normal urothelial
cells (27). The NMP22 test is
initially approved for surveillance of NMIBC in 1996. It utilizes
quantitative ELISA for detection, which demonstrates a sensitivity
of 69% and a specificity of 77% (28). In 2020, an additional
point-of-care (POC) test, BladderChek, developed by Matritech Inc.,
is also approved for use (sensitivity, 58%; specificity, 88%)
(29). The performance of NMP22
assays has been confirmed through several meta-analyses (30,31), demonstrating overall sensitivity
of 73 and specificity of 79% (sensitivity, 60% and specificity, 79%
for low-grade upper tract UC (UTUC); sensitivity and specificity,
both 79% for high-grade UTUC) (32). The expression of proteins is
dynamic and can vary based on factors including tumor stage and
grade, heterogeneity, size and presence of infections or
inflammation. Existing research indicates an association between
the sensitivity and specificity of NMP22 and tumor aggressiveness
(33-35). Recent studies have demonstrated
that NMP22 exhibits greater sensitivity, NPV and accuracy in
detecting recurrence compared with cytology (36,37). In low-grade BCa tumors, the
sensitivity of NMP22 is 81%, whereas cytology achieves a
sensitivity of 23%. Combination of NMP22 and cytology demonstrates
significantly higher sensitivity (91.63% vs. 81.83% vs. 53.96%) and
NPV (87.59% vs. 77.46% vs. 61.02%) than either NMP22 or cytology
alone (38,39).
The urinary tests for BTA, which include BTA stat
(Bion Diagnostic Sciences, Inc.) and BTA TRAK (Bard Diagnostics),
have obtained FDA approval as non-invasive diagnostic approaches
(40,41). BTA stat and BTA TRAK are
single-step immunochromatographic qualitative assays for diagnosis
and monitoring of BCa (42). BTA
stat test is a rapid test in which a urine sample is applied to a
test strip. This strip contains specific monoclonal antibodies that
bind to the targeted antigen if H-related protein (hCFHrp) is
detected (43). hCFHrp plays a
key role in regulating activity of the alternate complement
pathway, allowing malignant cells to escape from immune
surveillance. Complement factor H in the bloodstream has the
potential to interfere with the precision of BTA tests, resulting
in the occurrence of false positive outcomes (44). This effect is particularly
prominent in individuals who have benign urological conditions
(45). Sensitivity and
specificity of the BTA test in detecting BCa range from 57 to 83%
and 60 to 92%, respectively (40,46,47). Due to variability in its
specificity (29-86%), BTA is not recommended for the initial
diagnosis of BCa. However, it can be utilized as a component of
surveillance protocols to monitor changes in antigen levels over
time, particularly when combined with cystoscopy.
ImmunoCyt (UCyt+) assay has been approved by the FDA
since February 2000 as an approved supplementary tool for the
management of BCa alongside urine cytology and cystoscopy (60). ImmunoCyt test can identify
specific tumor-associated antigens in urine based on
immunofluorescence using monoclonal antibodies M344, LDQ10 and
19A211 (61). The sensitivity and
specificity of ImmunoCyt assay are 50-100% and 69-79%,
respectively, and the positive predictive value (PPV) and NPV are
26-67% and 91-96%, respectively (61,62).
A number of studies have examined the diagnostic
accuracy of ImmunoCyt and cytology, revealing notable differences
(55-58). ImmunoCyt exhibits lower
specificity (65.7% vs. 90.6%), positive (2.578 vs. 3.862) and
negative likelihood ratio (0.385 vs. 0.459) and area under the
curve (AUC) (0.791 vs. 0.824) compared with cytology (63). While ImmunoCyt demonstrates higher
sensitivity (72.5% vs. 56.6%), it may not possess the same level of
accuracy as cytology in relation to other factors such as
endogenous and exogenous interference, as well as the methods of
detection and sample handling. On the other hand, several studies
confirm that ImmunoCyt improves overall performance of cytology,
particularly for small (<1 cm), superficial and low histological
grade (tumor in situ, Ta, T1 and T2) tumors (64,65). Cytology tests demonstrate
sensitivity of 29% and NPV of 88%. When combined with ImmunoCyt,
these values improve to 84 and 95%, respectively (65). These results suggest the potential
utility of the ImmunoCyt test in decreasing the need for frequent
follow-up cystoscopies. This test is also suitable for continuous
monitoring of low-risk patients with BCa, thereby allowing early
detection of disease progression and prompt intervention when
necessary. ImmunoCyt test plays a key role in predicting painless
hematuria, surpassing the effectiveness of traditional cytology,
especially in cases where imaging and cystoscopy results are
negative (66).
A large number of protein-specific biomarkers or
molecular signatures are identified using proteomic techniques in
urine (67), blood serum
(68,69) and plasma (70,71). Advanced methodologies such as mass
spectrometry (MS) and machine learning have notably enhanced the
ability to identify these biomarkers, particularly in urine samples
(72-75). Studies have been performed to
determine potential prognostic or predictive value of multiple
protein candidates, including UBC Rapid immunoassay (CK8, CK18)
(IDL Biotech), CK20, cyclin dependent kinase inhibitor 2A (CDKN2A),
erb-B2 receptor tyrosine kinase 2 (ERBB2), phosphatase and tensin
homolog (PTEN), vascular endothelial growth factor (VEGF), matrix
metallopeptidase 9 (MMP-9) and Survivin, to enhance the assessment
of disease progression and risk of BCa recurrence (75-81). Similarly, a quantitative proteomic
approach using isotope tags for relative and absolute
quantification (iTRAQ) labeling has identified 21 candidate
proteins in urinary samples (82). Of these markers, bromodomain
testis associated (BRDT), calcyclin binding protein (CYBP),
glycyl-tRNA synthetase (GARS) and heparin binding growth factor
(HDGF) with a high discrimination ability in UC patients, have
shown substantially elevated expression in the UC cohort compared
with normal groups. The AUC values for the combined panel of these
biomarkers are 0.962 for distinguishing between UC and normal
groups, and 0.860 for differentiating between UC and control groups
(82). Machine learning has been
employed to identify various protein markers for constructing a BCa
diagnostic model, which comprises 14 markers found in serum and
urine supernatants, achieving 86% sensitivity and 82% specificity
and surpasses FISH in detecting low-risk BCa (64).
Use of ELISA and western blotting has enabled the
measurement and verification of specific proteins of interest in
urine samples obtained from patients with BCa (83). In one study, researchers analyzed
genome-wide gene and protein expression via quantitative PCR
(qPCR), western blotting and ELISA in 66 patients with BCa and 70
individuals without tumors of the urinary system (84). The results demonstrated a
significant increase in protein expression of MMP23B, a member of
the MMP family, in both urine and plasma compared with tissue
biopsies. There is an association between the level of urinary
MMP23B and risk stratification, suggesting MMP23B could be a
potential non-invasive biomarker for diagnosis of BCa (84,85).
Urine protein tests typically show greater
sensitivity than cytological analysis but lower specificity. For
example, the BTA assay exhibits a higher sensitivity (67% vs. 43%)
and a lower specificity (77% vs. 90%) compared with urine cytology
(86). To date, no single
biomarker has replaced cystoscopy and urine cytology in clinical
practice. These urinary tests are limited by lack of specificity
and high proportion of false positive results, with specificity
ranging from 50 to 80% and false-positive rates between 16.7 and
30.5% (87,88). False positives can be caused by
conditions such as inflammation, kidney and bladder stone disease
and hematuria (89). The
sensitivity of protein-based biomarkers typically increases with
the advancement in tumor stage or grade (90). To the best of our knowledge, there
are few prospective studies investigating urine biomarkers in
low-grade NMIBC and small papillary lesions and the available
cross-sectional studies appear to overestimate the sensitivity of
these biomarkers (91,92). In addition, the concentration of
potential protein biomarkers in urine is typically low, and
detection can be influenced by various factors, including sex, age,
hormonal status, diet and physical activity (93). Considering the limitations of
urinary protein-based markers, it is advisable to analyze these
markers alongside a panel of biomarkers or in combination with
clinical tests. An integrated approach is likely to enhance the
detection rate of BCa and improve effectiveness of screening
programs for high-risk populations.
Despite the benefits and potential value of protein
markers in predicting, diagnosing and determining the prognosis of
BCa, the field of urinary proteomics faces challenges due to lack
of standardized protocols for the collection, storage, processing
and analysis of samples (64).
This absence of standardization leads to inconsistencies and lack
of reproducibility in experimental results. Advances in proteomics
sequencing technology have enhanced the capability to generate
comprehensive datasets at the single-cell level, even with minimal
sample quantities (72,94). The incorporation of artificial
intelligence (AI) and machine learning into analysis of proteomics
data is expected to advance the early detection of superficial BCa
and enhance the precision of diagnosis.
BCa is a common malignant tumor characterized by
genomic abnormalities that influence tumorigenesis and progression
through the accumulation of mutational factors (95). In addition to protein biomarkers
released into urine by tumor cells, DNA-based analysis of genetic
material, such as point mutations, methylation patterns, copy
number variation (CNV) and microsatellite instability (MSI), as
well as the molecular characterization and fragmentation of
cell-free DNA (cfDNA), reveal an association between
genetic/epigenetic signatures and high mutational burden and
widespread variations affecting multiple chromosomes (15,96). Next-generation sequencing (NGS)
and machine learning approaches facilitate identification of the
most frequently mutated genes and methylated regions, as well as
the potential mechanisms of action in BCa (97). The systematic analysis of big
data, in conjunction with advancements in bioinformatics
technology, enhances assessment of the characteristics of driver
genes and sequence characterizations to establish associations
between cancer subtype, prognosis, grading, staging and treatment
responses. Consequently, it provides valuable insights and guidance
for early diagnosis of BCa, identification of therapeutic targets
and the monitoring of follow-up care.
Driver variants can be detected through molecular
testing and provide valuable information regarding the presence and
progression of UC. Numerous somatic hotspot mutations can be
identified utilizing diverse technological platforms, including
NGS, machine learning, real-time qPCR and Sanger sequencing
(98,99). Several commercially available
assays, such as UroSEEK (Memorial Sloan Kettering Cancer Center),
XpertBC (Cepheid) and Uromonitor-V2 (U-monitor, Porto, Portugal),
are employed for early detection and prognostic monitoring
(100-104). The aforementioned tests are
generally designed to evaluate combinations of common oncogenes and
tumor suppressor genes. For example, oncogenes such as Ras,
ERBB2, MDM2 proto-oncogene, cyclin D1 (CCND1),
myelocytomatosis proto-oncogene (C-MYC),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) and epidermal growth factor receptor (EGFR),
have been identified as potential diagnostic markers for BCa
(105). Mutations or expression
deletions in tumor suppressor genes, such as TP53,
p21, PTEN, fibroblast growth factor receptor 3
(FGFR3), serine/threonine-protein kinase 15 and fragile
histidine triad diadenosine triphosphatase (FHIT), have
shown promise as prognostic markers for BCa disease (106). Mutations in the FGFR3 and
PIK3CA genes are commonly found in superficial papillary
bladder tumors. The overall mutation rate of FGFR3 is 57.50%
(23/40), which is significantly higher compared with patients with
benign bladder disease (107).
The activation of the PI3K signaling pathway serves a key role in
promoting the aggressive behavior of tumor with FGFR3
mutation (108). Therefore,
evaluation of PIK3CA requires the concurrent assessment of
FGFR3 to determine its prognostic value, which holds
potential as a therapeutic strategy for BCa (109). Furthermore, p53 protein
expression is prevalent in BCa tissues, with a notable association
between p53 expression and the grade and stage of tumors. The
incidence of recurrence is notably higher in patients exhibiting
positive TP53 gene expression compared with those with
negative expression (110).
Alterations in TP53 gene expression may influence the
evolutionary and progressive aspects of BCa (111). Moreover, mutation classifiers
including TP53, PIK3CA and ataxia telangiectasia
mutated, have the capability to assess tumor mutational burden.
This assessment can subsequently guide therapeutic decisions
regarding use of immune checkpoint inhibitors (ICIs) across
different risk levels in BCa (112). Telomerase reverse transcriptase
(TERT) promoter mutations represent the most common genetic
alterations in the urinary system and have been identified in
70-80% of uroepithelial BCa cases. TERT promoter mutations
(C228T and C250T) occur in 50-70% and 8-15% of patients,
respectively (102,113). TERT promoter mutations
are demonstrated in urinary DNA, present high sensitivity and
specificity in detecting various types of UC. The overall
performance of urine supernatant cfDNA shows sensitivity of 80.7%
and a specificity of 96.6%, with NPV of 92.5% (114).
UroSEEK classifier, which integrates genetic
alterations from 11 commonly affected genes, exhibits a sensitivity
of 92.7% and a specificity of 90.7% for the diagnosis of UC
(115). Moreover, it has a high
accuracy rate of 91.8% in low-grade UC (116). Mutations in TERT,
FGFR3 and KRAS genes have a sensitivity of 93.3% and
a specificity of 80% for diagnosing BCa (117). A pilot study involving 16
patients with BCa revealed that gene mutations detected in both the
urine supernatant and sediment show a higher concordance with
cancerous tissue compared with those identified in plasma (118); AUC values are 0.94 in urine
supernatant (panel, TERT, FGFR3, TP53,
PIK3CA and KRAS) and 0.91 in urine sediment cohort
(panel, TERT, FGFR3, TP53, HRAS,
PIK3CA, KRAS and ERBB2), respectively. The
detection of somatic mutations in cfDNA in urine holds promise as a
diagnostic approach for identifying BCa in patients with hematuria
(118).
Several studies have provided evidence that BCa
transformation is frequently accompanied by widespread changes in
epigenetic landscape including DNA methylation and histone
modifications, which occur prior to formation of tumors (119,120). A comprehensive analysis of the
BCa epigenome has revealed epigenetic alterations in
methylation-specific PCR targeting orthodenticle homeobox 1
(OTX1), one cut homeobox 2 (ONECUT2) and twist family
bHLH transcription factor 1 (TWIST1) (121). AssureMDx (MDxHealth) assay is
developed to design a panel consisting of mutational statuses
(TERT, FGFR3, HRAS) and methylation patterns
(OTX1, ONECUT2, and TWIST1) in urinary
exfoliated cells (122).
Notably, there are distinct differences in DNA methylation patterns
between NMIBC and MIBC, with DNA hypomethylation being commonly
observed in low-grade, non-invasive uroepithelial tumors (123). Specifically, methylation levels
of insulin promoter factor 1 (IPF1), galanin receptor 1
(GALR1), T-cell acute lymphocytic leukemia 1 (TAL1),
proenkephalin (PENK) and tight junction protein 2
(TJP2), as determined through bisulfite sequencing, are
noticeably higher in MIBC compared with NMIBC samples (124). The continued presence of these
abnormal DNA methylation changes in urine samples following
surgical excision of the tumor indicates their potential as
biomarkers assessing the probability of early detection, tumor
recurrence and treatment efficacy (125). The aforementioned study
indicates the possibility of comprehensively studying the entire
process of tumor development, including precancerous lesions,
through the detection of methylation patterns in urinary tumor DNA
(utDNA) (126). Urinary DNA
methylation profiles are valuable in effectively stratifying the
risk of BCa. The sensitivity of these profiles for diagnosing
high-grade BCa is 100% (95% CI: 82.5-100%), while for low-grade
BCa, it is 62% (95% CI: 51.3-72.7%) (126,127).
Analysis of DNA methylation pattern can be utilized
to predict the likelihood of molecular residual disease (MRD).
Methylation-based recurrence monitoring models offer an advantage
over mutation-based recurrence monitoring by providing earlier
indications of MRD (128-130).
UCseek classifier (Beijing Institute of Genomics & Peking
University First Hospital), which uses methylation and CNV features
of urine samples to construct methylation models (118). This approach shows good
performance in accurate non-invasive diagnosis and recurrence
monitoring of UC. It demonstrates an accuracy of 91.8% for
low-grade UCs and recurrence monitoring accuracy (90.91% vs.
59.09%), which is better than that of cystoscopy (118). Compared with other urinary
biomarkers, Bladder Epicheck (Nucleix Ltd.) provides an advantage
in ruling out the recurrence of BCa, as its effectiveness is
uncompromised by previous or ongoing treatments (131). The reliability of hematuria
screening as an early indicator for diagnosing BCa faces challenges
due to variations in clinical presentation. The incidence of BCa is
between 10 and 20% in patients presenting with gross hematuria and
between 2 and 5% in individuals with microscopic hematuria
(132). The assessment of cancer
risk in populations with hematuria reveals promising outcomes when
using UriFind (AnchorDx Medical Company, Ltd.), a diagnostic tool
that leverages the methylation of the dual genes ONECUT2 and
vimentin. UriFind exhibits a diagnostic sensitivity of 91.2% and a
specificity of 85.7%, underscoring its strong performance (133). Methylation genes or
combinations, including TERT, FGFR3, TWIST and
orthodenticle homeobox 1 (OTX1), have been extensively
investigated but are not yet available for clinical use (134,135). Although urine methylation
studies in detecting MRD are still in the stage of large-scale
clinical observation (36,136,137),
urine testing may have advantages (such as non-invasive and
reproducible procedure, a low protein and cellular content, and
detection of multiple diseases and cancer type) compared with blood
testing.
Malignant tumor cells exhibit genetic instability
and frequently display copy number alteration (CNA) involving
entire chromosomes or segments. CNV serves a key role in the tumor
progression and metastasis of BCa; alternations can result in
upregulation of oncogenes or the loss of function of tumor
suppressor genes, promoting uncontrolled cell proliferation and
tumor progression (138). These
genetic abnormalities disrupt normal cellular function and enhance
the ability to proliferate, invade surrounding tissue and
metastasize. Studies have revealed that consistent CNV features are
identified in multiple cells of patients with UC (139,140). A total of 57 CNVs have been
identified in 131 cases of high-grade MIBC. Among these CNVs
identified by whole genome and RNA sequencing, notable copy number
deletions are observed in CDKN2A, retinoblastoma inhibitory gene,
phosphodiesterase 4D, FHIT and CREB binding protein, while copy
number gains are displayed in tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein zeta (YWHAZ), peroxisome
proliferator activated receptor gamma (PPARG), yes-associated
protein 1 (YAP1), MYC and E2F transcription factor 3 (E2F3)
(141). This analysis offers
perspective on mutations and regions of CNV across multiple
pathways that are frequently dysregulated in BCa. Further analysis
of exome sequencing data from tumor tissue obtained from 120
patients with UC highlighted notable disparities in CNV between
normal uroepithelial and tumor tissue and suggests that the
accumulation of mutations may not be adequate to drive
transformation of normal epithelium into malignant tumors (142). Studies have revealed a
noteworthy association between fibroblast growth factor receptor
substrate 2 CNV and protein expression in BCa, demonstrating
unfavorable prognosis characterized by decreased overall survival
and heightened risk of disease recurrence or progression (143,144). A novel diagnostic model, UroCAD
(Hongyuan Biotech, Inc.), has been developed using an optimized
low-coverage whole genome sequencing (WGS) technology. The test
demonstrates superiority over cytology as a non-invasive method for
diagnosis and monitoring of UC, exhibiting notably higher
sensitivity (80.4% vs. 33.9%) while maintaining comparable
specificity (94.9% vs. 100.0%) (145). Low-depth WGS has been performed
on a cohort of 168 patients diagnosed with urological tumors to
assess CNA; machine learning techniques have been used to develop
multidimensional algorithmic model UC Detector (Beijing Institute
of Genomics & Peking University First Hospital), designed to
identify 50 distinct CNA features associated with UC (sensitivity,
86.5%; specificity, 94.7%) (146).
Microsatellites, also referred to as short tandem
repeats, are repetitive DNA sequences ubiquitously present across
the human genome. These sequences are characterized by high levels
of polymorphic variability (147). MSI phenotype has importance in
BCa tumorigenesis and development due to its ability to cause the
insertion and deletion of repetitive sequences (148). There have been advancements in
analysis of genome-wide MSI status through NGS methods (149). Microsatellite analysis (MSA) not
only enables identification of valuable biomarker targets but also
facilitates detection of MSI-positive cancerous lesions for guiding
immunotherapy in BCa (150).
Studies on a cohort of 149 patients with 15 deficient mismatch
repair/high MSI (MSI-H) tumors have been performed and the FDA has
granted approval for utilization of MSI as a pan-cancer biomarker
for guiding pembrolizumab treatment (151,152). MSI-H status is associated with a
high mutational load, Lynch syndrome (LS) and a durable response to
ICIs in upper tract UC (153).
Additionally, MSI typing effectively distinguishes patients with
different prognoses, with those exhibiting MSI-H having a more
favorable prognosis compared to those with low microsatellite
instability (154).
An approach has been developed for detection of MSI
using amplicon-based NGS technology instead of tissue biopsy. This
approach involves mapping plasma circulating tumor DNA (ctDNA)
fragments to the genome (155).
Similarly, other groups have examined the association between MSI
and clinicopathological features of bladder or renal cell
carcinoma, which can be determined by detecting MSI in urine and
identifying targeted loss of heterozygosity (LOH) deletions during
tumor cell transformation (156,157). MSA can be an effective tool for
BCa stratification and evaluate the likelihood of recurrence. This
technique demonstrates a notable sensitivity in detecting both low-
and high-grade lesions. Specifically, it exhibits sensitivities of
67, 86 and 93% for G1, G2 and G3 lesions, respectively (157). Another study has explored use of
MSA assay for detecting recurrent transitional cell carcinoma of
the bladder in 21 patients; MSA assay is able to detect recurrence
4-6 months earlier than cystoscopy (158). The sensitivity of MSA in
detecting BCa tends to improve with the stage and grade, while the
accuracy of results can be affected by complications such as
inflammation. Combination of LOH analysis with other BCa diagnostic
methods, such as cytology or FISH, has demonstrated enhanced
accuracy compared with single assays (159). Integrating LOH and cytological
analysis of urine samples, in conjunction with cystoscopy, offers a
novel paradigm for monitoring recurrence in urinary tract
malignancies. Several clinical trials have confirmed that MSA
exhibits greater sensitivity than cytology in detecting and
predicting recurrent BCa (160,161). The sensitivity of MSA ranges
from 75 to 96%, whereas cytology shows a sensitivity range of
13-50% (162). Moreover, MSA has
high diagnostic performance, with an accuracy rate of 83.3% and
specificity of 100%, particularly in patients with superficial
disease or low-grade tumors (163). The performance is notably
superior to other diagnostic tests, such as cytology and BTA tests
and UBC tests (148,164).
Concentration of cfDNA in patients is significantly
higher compared with that found in a healthy population,
particularly in individuals with advanced and progressing disease
stages (165). Patients with
liver cancer show the highest concentration of cfDNA (46.0±35.6
ng/ml), while those with lung cancer exhibit the lowest
concentration (5.23±6.4 ng/ml). The concentrations of cfDNA are
significantly correlated with both the type and stage of cancer
(166). This makes urinary cfDNA
(ucfDNA) an ideal candidate for investigation of urological
malignancies, such as BCa and kidney cancer (167). Further studies have demonstrated
that the specific molecular characteristics, particularly
concentration and integrity of ucfDNA, may serve as potent
discriminators differentiating patients with BCa from healthy
controls (168,169). DNA originating from normal
apoptotic cells is typically highly fragmented, which distinguishes
it from the DNA of necrotic cancer cells, which maintains
structural integrity. The integrity of ucfDNA is >40-fold higher
in patients diagnosed with BCa than healthy controls (170). Investigations have been
conducted into variations in concentration of ucfDNA in relation to
BCa and specific benign urological conditions (171,172). The total quantity of ucfDNA
exhibits a notable discrepancy between pTa group (~40,000 ng for 25
BCa patients) and pT1-pT4 (around 360,000 ng for 40 BCa patients)
(165). These findings suggest
that ucfDNA could potentially serve as a promising biomarker for
distinguishing stages of BCa. In addition, patients with BCa
display ucfDNA concentrations >250 ng/ml. By contrast, <40%
of healthy individuals with negative cystoscopy results show ucfDNA
concentration levels >250 ng/ml (173). The levels of ucfDNA may vary
depending on number and size of tumors, grade, stage, presence of
urinary tract infection and leukocyturia. ucfDNA demonstrates
limited sensitivity in detection of poorly staged and graded BCa,
such as those in patients with pTa; therefore, there is a
continuing need for the development of standard methods for
quantification and amplification (174).
Distribution of ucfDNA fragments does not occur at
random, but rather demonstrates a patterned organization. Cells
undergoing apoptosis predominantly release ucfDNA that is highly
fragmented (185-200 bp). The size distribution of ucfDNA fragments
exhibits prominent peaks within the 40-120 bp spectrum, with a mode
value of 81 bp (175). Each peak
is regularly spaced by 10 bp intervals, suggesting a possible
transient defense against total degradation (176). ucfDNA fragments exhibit distinct
characteristics compared with those in plasma, including increased
degradation and decreased length (175). Despite the high fragmentation
observed in ucfDNA, it remains uncertain whether the profile of
these fragments accurately mirrors the inherent genomic
architecture. Owing to potential interference from cfDNA of
non-tumor origin, which may be released into the urine from the
systemic circulation, the concentration of ctDNA in urine becomes
diluted, resulting in reduced levels of detectable ctDNA. Long
degradation periods may lead to an abundance of short fragments of
ucfDNA. ucfDNA fragments >250 bp in length correspond to the
sequences of three specific oncogenes: c-Myc, human
epidermal growth factor receptor 2 and brain enriched myelin
associated protein 1 (177).
These oncogenes are frequently amplified in various stages of BCa,
which include premalignant conditions, primary invasive high-grade
and metastatic BCa (178).
Therefore, to enhance the capacity and precision of BCa detection
using this non-invasive marker, further exploration is warranted to
comprehend the precise mechanism and origin of ucfDNA fragments.
Evaluation of the relationship between ucfDNA content and tumors,
particularly through analysis of the ratio between longer and
shorter DNA fragments in conjunction with the concentration at a
specific fragment size may hold potential as an effective tool for
BCa diagnosis.
Fragmentation pattern of ucfDNA faces several
challenges, including a notable proportion of false positives and a
detection efficacy inferior to that of its blood-based counterparts
(179). Urine exhibits higher
activity levels of DNA enzymes compared with blood, thus elevating
the probability of cfDNA degradation (180,181). Secondly, the abundance of
microorganisms interferes with chemical and physical properties of
urine samples, as well as the accuracy of test results (182). Thirdly, enzymatic activities of
both deoxyribonuclease I and II demonstrate higher values in urine
than in blood (183). This leads
to a decreased half-life of the ucfDNA samples, ranging from 2.6 to
5.1 h. Consequently, the ucfDNA exhibits a heightened
susceptibility to degradation and extensive fragmentation. These
factors impede the extensive application of ucfDNA in BCa
detection. At present, morning urine samples are preferred due to
advantages including its higher concentration, reduced external
interference, decreased risk of contamination, and minimized
variation in urine composition for urine-based liquid biopsies
(184). To maintain the
integrity and stability of ucfDNA at room temperature, EDTA is used
(176,180). EDTA serves a pivotal role in
preventing the degradation of ucfDNA, thereby enhancing the
likelihood of accurate testing outcomes. Additionally, there is a
lack of standardized methodologies for both the preservation and
analysis of ucfDNA samples.
MicroRNAs (miRNAs or miRs), a class of small RNA
molecule, are characterized by their non-coding nature and size,
typically ranging from 18 to 25 bp. Aberrantly expressed microRNAs
serve as both oncogenes and tumor suppressors in different types of
malignant tumor (185). These
miRNAs regulate malignant biological properties of tumors,
including progression and metastasis of urinary tumors. Levels of
miRNA 15a, miRNA-200a (miR-200a) and miRNA-210-3p (miR-210-3p) in
the urine of patients with renal cell carcinoma (RCC) are increased
following surgical intervention (186). This is associated with tumor
size and suggests the potential utility of these miRNAs as
molecular markers for prognostic assessment and therapeutic
monitoring of RCC. The downregulation of miR-29b-3p and miR-31
expression is associated with clinicopathological features and
survival prognosis of patients with BCa (187,188). In addition, miRNA-10b (miR-10b)
is detected in urine at an early stage of BCa, suggesting its
potential for early screening and diagnosis. It exhibits a
specificity of 91.1% and sensitivity of 80.9%, which is higher than
that of urine cytology (specificity, 85%; sensitivity, 75%)
(189). A panel comprising
miR-141, miR-34b, miR-10b and miR-103 exhibits enhanced diagnostic
efficacy for BCa, achieving sensitivity of 75% and a specificity of
63.5% compared to the detection of individual miRNA (190). To evaluate the predictive
potential of miRNAs found in urine, a study has been performed to
identify differentially expressed miRNAs in bladder tumors through
transcriptome sequencing; miRNA-145 (miR-145) and miRNA-182
(miR-182) demonstrate superior diagnostic performance compared with
traditional methods such as urine cytology (sensitivity, 44%;
specificity, 85%) and cystoscope (sensitivity, 85-90%), achieving
sensitivity of 93% and a specificity of 86% (191). Despite these findings, the
diagnostic efficacy of individual or various combinations of miRNAs
has yet to be fully elucidated.
The role of circulating mRNAs, including mRNA,
circular RNA (circRNA) and non-coding RNA as markers in BCa has
garnered considerable interest (192-194). The urinary biomarker urothelial
cancer associated 1 (UCA1), a BCa-specific long non-coding RNA
(lncRNA), is associated with malignant characteristics of bladder
tumors (195) and serves a
crucial role in facilitating progression of BCa through the
regulation of glutamine-driven tricarboxylic acid cycle anaplerosis
(196). Meta-analysis indicates
that UCA1 exhibits a sensitivity of 84% and a specificity of 87%
for detection of BCa (197).
Transcriptomic data derived from 700 patient samples treated with
ICIs corroborates that a machine learning approach using
network-guided biomarkers enhances prediction of overall survival
in patients with BCa undergoing ICI therapy (198). Urinary exosomes serve as
abundant sources of lncRNAs such as prostate cancer associated
transcript 1 (PCAT-1), anti-ribosome protein intergenic lncRNA
(ANRIL), muskelin 1-antisense RNA (MKLN1-AS), tumor associated
lipid metabolism 1 (TALAM1) and titin-antisense 1 (TTN-AS1) that
exhibit notable enrichment and high expression in patients with BCa
compared to healthy controls (199). Studies report sensitivity
ranging from 43 to 96% and specificity between 48 and 90% (200,201). Additionally, the integrated AUC
is 0.87, indicating a strong overall diagnostic performance for
BCa. lncRNAs display the highest overall sensitivity, with
integrated sensitivity value of 78%. Conversely, miRNAs demonstrate
the highest overall specificity, achieving an integrated
specificity value of 81% (202,203). lncRNAs such as lymph node
metastasis-associated transcript 2 (LNMAT2) and brain cytoplasmic
RNA 1 (BCYRN1) are important factors in lymph node (LN) metastasis
of BCa and are associated with a poor prognosis (204,205). Furthermore, long intergenic
non-protein coding RNA 958 promotes LN metastasis, proliferation,
lymphatic vessel formation, distant metastasis and invasion in BCa
(206).
Urinary exosome-specific markers, including mRNA
kelch domain containing 7B, caspase 14 (CASP14), protease serine1
(PRSS1) and lncRNAs MIR205 host gene (MIR205HG) and growth arrest
specific 5 (GAS5), have been found to be associated with BCa stage,
grade and degree of hematuria (207). A model based on the combination
of these markers shows sensitivity of 88.5% and a specificity of
83.3% for diagnosing BCa at early stage (207). To date, the commercially
available mRNA-based urine tests for BCa are CxBladder (Pacific
Edge Ltd.) and Xpert BC (Cepheid, Sunnyvale, CA, USA) tests
(208). CxBladder uses reverse
transcription-qPCR to detect expression levels of several urinary
targets, including insulin like growth factor binding protein 5,
homeobox A13 (HOXA13), midkine (MDK), cyclin dependent kinase 1
(CDK1) and C-X-C motif chemokine receptor 2 (CXCR2). This approach
is suitable for excluding low-risk BCa with hematuria, diagnosing
the urinary tract-related disease and monitoring treatment
progress. The sensitivity of the CxBladder test is up to 82% in
patients with hematuria (209).
XpertBC targets mRNA signatures such as acute basophil leukemia 1,
corticotropin releasing hormone (CRH), insulin like growth factor 2
(IGF2), uroplakin 1B (UPK1B) and annexin A10 (ANXA10). It offers
simplicity, speed and automated detection, with sensitivity of
around 80% (210). Studies have
demonstrated that circRNAs are key regulators in BCa and prostate
and renal cancers, exerting their effects on proliferation,
metastasis, apoptosis, metabolism and drug resistance (211,212). By modulating expression of
miRNAs and associated proteins, circRNAs contribute to the complex
molecular landscape of cancers. For example, circ102336,
circ0008399 and circ0058063 have been found to exert effects on
drug resistance through the regulation of miRNAs expression and
associated proteins. Specifically, the sensitivity of
cisplatin-resistant BCa cells to cisplatin is enhanced by
modulating the expression levels and stability of circRNAs, either
through promotion or inhibition (194,213). In addition, circRNA lysine
demethylase 4C, circRNA itchy E3 ubiquitin protein ligase
(circITCH) and circRNA activin A receptor type 2A (circACVR2A)
enhance BCa invasion and metastasis via the miRNA-200bc-3p
(miR-200bc-3p)/zinc finger e-box binding homeobox 1 (ZEB1),
miRNA-17 (miR-17)/miRNA-224 (miR-224) and miRNA-626 (miR-626)/EYA
transcriptional coactivator and phosphatase 4 (EYA4) axes (214-216). circRNAs have a distinct
advantage in discovery of novel clinical diagnostic biomarkers due
to their increased stability compared with linear RNAs. Unlike
linear RNAs, circRNAs are resistant to nuclease degradation,
rendering them more resilient (217). A multi-omics analysis
incorporated mRNA, miRNA, lncRNA and methylation data, utilizing an
innovative clustering approach using 10 distinct machine learning
methods to develop risk prediction models and identify potential
biomarkers; these biomarkers could enhance prognosis and inform
immunotherapy strategies for MIBC (218).
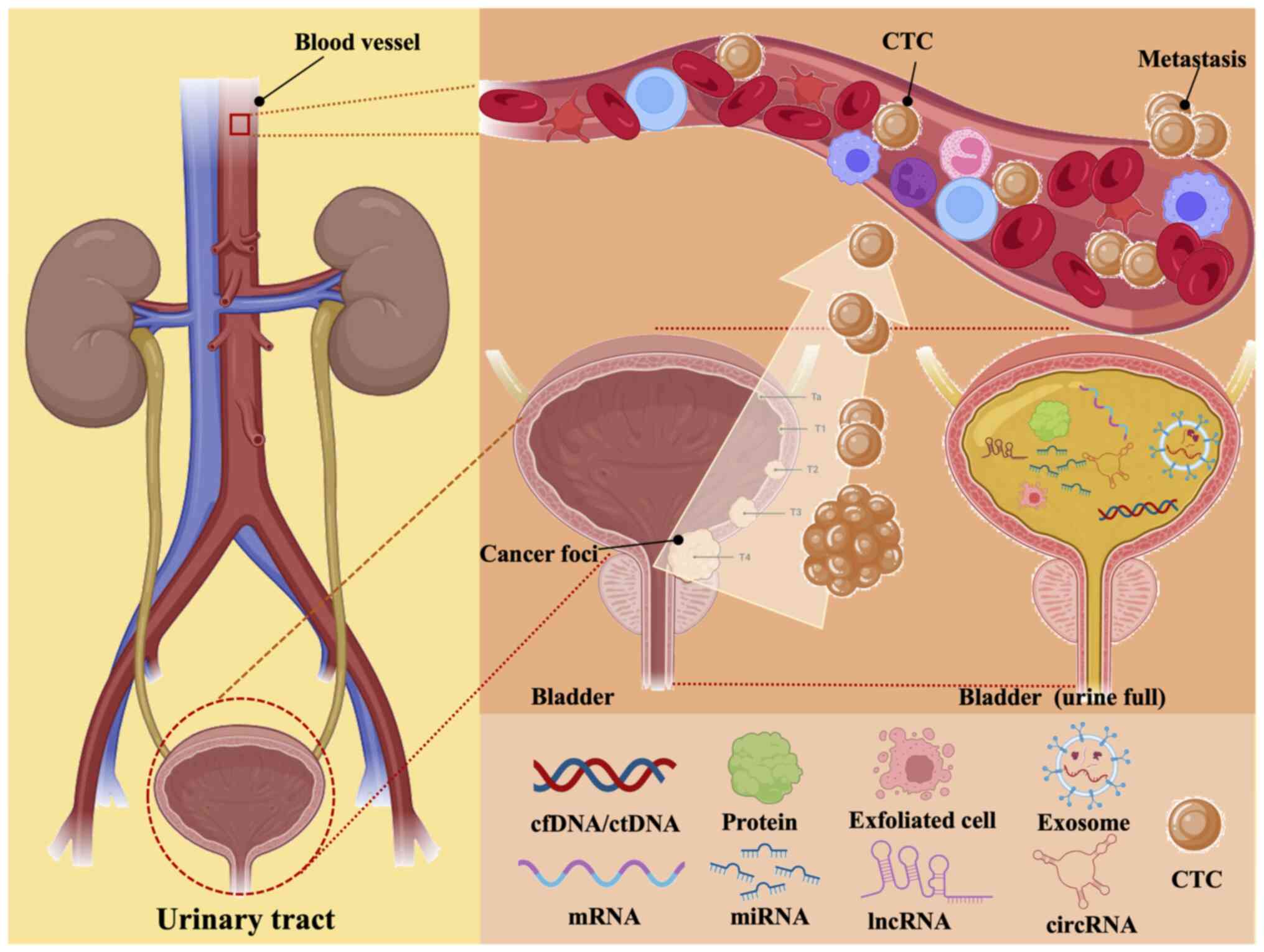
Urological tumors are characterized by a large
number of genetic variants in urine, making urine promising as a
carrier of genetic material such as DNA (cfDNA and exosome DNA) as
well as RNAs (Fig. 1). In recent
years, there has been a growing focus on investigating patterns of
point mutations, methylation changes, fragmentation and structural
variants (219,220). The aforementioned studies
support the hypothesis that novel genome type analysis can be used
not only as a diagnostic tool, but also for predicting the
likelihood of recurrence or progression following treatment in
clinical practice. European Association of Urology and FDA-approved
methods, such as NMP22, UroVysion, FGFR3, and microsatellite
assays in comparison to cystoscopy and cytology, demonstrate higher
sensitivity but lower specificity, particularly in cases of
low-grade, early or recurrent BCa (221). For example, FISH test does not
fully meet clinical standards and it has value in specific
scenarios including the initial diagnosis of patients experiencing
hematuria, recurrence monitoring following treatment and assessment
of patients with atypical cellular abnormalities (222). The specificity of most
biomarkers is <90%, with some <80%. The PPV of these markers
is attributed to their specificity and prevalence. While most
markers exhibit sensitivity ranging from 80 to 90% for high-grade
disease, the failure to detect 10-20% of high-grade tumors is
considered unacceptable.
The discovery of fetal genetic data in the urine of
pregnant patients was the first empirical evidence corroborating
the ability of cfDNA to traverse the glomerulus, subsequently
becoming a constituent of ucfDNA (223,224). Urine ctDNA can be derived from
two primary sources. Firstly, ctDNA is filtered through the
glomeruli, facilitating transfer from the bloodstream into the
urine. Secondly, ctDNA detected in urine can originate from tumor
cells directly shed by the urinary tract system (225). Studies have shown that both
urine supernatants and sediments exhibit a notably higher
cumulative mutation rate compared with DNA extracted from plasma
(181,226,227). The aforementioned findings
suggest consistency in the mutational data conveyed by utDNA and
ctDNA within ucfDNA. There are notable molecular and genetic
differences in ucfDNA that is derived from patients with BCa, which
reflect the genomic content of tumor cells. Ability to detect
genomic characterization in both blood (serum and plasma) and urine
(sediment and supernatant) indicates that liquid biopsy-based
biomarkers have potential for diagnosing and monitoring disease
conditions, including chronic disease and tumors (228).
Total cfDNA concentration in urine of patients with
cancer patients is higher compared with both the general population
and patients diagnosed with benign urological conditions (171). This increase is particularly
prominent in patients with advanced or progressive disease
recurrence. Studies on UC show that mutations detected in urine
samples are associated with those found in both plasma and tissue
samples (235,236). A total of 168 somatic mutations
have been identified in 25 cases of UC using targeted sequencing,
with approximately half of the mutations are identified in both the
urine supernatant and precipitates, whereas 2% of mutations
identified in the tumor are observed in the plasma samples
(237). Another study reports
high concordance between urine cfDNA and tissue DNA detection in
patients with UC with a sensitivity of 86.7% and a specificity of
99.3% (238). Consequently, it
is advisable to prioritize urine samples over plasma samples for
higher detection rates in patients with these conditions. Similar
findings are observed in PCa; methylation levels of glutathione
s-transferase pi 1, ras association domain family member 2
(RASSF2), histone cluster 1 H4 family member K
(HIST1H4K) and transcription factor AP-2 epsilon
(TFAP2E) are detected in both urine and plasma samples. The
aforementioned urinary methylation biomarkers with AUC ranging from
0.82 to 0.94, exhibit higher sensitivity to PCa compared with
plasma DNA (AUC, 0.30-0.53) (239). Evidence contrary to the
aforementioned results suggests that plasma DNA is more sensitive
to PCa than urine testing (240), whereas additional findings
propose there is no obvious difference (241). The sample size and criteria for
enrollment may not be sufficient to demonstrate differences in
performance between urine and blood samples. Differences in the
number and performance of urine and blood biomarkers suggest that
not all ctDNA found in both sample types originates from the same
tumor. It is more likely that plasma ctDNA originates from a
metastatic lesion, while changes in uDNA may indicate a primary
lesion in a urological tumor (242). Due to homeostatic mechanisms,
changes in blood are rapidly eliminated, only resulting in
substantial modifications at the point of decompensation. Overall,
the performance of biomarkers in urine and blood tests may be
influenced by histological grade, advanced pathological stage,
presence of carcinoma in situ, surgical procedure, marker
types, individual differences in background signal and
identification of cancer characteristics (243). Blood and urine markers may serve
as both complementary and independent assays, offering distinct
advantages and disadvantages depending on clinical application
scenario (Table II).
BCa imaging techniques such as ultrasound, computed
tomography (CT) and magnetic resonance imaging (MRI), along with
urine cytology, have emerged as non-invasive alternatives to
cystoscopy. However, there are limitations associated with these
procedures. For example, ultrasound exhibits relatively low
sensitivity and specificity, while CT and MRI have limited
capability in detecting small and flat lesions (244,245). Similarly, urine cytology
exhibits a high specificity but is accompanied by a low sensitivity
in its diagnostic capabilities. Furthermore, low-grade malignant
BCa often presents a high leakage rate, which can contribute to
false positive results in patients experiencing severe hematuria
and urinary tract infection (22). With technological advancements,
liquid biopsy utilizing urine is anticipated to emerge as a
dependable, precise and non-intrusive biomarker for diagnosis and
monitoring of BCa, offering improved personalized diagnosis and
treatment alternatives for patients.
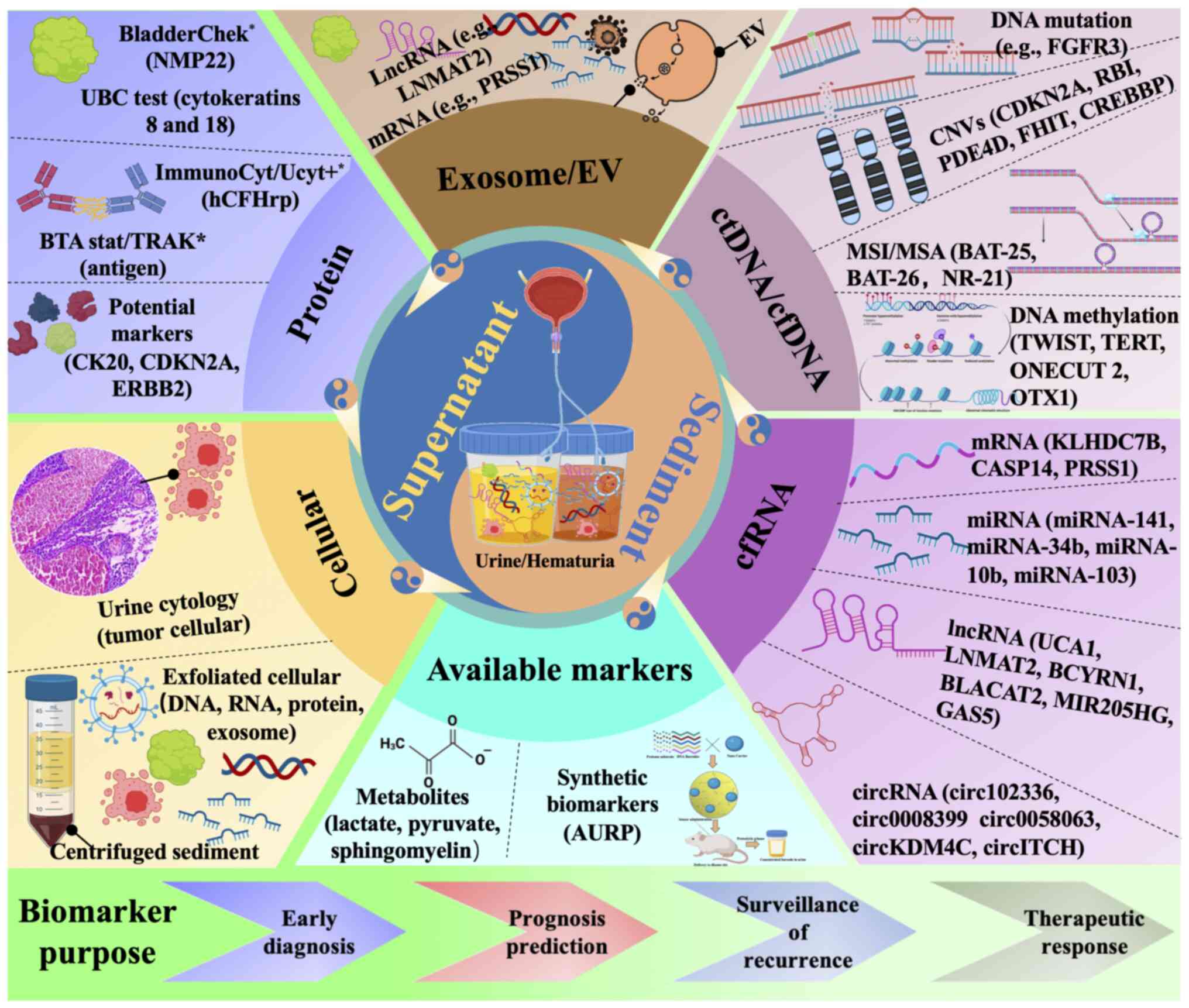
Over the past decade, research has primarily
focused on development of non-invasive biomarkers for BCa derived
from urine. These biomarkers originate from both urine supernatants
and sediment, and include tumor exfoliated cells, genetic material
(DNA and RNA), proteins, extracellular vesicles/exosomes, as well
as novel available markers currently being researched (Fig. 2) (246). Diagnostic tests NMP22, BTA,
ImmunoCyt, and UroVysion are used as adjuncts to cystoscopy because
they offer greater sensitivity than urine cytology. However, these
tests frequently lack of specificity and are susceptible to false
positive results, particularly in the context of benign urological
conditions (83). High-throughput
NGS technology has enhanced the identification of urinary
biomarkers originating from cancer cells. Consequently, assays such
as Cxbladder, XpertBC, UriFind, and Uromonitor have been developed
(208,247). This multi-target approach has
improved detection sensitivity and dynamic monitoring of tumor
progression or recurrence; however, the scope of these assays
remains inadequate to address the vast genomic heterogeneity of
NMIBC. The accuracy of clinical diagnosis and surveillance is
affected by potential genomic differences between primary and
residual lesions, as well as by false positives for somatic
mutations in cfDNA present in normal and benign patient cells.
While biomarker tests show promise, they should be used in
combination with cystoscopy and imaging.
Patients diagnosed with high-risk NMIBC are advised
to undergo cystoscopy every 3 months and Bacillus Calmette-Guérin
(BCG) is the standard initial treatment. Despite the stringent
surveillance protocol, ~50% of patients experience recurrence,
which results in a more unfavorable prognosis (248). The implementation of biomarker
testing alongside cystoscopy may support a decrease in the
frequency of surveillance cycles for cystoscopy. Urinary biomarkers
have potential as a diagnostic tool to rule out patients with
hematuria, enhance the diagnostic capabilities of cystoscopy for
patients with positive results, monitor post-treatment and assess
the risk of postoperative recurrence or progression following
initial intravesical therapy, such as BCG. These assessments guide
the necessity for a second line of treatment or radical surgery.
However, the current role of urinary biomarkers is limited and
requires additional support from prospective trials. The diagnostic
accuracy is influenced by the acceptable threshold for a positive
result and various sample processing methods. Further research is
needed to standardize sample collection (whole urine, supernatant,
pellet or extracellular vesicles) and processing protocols
(centrifugation techniques and selection of reference genes), as
well as validation of biomarker performance in clinical
practice.
The advent of high-throughput technologies, such as
NGS and MS, has accelerated the discovery of biomarkers (74,249,250). Use of these methodologies is
crucial in screening and identifying potential biomarkers by
employing large dataset analyses and machine learning algorithms.
These biomarkers, when coupled with bioinformatics and AI (machine
and deep learning) techniques, have been effectively validated for
the screening of potential drug targets (251). Likewise, bioinformatic
analytical approaches are key tools for the detection of highly
sensitive and disease-specific biomarkers, facilitating early
diagnosis and treatment of BCa (252).
In conclusion, liquid biopsy using urine holds
promise in BCa. Future research is expected to explore new
biomarkers and targets, using urine as a carrier and employing AI
techniques for marker analysis and data processing. For example,
application of molecular optical probes is intended to produce
synthetic biomarkers. This process involves the interaction of
these probes with disease biomarkers to produce artificial urinary
biomarkers. These artificial urine biomarker probes (AUBPs) are
designed to address limitations associated with the diagnostic
performance of endogenous markers. The development of AUBPs has the
potential to broaden the spectrum of biomarkers and diseases
detectable through urinalysis (253,254), facilitate signal amplification
and enable cost-effective follow-up monitoring. In conjunction with
the advancement of smart toilets, development of portable urine
biomarkers could potentially become a pivotal aspect of future BCa
monitoring and treatment. A portable analytical system employing
biosensors for real-time monitoring of urine has been developed,
offering both user-friendly operation and suitability for home
testing; this system exhibits a diagnostic sensitivity of 100% and
specificity of 92% (255).
Synthetic biomarker and urine metabolite analysis have the
capability to facilitate non-invasive disease detection and
monitoring in urine, offering high sensitivity and ability to
conduct multiplexing testing (256,257). The integration of
multidisciplinary approaches to diagnostic and follow-up protocols,
as well as clinical risk stratification, emphasizes the potential
role of these technologies for early detection, accurate staging
and targeted therapy for BCa.
Not applicable.
XW and DW conceived the study and wrote the
manuscript. XZ constructed figures and tables. MX and YH
constructed figures and tables. WQ and SC conceived the study and
revised the manuscript. All authors have read and approved the
final manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by Research Project on the
Industrial Application of Basic Research on Synthetic Biological
Devices to Intervene in Bladder Cancer and Shenzhen Academician
Expert Workstation (grant no. CJGJZD20200617102403009), Development
of Quantitative Gene Mutation Detection Technology for Targeted
Tumor Drugs (grant no. JSGG20201103153801005) and Liquid
Biopsy-Based Molecular Diagnostic Product Development Project for
Real-Time Tumor Detection (grant no. 2023B1111040002).
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Richters A, Aben KKH and Kiemeney LALM:
The global burden of urinary bladder cancer: An update. World J
Urol. 38:1895–1904. 2020. View Article : Google Scholar :
|
|
3
|
Tan WS, Rodney S, Lamb B, Feneley M and
Kelly J: Management of non-muscle invasive bladder cancer: A
comprehensive analysis of guidelines from the United States, Europe
and Asia. Cancer Treat Rev. 47:22–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teoh JY, Kamat AM, Black PC, Grivas P,
Shariat SF and Babjuk M: Recurrence mechanisms of
non-muscle-invasive bladder cancer-a clinical perspective. Nat Rev
Urol. 19:280–294. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Facchini G, Cavaliere C, Romis L, Mordente
S, Facchini S, Iovane G, Capasso M, D'Errico D, Liguori C, Formato
R, et al: Advanced/metastatic bladder cancer: Current status and
future directions. Eur Rev Med Pharmacol Sci. 24:11536–11552.
2020.PubMed/NCBI
|
|
6
|
Devlies W, de Jong JJ, Hofmann F, Bruins
HM, Zuiverloon TCM, Smith EJ, Yuan Y, van Rhijn BWG, Mostafid H,
Santesso N, et al: The diagnostic accuracy of cystoscopy for
detecting bladder cancer in adults presenting with haematuria: A
systematic review from the European Association of Urology
Guidelines Office. Eur Urol. 10:115–122. 2024.
|
|
7
|
Zuiverloon TCM, de Jong FC and Theodorescu
D: Clinical decision making in surveillance of non-muscle-invasive
bladder cancer: The evolving roles of urinary cytology and
molecular markers. Oncology (Williston Park). 31:855–862. 2017.
|
|
8
|
Chamie K, Litwin MS, Bassett JC, Daskivich
TJ, Lai J, Hanley JM, Konety BR and Saigal CS; Urologic Diseases in
America Project: Recurrence of high-risk bladder cancer: A
population-based analysis. Cancer. 119:3219–3227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rezaee ME, Lynch KE, Li Z, MacKenzie TA,
Seigne JD, Robertson DJ, Sirovich B, Goodney PP and Schroeck FR:
The impact of low-versus high-intensity surveillance cystoscopy on
surgical care and cancer outcomes in patients with high-risk
non-muscle-invasive bladder cancer (NMIBC). PLoS One.
15:e02304172020. View Article : Google Scholar
|
|
10
|
Burke DM, Shackley DC and O'Reilly PH: The
community-based morbidity of flexible cystoscopy. BJU Int.
89:347–349. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Der Aa MN, Steyerberg EW, Sen EF,
Zwarthoff EC, Kirkels WJ, van der Kwast TH and Essink-Bot ML:
Patients' perceived burden of cystoscopic and urinary surveillance
of bladder cancer: A randomized comparison. BJU Int. 101:1106–1110.
2008. View Article : Google Scholar
|
|
12
|
Matulewicz RS, DeLancey JO and Meeks JJ:
Cystoscopy. JAMA. 317:11872017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Talukdar S, Emdad L, Das SK, Sarkar D and
Fisher PB: Noninvasive approaches for detecting and monitoring
bladder cancer. Expert Rev Anticancer Ther. 15:283–294. 2015.
View Article : Google Scholar
|
|
14
|
Yaxley JP: Urinary tract cancers: An
overview for general practice. J Family Med Prim Care. 5:533–538.
2016. View Article : Google Scholar
|
|
15
|
Zhang X and Zhang Y: Bladder cancer and
genetic mutations. Cell Biochem Biophys. 73:65–69. 2015. View Article : Google Scholar
|
|
16
|
Thompson D, Lawrentschuk N and Bolton D:
New approaches to targeting epigenetic regulation in bladder
cancer. Cancers (Basel). 15:18562023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eissa S, Swellam M, Sadek M, Mourad MS, EI
Ahmady O and Khalifa A: Comparative evaluation of the nuclear
matrix protein, fibronectin, urinary bladder cancer antigen and
voided urine cytology in the detection of bladder tumors. J Urol.
168:465–469. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sale MS, Kulkarni VV, Kulkarni PV and
Patil CA: Efficacy of modified cell block cytology compared to fine
needle aspiration cytology for diagnostic oral cytopathology.
Biotech Histochem. 96:197–201. 2021. View Article : Google Scholar
|
|
19
|
Raskin RE and Meyer D: General Categories
of Cytologic Interpretation. Canine and Feline Cytology. A Color
Atlas and Interpretation Guide. 2nd edition. Elsevier; Amsterdam:
pp. 16–33. 2009
|
|
20
|
Sullivan PS, Chan JB, Levin MR and Rao J:
Urine cytology and adjunct markers for detection and surveillance
of bladder cancer. Am J Transl Res. 2:412–440. 2010.PubMed/NCBI
|
|
21
|
Xing J and Reynolds JP: Diagnostic
advances in urine cytology. Surg Pathol Clin. 11:601–610. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feiertag N, Barry E, Abramson M, Park JY,
Kovac E, Aboumohamed A, Watts K and Sankin A: Urine cytology rarely
escalates clinical management in the surveillance of
non-muscle-invasive bladder cancer. Clin Genitourin Cancer.
21:258–264. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lozano F, Raventós CX, Carrion A, Dinarés
C, Hernández J, Trilla E and Morote J: Xpert bladder cancer monitor
for the early detection of non-muscle invasive bladder cancer
recurrences: Could cystoscopy be substituted? Cancers (Basel).
15:36832023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Palou J, Rodríguez-Rubio F, Huguet J,
Segarra J, Ribal MJ, Alcaraz A and Villavicencio H: Multivariate
analysis of clinical parameters of synchronous primary superficial
bladder cancer and upper urinary tract tumor. J Urol. 174:859–861;
discussion 861. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raitanen MP, Aine R, Rintala E, Kallio J,
Rajala P, Juusela H and Tammela TL; FinnBladder Group: Differences
between local and review urinary cytology in diagnosis of bladder
cancer. An interobserver multicenter analysis. Eur Urol.
41:284–289. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oliveira MC, Caires HR, Oliveira MJ, Fraga
A, Vasconcelos MH and Ribeiro R: Urinary biomarkers in bladder
cancer: Where do we stand and potential role of extracellular
vesicles. Cancers (Basel). 12:14002020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kundal VK, Pandith AA, Hamid A, Shah A,
Kundal R and Wani SM: Role of NMP22 bladder check test in early
detection of bladder cancer with recurrence. Asian Pacific J Cancer
Prev. 11:1279–1282. 2010.
|
|
28
|
Soloway MS, Briggman JV, Carpinito GA,
Chodak GW, Church PA, Lamm DL, Lange P, Messing E, Pasciak RM,
Reservitz GB, et al: Use of a new tumor marker, urinary NMP22, in
the detection of occult or rapidly recurring transitional cell
carcinoma of the urinary tract following surgical treatment. J
Urol. 156:363–367. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miyake M, Goodison S, Giacoia EG, Rizwani
W, Ross S and Rosser CJ: Influencing factors on the NMP-22 urine
assay: An experimental model. BMC Urol. 12:232012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang Q, Zhang G, Li W, Wang J and Sheng
S: Comparison of the diagnostic performance of fluorescence in situ
hybridization (Fish), nuclear matrix protein 22 (nmp22), and their
combination model in bladder carcinoma detection: A systematic
review and meta-analysis. Onco Targets Ther. 12:349–358. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Y, Zhang T, Li X, Yu F, Yu H and Shao
S: Urine biomarkers for the diagnosis of bladder cancer: A network
meta-analysis. Urol J. 18:623–632. 2021.PubMed/NCBI
|
|
32
|
Gong YW, Wang YR, Fan GR, Niu Q, Zhao YL,
Wang H, Svatek R, Rodriguez R and Wang ZP: Diagnostic and
prognostic role of BTA, NMP22, survivin and cytology in urothelial
carcinoma. Transl Cancer Res. 10:3192–3205. 2021. View Article : Google Scholar
|
|
33
|
Egawa S and Kuruma H: Search for
biomarkers of aggressiveness in bladder cancer. Eur Urol. 50:20–22.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang X, Cao Y, Liu J, Wang S, Yang Y and
Du P: The diagnostic and prognostic value of nuclear matrix protein
22 in bladder cancer. Transl Cancer Res. 9:7174–7182. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Frantzi M, Makridakis M and Vlahou A:
Biomarkers for bladder cancer aggressiveness. Curr Opin Urol.
22:390–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Białek Ł, Bilski K, Dobruch J, Krajewski
W, Szydełko T, Kryst P and Poletajew S: Non-invasive biomarkers in
the diagnosis of upper urinary tract urothelial carcinoma-A
systematic review. Cancers (Basel). 14:15202022. View Article : Google Scholar
|
|
37
|
Ecke TH, Scislowski M, Hassan N, Saura M,
Hallmann S, Koch S, Vuolle S, Malakoutikhah M, Kopra K and Härmä H:
Luminophore chemistry for detection of urinary bladder
cancer-comparison to cytology and urinary rapid tests (BTA
stat®, NMP22® BladderChek® and
UBC® Rapid Test). Anticancer Res. 42:5249–5256. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar A, Kumar R and Gupta NP: Comparison
of NMP22 BladderChek test and urine cytology for the detection of
recurrent bladder cancer. Jpn J Clin Oncol. 36:172–175. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sajid MT, Zafar MR, Ahmad H, Ullah S,
Mirza ZI and Shahzad K: Diagnostic accuracy of NMP 22 and urine
cytology for detection of transitional cell carcinoma urinary
bladder taking cystoscopy as gold standard. Pak J Med Sci.
36:705–710. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sarosdy MF, Hudson MA, Ellis WJ, Soloway
MS, deVere White R, Sheinfeld J, Jarowenko MV, Schellhammer PF,
Schervish EW, Patel JV, et al: Improved detection of recurrent
bladder cancer using the bard BTA stat test. Urology. 50:349–353.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thomas L, Leyh H, Marberger M, Bombardieri
E, Bassi P, Pagano F, Pansadoro V, Sternberg CN, Boccon-Gibod L,
Ravery V, et al: Multicenter trial of the quantitative BTA TRAK
assay in the detection of bladder cancer. Clin Chem. 45:472–477.
1999.PubMed/NCBI
|
|
42
|
Heicappell R, Wettig IC, Schostak M,
Muller M, Steiner U, Sauter T and Miller K: Quantitative detection
of human complement factor H-related protein in transitional cell
carcinoma of the urinary bladder. Eur Urol. 35:81–87. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Raitanen MP, Marttila T, Kaasinen E,
Rintala E, Aine R and Tammela TL: Sensitivity of human complement
factor H related protein (BTA stat) test and voided urine cytology
in the diagnosis of bladder cancer. J Urol. 163:1689–1692. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Heicappell R, Müller M, Fimmers R and
Miller K: Qualitative determination of urinary human complement
factor H-related protein (hcfHrp) in patients with bladder cancer,
healthy controls, and patients with benign urologic disease. Urol
Int. 65:181–184. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Raitanen MP, Marttila T, Nurmi M, Ala-Opas
M, Nieminen P, Aine R and Tammela TL; Finnbladder Group: Human
complement factor H related protein test for monitoring bladder
cancer. J Urol. 165:374–377. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyake M, Goodison S, Rizwani W, Ross S,
Bart Grossman H and Rosser CJ: Urinary BTA: Indicator of bladder
cancer or of hematuria. World J Urol. 30:869–873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ecke TH, Arndt C, Stephan C, Hallmann S,
Lux O, Otto T, Ruttloff J and Gerullis H: Preliminary results of a
multicentre study of the UBC rapid test for detection of urinary
bladder cancer. Anticancer Res. 35:2651–2655. 2015.PubMed/NCBI
|
|
48
|
Reid-Nicholson MD, Ramalingam P, Adeagbo
B, Cheng N, Peiper SC and Terris MK: The use of Urovysion
fluorescence in situ hybridization in the diagnosis and
surveillance of non-urothelial carcinoma of the bladder. Mod
Pathol. 22:119–127. 2009. View Article : Google Scholar
|
|
49
|
Dalquen P, Kleiber B, Grilli B, Herzog M,
Bubendorf L and Oberholzer M: DNA image cytometry and fluorescence
in situ hybridization for noninvasive detection of urothelial
tumors in voided urine. Cancer. 96:374–379. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou H, Yan Y and Song L: The clinical
application of fluorescence in situ hybridization in diagnosing
urothelial carcinoma. Clin Lab. 62:2001–2009. 2016. View Article : Google Scholar
|
|
51
|
Ainthachot S, Sa-ngiamwibool P, Thanee M,
Watcharadetwittaya S, Chamgramol Y, Pairojkul C and Deenonpoe R:
Chromosomal aberrations, visualized using UroVysion®
fluorescence in-situ hybridization assay, can predict poor
prognosis in formalin-fixed paraffin-embedded tissues of
cholangiocarcinoma patients. Hum Pathol. 126:31–44. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Stadler WM and Olopade OI: The 9p21 region
in bladder cancer cell lines: Large homozygous deletions inactivate
the CDKN2, CDKN2B and MTAP genes. Urol Res. 24:239–244. 1996.
View Article : Google Scholar
|
|
53
|
Cairns P, Tokino K, Eby Y and Sidransky2
D: Homozygous deletions of 9p21 in primary human bladder tumors
detected by comparative multiplex polymerase chain reaction. Cancer
Res. 54:1422–1424. 1994.PubMed/NCBI
|
|
54
|
Moonen PM, Merkx GF, Peelen P, Karthaus
HF, Smeets DF and Witjes JA: UroVysion compared with cytology and
quantitative cytology in the surveillance of non-muscle-invasive
bladder cancer. Eur Urol. 51:1275–1280. 2007. View Article : Google Scholar
|
|
55
|
Pycha S, Trenti E, Mian C, Schwienbacher
C, Hanspeter E, Palermo M, Pycha A, Danuser H and D'Elia C:
Diagnostic value of Xpert® BC detection, bladder
epicheck®, Urovysion® FISH and cytology in
the detection of upper urinary tract urothelial carcinoma. World J
Urol. 41:1323–1328. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Halling KC, King W, Sokolova IA, Meyer RG,
Burkhardt HM, Halling AC, Cheville JC, Sebo TJ, Ramakumar S,
Stewart CS, et al: A comparison of cytology and fluorescence in
situ hybridization for the detection of urothelial carcinoma. J
Urol. 164:1768–1775. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Halling KC and Kipp BR: Bladder cancer
detection using FISH (UroVysion assay). Adv Anat Pathol.
15:279–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Laudadio J, Keane TE, Reeves HM, Savage
SJ, Hoda RS, Lage JM and Wolff DJ: Fluorescence in situ
hybridization for detecting transitional cell carcinoma:
Implications for clinical practice. BJU Int. 96:1280–1285. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Youssef RF, Schlomer BJ, Ho R, Sagalowsky
AI, Ashfaq R and Lotan Y: Role of fluorescence in situ
hybridization in bladder cancer surveillance of patients with
negative cytology. Urol Oncol. 30:273–277. 2012. View Article : Google Scholar
|
|
60
|
Messing EM, Teot L, Korman H, Underhill E,
Barker E, Stork B, Qian J and Bostwick DG: Performance of urine
test in patients monitored for recurrence of bladder cancer: A
multicenter study in the United States. J Urol. 174:1238–1241.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fradet Y and Lockhard C: Performance
characteristics of a new monoclonal antibody test for bladder
cancer: ImmunoCyt trade mark. Can J Urol. 4:400–405. 1997.
|
|
62
|
Yafi FA, Brimo F, Steinberg J, Aprikian
AG, Tanguay S and Kassouf W: Prospective analysis of sensitivity
and specificity of urinary cytology and other urinary biomarkers
for bladder cancer. Urol Oncol. 33:66.e25–e31. 2015. View Article : Google Scholar
|
|
63
|
He H, Han C, Hao L and Zang G: ImmunoCyt
test compared to cytology in the diagnosis of bladder cancer: A
meta-analysis. Oncol Lett. 12:83–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kong T, Qu Y, Zhao T, Niu Z, Lv X, Wang Y,
Ding Q, Wei P, Fu J, Wang L, et al: Identification of novel protein
biomarkers from the blood and urine for the early diagnosis of
bladder cancer via proximity extension analysis. J Transl Med.
22:3142024. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Têtu B, Tiguert R, Harel F and Fradet Y:
ImmunoCyt/uCyt+TM improves the sensitivity of urine
cytology in patients followed for urothelial carcinoma. Mod Pathol.
18:83–89. 2005. View Article : Google Scholar
|
|
66
|
Schmitz-Dräger BJ, Beiche B, Tirsar LA,
Schmitz-Dräger C, Bismarck E and Ebert T: Immunocytology in the
assessment of patients with asymptomatic microhaematuria. Eur Urol.
51:1582–1588. 2007. View Article : Google Scholar
|
|
67
|
Alberice JV, Amaral AF, Armitage EG,
Lorente JA, Algaba F, Carrilho E, Márquez M, García A, Malats N and
Barbas C: Searching for urine biomarkers of bladder cancer
recurrence using a liquid chromatography-mass spectrometry and
capillary electrophoresis-mass spectrometry metabolomics approach.
J Chromatogr A. 1318:163–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gómez BB, López-Cortés R, Casas-Nebra FJ,
Vázquez-Estévez S, Pérez-Fentes D, Chantada-Vázquez MDP, Bravo SB
and Núñez C: Detection of circulating serum protein biomarkers of
non-muscle invasive bladder cancer after protein corona-silver
nanoparticles analysis by SWATH-MS. Nanomaterials (Basel).
11:23842021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Schwamborn K, Krieg RC, Grosse J, Reulen
N, Weiskirchen R, Knuechel R, Jakse G and Henkel C: Serum proteomic
profiling in patients with bladder cancer. Eur Urol. 56:989–996.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Nedjadi T, Benabdelkamal H, Albarakati N,
Masood A, Al-Sayyad A, Alfadda AA, Alanazi IO, Al-Ammari A and
Al-Maghrabi J: Circulating proteomic signature for detection of
biomarkers in bladder cancer patients. Sci Rep. 10:109992020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Thomas S, Hao L, Ricke WA and Li L:
Biomarker discovery in mass spectrometry-based urinary proteomics.
Proteomics Clin Appl. 10:358–370. 2016. View Article : Google Scholar :
|
|
72
|
Ahn JH, Kang CK, Kim EM, Kim AR and Kim A:
Proteomics for early detection of non-muscle-invasive bladder
cancer: Clinically useful urine protein biomarkers. Life (Basel).
12:3952022.PubMed/NCBI
|
|
73
|
Wang YT, Shi T, Srivastava S, Kagan J, Liu
T and Rodland KD: Proteomic analysis of exosomes for discovery of
protein biomarkers for prostate and bladder cancer. Cancers
(Basel). 12:23352020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ren R, Wang H, Xie L, Muthupandian S and
Yang X: Identify potential urine biomarkers for bladder cancer
prognosis using NGS data analysis and experimental validation. Appl
Biochem Biotechnol. 195:2947–2964. 2023. View Article : Google Scholar
|
|
75
|
Jazayeri MH, Aghaie T, Nedaeinia R, Manian
M and Nickho H: Rapid noninvasive detection of bladder cancer using
survivin antibody-conjugated gold nanoparticles (GNPs) based on
localized surface plasmon resonance (LSPR). Cancer Immunol
Immunother. 69:1833–1840. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kumar B, Koul S, Petersen J, Khandrika L,
Hwa JS, Meacham RB, Wilson S and Koul HK: p38 Mitogen-activated
protein kinase-driven MAPKAPK2 regulates invasion of bladder cancer
by modulation of MMP-2 and MMP-9 activity. Cancer Res. 70:832–841.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Koshkin VS, Boyiddle C, Schwartz N, Yu J,
Yu KS, Kang A, Bloudek L, Fang Q, Schafer JM, Baker AF, et al:
Systematic literature review and testing of HER2 status in
urothelial carcinoma (UC). J Clin Oncol. 41:556. 2023. View Article : Google Scholar
|
|
78
|
Zhang HH, Qi F, Zu XB, Cao YH, Miao JG, Xu
L and Qi L: A proteomic study of potential VEGF-C-associated
proteins in bladder cancer T24 cells. Med Sci Monit.
18:BR441–BR449. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Z, Xu H, Ji J, Shi X, Lyu J, Zhu Y,
Yu H and Wang F: Heterogeneity of PTEN and PPAR-γ in cancer and
their prognostic application to bladder cancer. Exp Ther Med.
18:3177–3183. 2019.PubMed/NCBI
|
|
80
|
Chen Z, Guo Y, Zhao D, Zou Q, Yu F, Zhang
L and Xu L: Comprehensive analysis revealed that CDKN2A is a
biomarker for immune infiltrates in multiple cancers. Front Cell
Dev Biol. 9:8082082021. View Article : Google Scholar
|
|
81
|
Mi Y, Zhao Y, Shi F, Zhang M, Wang C and
Liu X: Diagnostic accuracy of urine cytokeratin 20 for bladder
cancer: A meta-analysis. Asia Pac J Clin Oncol. 15:e11–e19. 2019.
View Article : Google Scholar
|
|
82
|
Chen CJ, Chou CY, Shu KH, Chen HC, Wang
MC, Chang CC, Hsu BG, Wu MS, Yang YL, Liao WL, et al: Discovery of
novel protein biomarkers in urine for diagnosis of urothelial
cancer using iTRAQ proteomics. J Proteome Res. 20:2953–2963. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tsai CH, Chen YT, Chang YH, Hsueh C, Liu
CY, Chang YS, Chen CL and Yu JS: Systematic verification of bladder
cancer-associated tissue protein biomarker candidates in clinical
urine specimens. Oncotarget. 9:30731–30747. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Allione A, Pardini B, Viberti C, Giribaldi
G, Turini S, Di Gaetano C, Guarrera S, Cordero F, Oderda M, Allasia
M, et al: MMP23B expression and protein levels in blood and urine
are associated with bladder cancer. Carcinogenesis. 39:1254–1263.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Gobin E, Bagwell K, Wagner J, Mysona D,
Sandirasegarane S, Smith N, Bai S, Sharma A, Schleifer R and She
JX: A pan-cancer perspective of matrix metalloproteases (MMP) gene
expression profile and their diagnostic/prognostic potential. BMC
Cancer. 19:5812019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lokeshwar VB, Schroeder GL, Selzer MG,
Hautmann SH, Posey JT, Duncan RC, Watson R, Rose L, Markowitz S and
Soloway MS: Bladder tumor markers for monitoring recurrence and
screening comparison of hyaluronic acid-hyaluronidase and BTA-stat
tests. Cancer. 95:61–72. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lokeshwar VB, Habuchi T, Grossman HB,
Murphy WM, Hautmann SH, Hemstreet GP III, Bono AV, Getzenberg RH,
Goebell P, Schmitz-Dräger BJ, et al: Bladder tumor markers beyond
cytology: International consensus panel on bladder tumor markers.
Urology. 66(Suppl 1): S35–S63. 2005. View Article : Google Scholar
|
|
88
|
Friedrich MG, Hellstern A, Toma MI,
Hammerer P and Huland H: Are false-positive urine markers for the
detection of bladder carcinoma really wrong or do they predict
tumor recurrence? Eur Urol. 43:146–151. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Todenhöfer T, Hennenlotter J, Khs U, Tews
V, Gakis G, Aufderklamm S, Stenzl A and Schwentner C: Influence of
urinary tract instrumentation and inflammation on the performance
of urine markers for the detection of bladder cancer. Urology.
79:620–624. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li A, Zhang T, Zheng M, Liu Y and Chen Z:
Exosomal proteins as potential markers of tumor diagnosis. J
Hematol Oncol. 10:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Song JB, Morrissey JJ, Mobley JM,
Figenshau KG, Vetter JM, Bhayani SB, Kharasch ED and Figenshau RS:
Urinary aquaporin 1 and perilipin 2: Can these novel markers
accurately characterize small renal masses and help guide patient
management? Int J Urol. 26:260–265. 2019. View Article : Google Scholar
|
|
92
|
Benderska-Söder N, Hovanec J, Pesch B,
Goebell PJ, Roghmann F, Noldus J, Rabinovich J, Wichert K,
Gleichenhagen J, Käfferlein HU, et al: Toward noninvasive follow-up
of low-risk bladder cancer-Rationale and concept of the UroFollow
trial. Urol Oncol. 38:886–895. 2020. View Article : Google Scholar
|
|
93
|
Aitekenov S, Gaipov A and Bukasov R:
Review: Detection and quantification of proteins in human urine.
Talanta. 223:1217182021. View Article : Google Scholar
|
|
94
|
Goecks J, Jalili V, Heiser LM and Gray JW:
How machine learning will transform biomedicine. Cell. 181:92–101.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li Y, Sun L, Guo X, Mo N, Zhang J and Li
C: Frontiers in bladder cancer genomic research. Front Oncol.
11:6707292021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sarhadi VK and Armengol G: Molecular
biomarkers in cancer. Biomolecules. 12:10212022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shi R, Wang X, Wu Y, Xu B, Zhao T, Trapp
C, Wang X, Unger K, Zhou C, Lu S, et al: APOBEC-mediated
mutagenesis is a favorable predictor of prognosis and immunotherapy
for bladder cancer patients: evidence from pan-cancer analysis and
multiple databases. Theranostics. 12:4181–4199. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Le NQK, Kha QH, Nguyen VH, Chen YC, Cheng
SJ and Chen CY: Machine learning-based radiomics signatures for
egfr and kras mutations prediction in non-small-cell lung cancer.
Int J Mol Sci. 22:92542021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Gao J, Wu H, Shi X, Huo Z, Zhang J and
Liang Z: Comparison of next-generation sequencing, quantitative
PCR, and sanger sequencing for mutation profiling of EGFR, KRAS,
PIK3CA and BRAF in clinical lung tumors. Clin Lab. 62:689–696.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chauhan PS, Shiang A, Alahi I, Sundby RT,
Feng W, Gungoren B, Nawaf C, Chen K, Babbra RK, Harris PK, et al:
Urine cell-free DNA multi-omics to detect MRD and predict survival
in bladder cancer patients. NPJ Precis Oncol. 7:62023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sieverink CA, Batista RPM, Prazeres HJM,
Vinagre J, Sampaio C, Leão RR, Máximo V, Witjes JA and Soares P:
Clinical validation of a urine test (uromonitor-V2®) for
the surveillance of non-muscle-invasive bladder cancer patients.
Diagnostics (Basel). 10:7452020. View Article : Google Scholar
|
|
102
|
Hosen MI, Sheikh M, Zvereva M, Scelo G,
Forey N, Durand G, Voegele C, Poustchi H, Khoshnia M, Roshandel G,
et al: Urinary TERT promoter mutations are detectable up to 10
years prior to clinical diagnosis of bladder cancer: Evidence from
the golestan cohort study. EBioMedicine. 53:1026432020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Soputro NA, Gracias DN, Dias BH, Nzenza T,
O'Connell H and Sethi K: Utility of urinary biomarkers in primary
haematuria: Systematic review and meta-analysis. BJUI Compass.
3:334–343. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Springer SU, Chen CH, Rodriguez Pena MDC,
Li L, Douville C, Wang Y, Cohen JD, Taheri D, Silliman N, Schaefer
J, et al: Non-invasive detection of urothelial cancer through the
analysis of driver gene mutations and aneuploidy. Elife.
7:e321432018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Baffa R, Letko J, McClung C, LeNoir J,
Vecchione A and Gomella LG: Molecular genetics of bladder cancer:
Targets for diagnosis and therapy. J Exp Clin Cancer Res.
25:145–160. 2006.PubMed/NCBI
|
|
106
|
Mertens LS, Claps F, Mayr R, Bostrom PJ,
Shariat SF, Zwarthoff EC, Boormans JL, Abas C, van Leenders GJLH,
Götz S, et al: Prognostic markers in invasive bladder cancer: FGFR3
mutation status versus P53 and KI-67 expression: A multi-center,
multi-laboratory analysis in 1058 radical cystectomy patients. Urol
Oncol. 40:110.e1–110.e9. 2022. View Article : Google Scholar
|
|
107
|
Kim YS, Kim K, Kwon GY, Lee SJ and Park
SH: Fibroblast growth factor receptor 3 (FGFR3) aberrations in
muscle-invasive urothelial carcinoma. BMC Urol. 18:682018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Calderaro J, Rebouissou S, De Koning L,
Masmoudi A, Hérault A, Dubois T, Maille P, Soyeux P, Sibony M, de
la Taille A, et al: PI3K/AKT pathway activation in bladder
carcinogenesis. Int J Cancer. 134:1776–1784. 2014. View Article : Google Scholar
|
|
109
|
Hao L, Fang J, Xu R, Liu S, Luo G and Wang
X: Significance of the FGFR3 mutation in Chinese patients with
bladder cancer. Transl Androl Urol. 12:761–769. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Sakr SA, Mahran HA, Fahmy AM, El-Kholy MA
and Meawad M: Expression of c-erb-B2 gene in bladder cancer of
Egyptian patients and its correlation with p53 and bcl-2. Biomed
Pharmacother. 76:73–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
George B, Datar RH, Wu L, Cai J, Patten N,
Beil SJ, Groshen S, Stein J, Skinner D, Jones PA and Cote RJ: p53
gene and protein status: The role of p53 alterations in predicting
outcome in patients with bladder cancer. J Clin Oncol.
25:5352–5358. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Landolfi JA and Terio KA: Transitional
cell carcinoma in fishing cats (Prionailurus viverrinus): Pathology
and expression of cyclooxygenase-1, -2, and p53. Vet Pathol.
43:674–681. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Pakmanesh H, Anvari O, Forey N, Weiderpass
E, Malekpourafshar R, Iranpour M, Shahesmaeili A, Ahmadi N,
Bazrafshan A, Zendehdel K, et al: TERT promoter mutations as simple
and non-invasive urinary biomarkers for the detection of urothelial
bladder cancer in a high-risk region. Int J Mol Sci. 23:143192022.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Avogbe PH, Manel A, Vian E, Durand G,
Forey N, Voegele C, Zvereva M, Hosen MI, Meziani S, De Tilly B, et
al: Urinary TERT promoter mutations as non-invasive biomarkers for
the comprehensive detection of urothelial cancer. EBioMedicine.
44:431–438. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Eich ML, Rodriguez Pena MDC, Springer SU,
Taheri D, Tregnago AC, Salles DC, Bezerra SM, Cunha IW, Fujita K,
Ertoy D, et al: Incidence and distribution of UroSEEK gene panel in
a multi-institutional cohort of bladder urothelial carcinoma. Mod
Pathol. 32:1544–1550. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Wang P, Shi Y, Zhang J, Shou J, Zhang M,
Zou D, Liang Y, Li J, Tan Y, Zhang M, et al: UCseek: Ultrasensitive
early detection and recurrence monitoring of urothelial carcinoma
by shallow-depth genome-wide bisulfite sequencing of urinary
sediment DNA. EBioMedicine. 89:1044372023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ramos P, Brás JP, Dias C, Bessa-Gonçalves
M, Botelho F, Silva J, Silva C and Pacheco-Figueiredo L:
Uromonitor: Clinical validation and performance assessment of a
urinary biomarker within the surveillance of patients with
non-muscle-invasive bladder cancer patients. J Urol. Nov
19–2024.Epub ahead of print. View Article : Google Scholar
|
|
118
|
Ou Z, Li K, Yang T, Dai Y, Chandra M, Ning
J, Wang Y, Xu R, Gao T, Xie Y, et al: Detection of bladder cancer
using urinary cell-free DNA and cellular DNA. Clin Transl Med.
9:42020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhang T, Du E, Liu Y, Cheng J, Zhang Z, Xu
Y, Qi S and Chen Y: Anticancer effects of zinc oxide nanoparticles
through altering the methylation status of histone on bladder
cancer cells. Int J Nanomedicine. 15:1457–1468. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Porten SP: Epigenetic alterations in
bladder cancer. Curr Urol Rep. 19:1022018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
van Kessel KEM, Van Neste L, Lurkin I,
Zwarthoff EC and Van Criekinge W: Evaluation of an epigenetic
profile for the detection of bladder cancer in patients with
hematuria. J Urol. 195:601–607. 2016. View Article : Google Scholar
|
|
122
|
Sathianathen NJ, Butaney M, Weight CJ,
Kumar R and Konety BR: Urinary biomarkers in the evaluation of
primary hematuria: A systematic review and meta-analysis. Bladder
Cancer. 4:353–363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Tripathi K, Goel A, Singhai A and Garg M:
Promoter hypomethylation as potential confounder of Ras gene
overexpression and their clinical significance in subsets of
urothelial carcinoma of bladder. Mol Biol Rep. 48:2183–2199. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Wolff EM, Chihara Y, Pan F, Weisenberger
DJ, Siegmund KD, Sugano K, Kawashima K, Laird PW, Jones PA and
Liang G: Unique DNA methylation patterns distinguish noninvasive
and invasive urothelial cancers and establish an epigenetic field
defect in premalignant tissue. Cancer Res. 70:8169–8178. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Locke WJ, Guanzon D, Ma C, Liew YJ,
Duesing KR, Fung KYC and Ross JP: DNA methylation cancer
biomarkers: Translation to the clinic. Front Genet. 10:11502019.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Xiao Y, Ju L, Qian K, Jin W, Wang G, Zhao
Y, Jiang W, Liu N, Wu K, Peng M, et al: Non-invasive diagnosis and
surveillance of bladder cancer with driver and passenger DNA
methylation in a prospective cohort study. Clin Transl Med.
12:e10082022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Shindo T, Shimizu T, Nojima M, Niinuma T,
Maruyama R, Kitajima H, Kai M, Itoh N, Suzuki H and Masumori N:
Evaluation of urinary DNA methylation as a marker for recurrent
bladder cancer: A 2-center prospective study. Urology. 113:71–78.
2018. View Article : Google Scholar
|
|
128
|
Ko K, Kananazawa Y, Yamada T, Kakinuma D,
Matsuno K, Ando F, Kuriyama S, Matsuda A and Yoshida H: Methylation
status and long-fragment cell-free DNA are prognostic biomarkers
for gastric cancer. Cancer Med. 10:2003–2012. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lutfi A, Afghan MK, Swed B and Kasi PM:
False-positive liquid biopsy assays secondary to overlapping
aberrant methylation from non-cancer disease states. Case Rep
Oncol. 16:1536–1541. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Shields MD, Chen K, Dutcher G, Patel I and
Pellini B: Making the rounds: Exploring the role of circulating
tumor DNA (ctDNA) in non-small cell lung cancer. Int J Mol Sci.
23:90062022. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
D'Andrea D, Soria F, Zehetmayer S, Gust
KM, Korn S, Witjes JA and Shariat SF: Diagnostic accuracy, clinical
utility and influence on decision-making of a methylation urine
biomarker test in the surveillance of non-muscle-invasive bladder
cancer. BJU Int. 123:959–967. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Tan WS, Feber A, Sarpong R, Khetrapal P,
Rodney S, Jalil R, Mostafid H, Cresswell J, Hicks J, Rane A, et al:
Who should be investigated for haematuria? Results of a
contemporary prospective observational study of 3556 patients. Eur
Urol. 74:10–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Davalos V and Esteller M: Cancer
epigenetics in clinical practice. CA Cancer J Clin. 73:376–424.
2023. View Article : Google Scholar
|
|
134
|
Wu J, Lin Y, Yang K, Liu X, Wang H, Yu T,
Tao R, Guo J, Chen L, Cheng H, et al: Clinical effectiveness of a
multitarget urine DNA test for urothelial carcinoma detection: A
double-blinded, multicenter, prospective trial. Mol Cancer.
23:572024. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Maas M, Todenhöfer T and Black PC: Urine
biomarkers in bladder cancer-current status and future
perspectives. Nat Rev Urol. 20:597–614. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Iisager L, Ahrenfeldt J, Keller AK,
Nielsen TK, Fristrup N and Lyskjær I: KIDNEY-PAGER: Analysis of
circulating tumor DNA as a biomarker in renal cancer-an
observational trial. Study protocol. Acta Oncol. 63:51–55. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Schardt J, Roth B and Seiler R: Forty
years of cisplatin-based chemotherapy in muscle-invasive bladder
cancer: Are we understanding how, who and when? World J Urol.
37:1759–1765. 2019. View Article : Google Scholar
|
|
138
|
Soave A, Kluwe L, Yu H, Rink M, Gild P,
Vetterlein MW, Marks P, Sauter G, Fisch M, Meyer CP, et al: Copy
number variations in primary tumor, serum and lymph node metastasis
of bladder cancer patients treated with radical cystectomy. Sci
Rep. 10:215622020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Soave A, Chun FK, Hillebrand T, Rink M,
Weisbach L, Steinbach B, Fisch M, Pantel K and Schwarzenbach H:
Copy number variations of circulating, cell-free DNA in urothelial
carcinoma of the bladder patients treated with radical cystectomy:
A prospective study. Oncotarget. 8:56398–56407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Keck B, Ellmann C, Stoehr R, Weigelt K,
Goebell PJ, Kunath F, Taubert H, Hartmann A, Wullich B and Wach S:
Comparative genomic hybridization shows complex genomic changes of
plasmacytoid urothelial carcinoma. Urol Oncol. 32:1234–1239. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of urothelial bladder
carcinoma. Nature. 507:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Li R, Du Y, Chen Z, Xu D, Lin T, Jin S,
Wang G, Liu Z, Lu M, Chen X, et al: Macroscopic somatic clonal
expansion in morphologically normal human urothelium. Science.
370:82–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Luo T, Yi X and Si W: Identification of
miRNA and genes involving in osteosarcoma by comprehensive analysis
of microRNA and copy number variation data. Oncol Lett.
14:5427–5433. 2017.PubMed/NCBI
|
|
144
|
Wu S, Ou T, Xing N, Lu J, Wan S, Wang C,
Zhang X, Yang F, Huang Y and Cai Z: Whole-genome sequencing
identifies ADGRG6 enhancer mutations and FRS2 duplications as
angiogenesis-related drivers in bladder cancer. Nat Commun.
10:7202019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Zeng S, Ying Y, Xing N, Wang B, Qian Z,
Zhou Z, Zhang Z, Xu W, Wang H, Dai L, et al: Noninvasive detection
of urothelial carcinoma by cost-effective low-coverage whole-genome
sequencing from urine-exfoliated cell DNA. Clin Cancer Res.
26:5646–5654. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Ge G, Peng D, Guan B, Zhou Y, Gong Y, Shi
Y, Hao X, Xu Z, Qi J, Lu H, et al: Urothelial carcinoma detection
based on copy number profiles of urinary cell-free DNA by shallow
whole-genome sequencing. Clin Chem. 66:188–198. 2020. View Article : Google Scholar
|
|
147
|
Ellegren H: Microsatellites: Simple
sequences with complex evolution. Nat Rev Genet. 5:435–445. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Moon C, Gordon M, Moon D and Reynolds T:
Microsatellite instability analysis (MSA) for bladder cancer: Past
history and future directions. Int J Mol Sci. 22:128642021.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zheng K, Wan H, Zhang J, Shan G, Chai N,
Li D, Fang N, Liu L, Zhang J, Du R, et al: A novel NGS-based
microsatellite instability (MSI) status classifier with 9 loci for
colorectal cancer patients. J Transl Med. 18:2152020. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zheng J, Miao F, Wang Z, Ma Y, Lin Z, Chen
Y, Kong X, Wang Y, Zhuang A, Wu T and Li W: Identification of MDM2
as a prognostic and immunotherapeutic biomarker in a comprehensive
pan-cancer analysis: A promising target for breast cancer, bladder
cancer and ovarian cancer immunotherapy. Life Sci. 327:1218322023.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Malapelle U, Parente P, Pepe F, De Luca C,
Pisapia P, Sgariglia R, Nacchio M, Gragnano G, Russo G, Conticelli
F, et al: Evaluation of micro satellite instability and mismatch
repair status in different solid tumors: A multicenter analysis in
a real world setting. Cells. 10:18782021. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Kasi PM, Bucheit LA, Liao J, Starr J,
Barata P, Klempner SJ, Gandara D, Shergill A, Madeira da Silva L,
Weipert C, et al: Pan-cancer prevalence of microsatellite
instability-high (MSI-H) identified by circulating tumor DNA and
associated real-world clinical outcomes. JCO Precis Oncol.
7:e23001182023. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Ma YT, Hua F, Zhong XM, Xue YJ, Li J, Nie
YC, Zhang XD, Ma JW, Lin CH, Zhang HZ, et al: Clinicopathological
characteristics, molecular landscape, and biomarker landscape for
predicting the efficacy of PD-1/PD-L1 inhibitors in Chinese
population with mismatch repair deficient urothelial carcinoma: A
real-world study. Front Immunol. 14:12690972023. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chang L, Chang M, Chang HM and Chang F:
Microsatellite instability: A predictive biomarker for cancer
immunotherapy. Appl Immunohistochem Mol Morphol. 26:e15–e21. 2018.
View Article : Google Scholar
|
|
155
|
Hirotsu Y, Nagakubo Y, Amemiya K, Oyama T,
Mochizuki H and Omata M: Microsatellite instability status is
determined by targeted sequencing with MSIcall in 25 cancer types.
Clin Chim Acta. 502:207–213. 2020. View Article : Google Scholar
|
|
156
|
Zekri AR, Khaled HM, Mohammed MB, Diab FM,
Abdellateif MS, El Deeb S, Badr AM, Mohanad M, Abdallah SO and
Bahnassy AA: Microsatellite instability profiling in Egyptian
bladder cancer patients: A pilot study. Curr Probl Cancer.
43:1004722019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Awadalla A, Harraz AM, Abol-Enein H,
Laymon M, Ahmed AE, Abdel-Rahim M, Zekri AN and Shokeir AA:
Prognostic influence of microsatellite alterations of
muscle-invasive bladder cancer treated with radical cystectomy.
Urol Oncol. 40:64.e9–64.e15. 2022. View Article : Google Scholar
|
|
158
|
Steiner G, Schoenberg MP, Linn JF, Mao L
and Sidransky D: Detection of bladder cancer recurrence by
microsatellite analysis of urine. Nat Med. 3:621–624. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Wild PJ, Fuchs T, Stoehr R, Zimmermann D,
Frigerio S, Padberg B, Steiner I, Zwarthoff EC, Burger M, Denzinger
S, et al: Detection of urothelial bladder cancer cells in voided
urine can be improved by a combination of cytology and standardized
microsatellite analysis. Cancer Epidemiol Biomarkers Prev.
18:1798–1806. 2009. View Article : Google Scholar
|
|
160
|
Carpagnano GE, Costantino E, Palladino GP,
Lacedonia D, Martinelli D, Orlando S and Foschino-Barbaro MP:
Microsatellite alterations and cell-free dna analysis: Could they
increase the cytology sensitivity in the diagnosis of malignant
pleural effusion? Rejuvenation Res. 15:265–273. 2012. View Article : Google Scholar
|
|
161
|
Wagner SJ, Reisenbüchler D, West NP,
Niehues JM, Zhu J, Foersch S, Veldhuizen GP, Quirke P, Grabsch HI,
van den Brandt PA, et al: Transformer-based biomarker prediction
from colorectal cancer histology: A large-scale multicentric study.
Cancer Cell. 41:1650–1661. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Van Rhijn BW, Van Der Poel HG and Van Der
Kwast TH: Urine markers for bladder cancer surveillance: A
systematic review. Eur Urol. 47:736–748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Bartoletti R, Dal Canto M, Cai T, Piazzini
M, Travaglini F, Gavazzi A and Rizzo M: Early diagnosis and
monitoring of superficial transitional cell carcinoma by
microsatellite analysis on urine sediment. Oncol Rep. 13:531–537.
2005.PubMed/NCBI
|
|
164
|
Frigerio S, Padberg BC, Strebel RT,
Lenggenhager DM, Messthaler A, Abdou MT, Moch H and Zimmermann DR:
Improved detection of bladder carcinoma cells in voided urine by
standardized microsatellite analysis. Int J Cancer. 121:329–338.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Brisuda A, Pazourkova E, Soukup V, Horinek
A, Hrbáček J, Capoun O, Svobodova I, Pospisilova S, Korabecna M,
Mares J, et al: Urinary cell-free DNA quantification as
non-invasive biomarker in patients with bladder cancer. Urol Int.
96:25–31. 2016. View Article : Google Scholar
|
|
166
|
Bryzgunova OE, Konoshenko MY and Laktionov
PP: Concentration of cell-free DNA in different tumor types. Expert
Rev Mol Diagn. 21:63–75. 2021. View Article : Google Scholar
|
|
167
|
Green EA, Li R, Albiges L, Choueiri TK,
Freedman M, Pal S, Dyrskjøt L and Kamat AM: Clinical utility of
cell-free and circulating tumor DNA in kidney and bladder cancer: A
critical review of current literature. Eur Urol Oncol. 4:893–903.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Ralla B, Stephan C, Meller S, Dietrich D,
Kristiansen G and Jung K: Nucleic acid-based biomarkers in body
fluids of patients with urologic malignancies. Crit Rev Clin Lab
Sci. 51:200–231. 2014. View Article : Google Scholar
|
|
169
|
Li P, Ning J, Luo X, Du H, Zhang Q, Zhou
G, Du Q, Ou Z, Wang L and Wang Y: New method to preserve the
original proportion and integrity of urinary cell-free DNA. J Clin
Lab Anal. 33:e226682019. View Article : Google Scholar
|
|
170
|
Tse RT, Zhao H, Wong CY, Cheng CK, Kong
AW, Peng Q, Chiu PK, Ng CF and Teoh JY: Urinary cell-free DNA in
bladder cancer detection. Diagnostics (Basel). 11:3062021.
View Article : Google Scholar :
|
|
171
|
Augustus E, van Casteren K, Sorber L, van
Dam P, Roeyen G, Peeters M, Vorsters A, Wouters A, Raskin J, Rolfo
C, et al: The art of obtaining a high yield of cell-free DNA from
urine. PLoS One. 15:e02310582020. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Piao XM, Jeong P, Kim YH, Byun YJ, Xu Y,
Kang HW, Ha YS, Kim WT, Lee JY, Woo SH, et al: Urinary cell-free
microRNA biomarker could discriminate bladder cancer from benign
hematuria. Int J Cancer. 144:380–388. 2019. View Article : Google Scholar
|
|
173
|
Zancan M, Galdi F, Di Tonno F, Mazzariol
C, Orlando C, Malentacchi F, Agostini M, Maran M, Del Bianco P,
Fabricio AS, et al: Evaluation of cell-free DNA in urine as a
marker for bladder cancer diagnosis. Int J Biol Markers.
24:147–155. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
do Nascimento Alves SIPM, Lavalhegas
Hallack M, Moreira Perez M, da Costa Aguiar Alves B, da Silva LH
and Afonso Fonseca FL: Application of the Z-scan technique for the
detection of CFCDNA (cell-free circulating DNA) and urine DNA
(uDNA) in patients with bladder cancer. Photodiagnosis Photodyn
Ther. 26:131–133. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Nel I, Münch C, Shamkeeva S, Heinemann ML,
Isermann B and Aktas B: The challenge to stabilize, extract and
analyze urinary cell-free DNA (ucfDNA) during clinical routine.
Diagnostics (Basel). 13:36702023. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Markus H, Zhao J, Contente-Cuomo T,
Stephens MD, Raupach E, Odenheimer-Bergman A, Connor S, McDonald
BR, Moore B, Hutchins E, et al: Analysis of recurrently protected
genomic regions in cell-free DNA found in urine. Sci Transl Med.
13:aaz30882021. View Article : Google Scholar
|
|
177
|
Casadio V, Calistri D, Salvi S, Gunelli R,
Carretta E, Amadori D, Silvestrini R and Zoli W: Urine cell-free
DNA integrity as a marker for early prostate cancer diagnosis: A
pilot study. Biomed Res Int. 2013:2704572013. View Article : Google Scholar
|
|
178
|
Casadio V, Calistri D, Tebaldi M,
Bravaccini S, Gunelli R, Martorana G, Bertaccini A, Serra L, Scarpi
E, Amadori D, et al: Urine Cell-Free DNA integrity as a marker for
early bladder cancer diagnosis: Preliminary data. Urol Oncol.
31:1744–1750. 2013. View Article : Google Scholar
|
|
179
|
Nuzzo PV, Berchuck JE, Korthauer K, Spisak
S, Nassar AH, Abou Alaiwi S, Chakravarthy A, Shen SY, Bakouny Z,
Boccardo F, et al: Detection of renal cell carcinoma using plasma
and urine cell-free DNA methylomes. Nat Med. 26:1041–1043. 2020.
View Article : Google Scholar
|
|
180
|
Zhou Q, Liu F, Guo L, Chen R, Yuan X, Li
C, Shu L, Liu H, Zhou Y, Wu Y, et al: A novel urine cell-free DNA
preservation solution and its application in kidney
transplantation. Nephrology (Carlton). 26:684–691. 2021. View Article : Google Scholar
|
|
181
|
Mouliere F, Smith CG, Heider K, Su J, van
der Pol Y, Thompson M, Morris J, Wan JCM, Chandrananda D, Hadfield
J, et al: Fragmentation patterns and personalized sequencing of
cell-free DNA in urine and plasma of glioma patients. EMBO Mol Med.
13:e128812021. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Chang A, Mzava O, Lenz JS, Cheng AP,
Burnham P, Motley ST, Bennett C, Connelly JT, Dadhania DM,
Suthanthiran M, et al: Measurement biases distort cell-free DNA
fragmentation profiles and define the sensitivity of metagenomic
cell-free DNA sequencing assays. Clin Chem. 68:163–171. 2022.
View Article : Google Scholar :
|
|
183
|
Yasuda T, Nadano D, Awazu S and Kishi K:
Human urine deoxyribonuclease II (DNase II) isoenzymes: A novel
immunoaffinity purification, biochemical multiplicity, genetic
heterogeneity and broad distribution among tissues and body fluids.
Biochim Biophys Acta. 1119:185–193. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Nardelli C, Aveta A, Pandolfo SD, Tripodi
L, Russo F, Imbimbo C, Castaldo G and Pastore L: Microbiome
profiling in bladder cancer patients using the first-morning urine
sample. Eur Urol Open Sci. 59:18–26. 2023. View Article : Google Scholar
|
|
185
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
186
|
Elballal MS, Sallam AM, Elesawy AE, Shahin
RK, Midan HM, Elrebehy MA, Elazazy O, El-Boghdady RM, Blasy SH,
Amer NM, et al: miRNAs as potential game-changers in renal cell
carcinoma: Future clinical and medicinal uses. Pathol Res Pract.
245:1544392023. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Xie Y, Ma X, Chen L, Li H, Gu L, Gao Y,
Zhang Y, Li X, Fan Y, Chen J and Zhang X: MicroRNAs with prognostic
significance in bladder cancer: A systematic review and
meta-analysis. Sci Rep. 7:56192017. View Article : Google Scholar
|
|
188
|
Pan D, Du Y, Li R, Shen A, Liu X, Li C and
Hu B: miR-29b-3p increases radiosensitivity in stemness cancer
cells via modulating oncogenes axis. Front Cell Dev Biol.
9:7410742021. View Article : Google Scholar :
|
|
189
|
Eissa S, Matboli M, Hegazy MG, Kotb YM and
Essawy NO: Evaluation of urinary microRNA panel in bladder cancer
diagnosis: Relation to bilharziasis. Transl Res. 165:731–739. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Nekoohesh L, Modarressi MH, Mowla SJ,
Sadroddiny E, Etemadian M, Afsharpad M, Zolfaghari F, Barzegari M,
Saffari M, Oskooei VK, et al: Expression profile of miRNAs in urine
samples of bladder cancer patients. Biomark Med. 12:1311–1321.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Suarez-Cabrera C, Estudillo L, Ramón-GilE,
Martínez-Fernández M, Peral J, Rubio C, Lodewijk I, Martínde
Bernardo Á, García-Escudero R, Villacampa F, et al: BlaDimiR: A
urine-based miRNA score for accurate bladder cancer diagnosis and
follow-up. Eur Urol. 82:663–667. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Li HJ, Gong X, Li ZK, Qin W, He CX, Xing
L, Zhou X, Zhao D and Cao HL: Role of long non-coding RNAs on
bladder cancer. Front Cell Dev Biol. 9:6726792021. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Wang X, Song D, Zhu B, Jin Y, Cai C and
Wang Z: Urinary exosomal mRNA as a biomarker for the diagnosis of
bladder cancer. Anticancer Drugs. 35:362–370. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhang ZH, Wang Y, Zhang Y, Zheng SF, Feng
T, Tian X, Abudurexiti M, Wang ZD, Zhu WK, Su JQ, et al: The
function and mechanisms of action of circular RNAs in urologic
cancer. Mol Cancer. 22:612023. View Article : Google Scholar
|
|
195
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar
|
|
196
|
Zhao H, Wu W, Li X and Chen W: Long
noncoding RNA UCA1 promotes glutamine-driven anaplerosis of bladder
cancer by interacting with hnRNP I/L to upregulate GPT2 expression.
Transl Oncol. 17:1013402022. View Article : Google Scholar
|
|
197
|
Wang Z, Wang X, Zhang D, Yu Y, Cai L and
Zhang C: Long non-coding RNA urothelial carcinoma-associated 1 as a
tumor biomarker for the diagnosis of urinary bladder cancer. Tumor
Biol. 39:10104283177099902017. View Article : Google Scholar
|
|
198
|
Kong JH, Ha D, Lee J, Kim I, Park M, Im
SH, Shin K and Kim S: Network-based machine learning approach to
predict immunotherapy response in cancer patients. Nat Commun.
13:37032022. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Abbastabar M, Sarfi M, Golestani A, Karimi
A, Pourmand G and Khalili E: Tumor-derived urinary exosomal long
non-coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI
J. 19:301–310. 2020.PubMed/NCBI
|
|
200
|
Bao Z, Zhang W and Dong D: A potential
prognostic lncRNA signature for predicting survival in patients
with bladder urothelial carcinoma. Oncotarget. 8:10485–10497. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Zhao L, Li J, Xue Z and Wang J: Exosomal
noncoding RNAs as noninvasive biomarkers in bladder cancer: A
diagnostic meta-analysis. Clin Transl Oncol. 26:1497–1507. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Bian B, Li L, Ke X, Chen H, Liu Y, Zheng
N, Zheng Y, Ma Y, Zhou Y, Yang J, et al: Urinary exosomal long
non-coding RNAs as noninvasive biomarkers for diagnosis of bladder
cancer by RNA sequencing. Front Oncol. 12:9763292022. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Yu Y, Zhang W, Li A, Chen Y, Ou Q, He Z,
Zhang Y, Liu R, Yao H and Song E: Association of long noncoding RNA
biomarkers with clinical immune subtype and prediction of
immunotherapy response in patients with cancer. JAMA Netw Open.
3:e2021492020. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Lin YH: The effects of intracellular and
exosomal ncRNAs on cancer progression. Cancer Gene Ther.
30:1587–1597. 2023. View Article : Google Scholar
|
|
205
|
Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu
H, Zhong G, Li Y, Li J, Huang J, et al: Exosomal long noncoding RNA
LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin
Invest. 130:404–421. 2020. View Article : Google Scholar
|
|
206
|
He W, Zhong G, Jiang N, Wang B, Fan X,
Chen C, Chen X, Huang J and Lin T: Long noncoding RNA BLACAT2
promotes bladder cancer-associated lymphangiogenesis and lymphatic
metastasis. J Clin Invest. 128:861–875. 2018. View Article : Google Scholar
|
|
207
|
Huang H, Du J, Jin B, Pang L, Duan N,
Huang C, Hou J, Yu W, Hao H and Li H: Combination of urine exosomal
mRNAs and lncRNAs as novel diagnostic biomarkers for bladder
cancer. Front Oncol. 11:6672122021. View Article : Google Scholar :
|
|
208
|
Laukhtina E, Shim SR, Mori K, D'Andrea D,
Soria F, Rajwa P, Mostafaei H, Compérat E, Cimadamore A, Moschini
M, et al: Diagnostic accuracy of novel urinary biomarker tests in
non-muscle-invasive bladder cancer: A systematic review and network
meta-analysis. Eur Urol Oncol. 4:927–942. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Li KD, Chu CE, Patel M, Meng MV, Morgan TM
and Porten SP: Cxbladder monitor testing to reduce cystoscopy
frequency in patients with bladder cancer. Urol Oncol.
41:326.e1–326.e8. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Fasulo V, Paciotti M, Lazzeri M, Contieri
R, Casale P, Saita A, Lughezzani G, Diana P, Frego N, Avolio PP, et
al: Xpert bladder cancer monitor may avoid cystoscopies in patients
under 'active surveillance' for recurrent bladder cancer (BIAS
Project): Longitudinal cohort study. Front Oncol. 12:8328352022.
View Article : Google Scholar
|
|
211
|
Tortora F, La Civita E, Trivedi P,
Febbraio F, Terracciano D and Cimmino A: Emerging RNA-based
therapeutic and diagnostic options: Recent advances and future
challenges in genitourinary cancers. Int J Mol Sci. 24:46012023.
View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Yang L, Zou X, Zou J and Zhang G:
Functions of circular RNAs in bladder, prostate and renal cell
cancer (Review). Mol Med Rep. 23:3072021. View Article : Google Scholar
|
|
213
|
Jiang YK, Shuai YJ, Ding HM, Zhang H,
Huang C, Wang L, Sun JY, Wei WJ, Xiao XY and Jiang GS: Erratum to:
ARID1A inactivation increases expression of circ0008399 and
promotes cisplatin resistance in bladder cancer. Curr Med Sci.
43:12602023. View Article : Google Scholar
|
|
214
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar
|
|
215
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar
|
|
216
|
Ma X, Ying Y, Sun J, Xie H, Li J, He L,
Wang W, Chen S, Shen H, Yi J, et al: circKDM4C enhances bladder
cancer invasion and metastasis through miR-200bc-3p/ZEB1 axis. Cell
Death Discov. 7:3652021. View Article : Google Scholar
|
|
217
|
Liang Z, Guo W, Fang S, Zhang Y, Lu L, Xu
W and Qian H: CircRNAs: Emerging bladder cancer biomarkers and
targets. Front Oncol. 10:6064852021. View Article : Google Scholar :
|
|
218
|
Chu G, Ji X, Wang Y and Niu H: Integrated
multiomics analysis and machine learning refine molecular subtypes
and prognosis for muscle-invasive urothelial cancer. Mol Ther
Nucleic Acids. 33:110–126. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Harsanyi S, Novakova ZV, Bevizova K,
Danisovic L and Ziaran S: Biomarkers of bladder cancer: Cell-free
DNA, epigenetic modifications and non-coding RNAs. Int J Mol Sci.
23:132062022. View Article : Google Scholar :
|
|
220
|
Martin-Way D, Puche-Sanz I, Cozar JM,
Zafra-Gomez A, Gomez-Regalado MDC, Morales-Alvarez CM, Hernandez
AF, Martinez-Gonzalez LJ and Alvarez-Cubero MJ: Genetic variants of
antioxidant enzymes and environmental exposures as molecular
biomarkers associated with the risk and aggressiveness of bladder
cancer. Sci Total Environ. 843:1569652022. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Babjuk M, Burger M, Capoun O, Cohen D,
Compérat EM, Dominguez Escrig JL, Gontero P, Liedberg F,
Masson-Lecomte A, Mostafid AH, et al: European association of
urology guidelines on non-muscle-invasive bladder cancer (Ta, T1,
and carcinoma in situ). Eur Urol. 81:75–94. 2022. View Article : Google Scholar
|
|
222
|
Chen AA and Grasso M: Is there a role for
FISH in the management and surveillance of patients with upper
tract transitional-cell carcinoma? J Endourol. 22:1371–1374. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Koide K, Sekizawa A, Iwasaki M, Matsuoka
R, Honma S, Farina A, Saito H and Okai T: Fragmentation of
cell-free fetal DNA in plasma and urine of pregnant women. Prenat
Diagn. 25:604–607. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Wong FC and Lo YM: Prenatal diagnosis
innovation: Genome sequencing of maternal plasma. Annu Rev Med.
32:419–432. 2016. View Article : Google Scholar
|
|
225
|
Tivey A, Church M, Rothwell D, Dive C and
Cook N: Circulating tumour DNA-looking beyond the blood. Nat Rev
Clin Oncol. 19:600–612. 2022. View Article : Google Scholar
|
|
226
|
Wu Z, Yang Z, Li CS, Zhao W, Liang ZX, Dai
Y, Zhu Q, Miao KL, Cui DH and Chen LA: Differences in the genomic
profiles of cell-free DNA between plasma, sputum, urine, and tumor
tissue in advanced NSCLC. Cancer Med. 8:910–919. 2019. View Article : Google Scholar
|
|
227
|
Lo YM, Chan KC, Sun H, Chen EZ, Jiang P,
Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR and Chiu RW: Maternal
plasma DNA sequencing reveals the genome-wide genetic and
mutational profile of the fetus. Sci Transl Med. 2:61ra912010.
View Article : Google Scholar
|
|
228
|
Chen G, Jia G, Chao F, Xie F, Zhang Y, Hou
C, Huang Y, Tang H, Yu J, Zhang J, et al: Urine- and blood-based
molecular profiling of human prostate cancer. Front Oncol.
12:7597912022. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Saha MK, Massicotte-Azarniouch D, Reynolds
ML, Mottl AK, Falk RJ, Jennette JC and Derebail VK: Glomerular
hematuria and the utility of urine microscopy: A review. Am J
Kidney Dis. 80:383–392. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
He Y, Pan C, Zhang Y, Lv M and Yang B:
Nomogram for customized recurrence prediction in primary
non-muscle-invasive bladder cancer based on routine blood and urine
parameters. BMC Urol. 24:672024. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Teixeira-Marques A, Lourenço C, Oliveira
MC, Henrique R and Jerónimo C: Extracellular vesicles as potential
bladder cancer biomarkers: Take it or leave it? Int J Mol Sci.
24:67572023. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Latosinska A, Frantzi M, Vlahou A and
Mischak H: Clinical applications of capillary electrophoresis
coupled to mass spectrometry in biomarker discovery: Focus on
bladder cancer. Proteomics Clin Appl. 7:779–793. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Claps F, Rossin G, van Rhijn BWG, Mir MC,
Mertens LS, Ongaro L, Traunero F, Iachimovsky AI, Piasentin A,
Vedovo F, et al: The utility of inflammatory serum markers in the
assessment of perioperative morbidity after radical cystectomy for
bladder cancer. Medicina (Kaunas). 59:9262023. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
López-Cortés R, Gómez BB, Vázquez-Estévez
S, Pérez-Fentes D and Núñez C: Blood-based protein biomarkers in
bladder urothelial tumors. J Proteomics. 247:1043292021. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Lee DH, Yoon H, Park S, Kim JS, Ahn YH,
Kwon K, Lee D and Kim KH: Urinary exosomal and cell-free DNA
detects somatic mutation and copy number alteration in urothelial
carcinoma of bladder. Sci Rep. 8:147072018. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Kobayashi M, Abe H, Arai K, Murakami S and
Kamai T: Circulating tumor cells and cell-free tumor DNA analyses
in urothelial cancer using the LiquidBiopsy platform. Curr Urol.
16:99–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Hirotsu Y, Yokoyama H, Amemiya K, Hagimoto
T, Daimon H, Hosaka K, Oyama T, Mochizuki H and Omata M: Genomic
profile of urine has high diagnostic sensitivity compared to
cytology in non-invasive urothelial bladder cancer. Cancer Sci.
110:3235–3243. 2019. View Article : Google Scholar
|
|
238
|
Zhang R, Zang J, Xie F, Zhang Y, Wang Y,
Jing Y, Zhang Y, Chen Z, Shahatiaili A, Cai MC, et al: Urinary
molecular pathology for patients with newly diagnosed urothelial
bladder cancer. J Urol. 206:873–884. 2021. View Article : Google Scholar
|
|
239
|
Payne SR, Serth J, Schostak M, Kamradt J,
Strauss A, Thelen P, Model F, Day JK, Liebenberg V, Morotti A, et
al: DNA methylation biomarkers of prostate cancer: Confirmation of
candidates and evidence urine is the most sensitive body fluid for
non-invasive detection. Prostate. 69:1257–1269. 2009. View Article : Google Scholar
|
|
240
|
Jerónimo C, Usadel H, Henrique R, Silva C,
Oliveira J, Lopes C and Sidransky D: Quantitative GSTP1
hypermethylation in bodily fluids of patients with prostate cancer.
Urology. 60:1131–1135. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Goessl C, Müller M, Heicappell R, Krause H
and Miller K: DNA-based detection of prostate cancer in blood,
urine, and ejaculates. Ann N Y Acad Sci. 945:51–58. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Smith CG, Moser T, Mouliere F,
Field-Rayner J, Eldridge M, Riediger AL, Chandrananda D, Heider K,
Wan JCM, Warren AY, et al: Comprehensive characterization of
cell-free tumor DNA in plasma and urine of patients with renal
tumors. Genome Med. 12:232020. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Christensen E, Nordentoft I,
Birkenkamp-Demtröder K, Elbæk SK, Lindskrog SV, Taber A, Andreasen
TG, Strandgaard T, Knudsen M, Lamy P, et al: Cell-free urine and
plasma DNA mutational analysis predicts neoadjuvant chemotherapy
response and outcome in patients with muscle-invasive bladder
cancer. Clin Cancer Res. 29:1582–1591. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Crozier J, Papa N, Perera M, Ngo B, Bolton
D, Sengupta S and Lawrentschuk N: Comparative sensitivity and
specificity of imaging modalities in staging bladder cancer prior
to radical cystectomy: A systematic review and meta-analysis. World
J Urol. 37:667–690. 2019. View Article : Google Scholar
|
|
245
|
Bandyk MG, Gopireddy DR, Lall C, Balaji KC
and Dolz J: MRI and CT bladder segmentation from classical to deep
learning based approaches: Current limitations and lessons. Comput
Biol Med. 134:1044722021. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Humayun-Zakaria N, Ward DG, Arnold R and
Bryan RT: Trends in urine biomarker discovery for urothelial
bladder cancer: DNA, RNA, or protein? Transl Androl Urol.
10:2787–2808. 2021. View Article : Google Scholar
|
|
247
|
Sun T, Hutchinson L, Tomaszewicz K,
Caporelli ML, Meng X, McCauley K, Fischer AH, Cosar EF and Cornejo
KM: Diagnostic value of a comprehensive, urothelial
carcinoma-specific next-generation sequencing panel in urine
cytology and bladder tumor specimens. Cancer Cytopathol.
129:537–547. 2021. View Article : Google Scholar
|
|
248
|
Claps F, Pavan N, Ongaro L, Tierno D,
Grassi G, Trombetta C, Tulone G, Simonato A, Bartoletti R, Mertens
LS, et al: BCG-unresponsive non-muscle-invasive bladder cancer:
Current treatment landscape and novel emerging molecular targets.
Int J Mol Sci. 24:125962023. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Frantzi M, Latosinska A, Flühe L, Hupe MC,
Critselis E, Kramer MW, Merseburger AS, Mischak H and Vlahou A:
Developing proteomic biomarkers for bladder cancer: Towards
clinical application. Nat Rev Urol. 12:317–330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Xu X, Huang F, Cao M, Chen X, Wang H,
Jiang H, Yu Y, Shen M, Yang Y, Wang B, et al: Cross-platform
comparison of next-generation sequencing and matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry for
detecting KRAS/NRAS/BRAF/PIK3CA mutations in cfDNA from metastatic
colorectal cancer patients. J Clin Lab Anal. 35:e238182021.
View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Raina R, Nada A, Shah R, Aly H, Kadatane
S, Abitbol C, Aggarwal M, Koyner J, Neyra J and Sethi SK:
Artificial intelligence in early detection and prediction of
pediatric/neonatal acute kidney injury: Current status and future
directions. Pediatr Nephrol. 39:2309–2324. 2024. View Article : Google Scholar
|
|
252
|
Lima T, Henrique R, Vitorino R and
Fardilha M: Bioinformatic analysis of dysregulated proteins in
prostate cancer patients reveals putative urinary biomarkers and
key biological pathways. Med Oncol. 38:92021. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Cheng P, Wang R, He S, Yan P, Huang H,
Chen J, Shen J and Pu K: Artificial Urinary biomarkers for early
diagnosis of acute renal allograft rejection. Angew Chem Int Ed
Engl. 62:e2023065392023. View Article : Google Scholar
|
|
254
|
Xu C, Xu M, Hu Y, Liu J, Cheng P, Zeng Z
and Pu K: Ingestible artificial urinary biomarker probes for urine
test of gastrointestinal cancer. Adv Mater. 36:e23140842024.
View Article : Google Scholar
|
|
255
|
Yang Y, Wang J, Huang W, Wan G, Xia M,
Chen D, Zhang Y, Wang Y, Guo F, Tan J, et al: Integrated urinalysis
devices based on interface-engineered field-effect transistor
biosensors incorporated with electronic circuits. Adv Mater.
34:e22032242022. View Article : Google Scholar
|
|
256
|
Hao L, Zhao RT, Welch NL, Tan EKW, Zhong
Q, Harzallah NS, Ngambenjawong C, Ko H, Fleming HE, Sabeti PC and
Bhatia SN: CRISPR-Cas-amplified urinary biomarkers for multiplexed
and portable cancer diagnostics. Nat Nanotechnol. 18:798–807. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Shi H, Li X, Zhang Q, Yang H and Zhang X:
Discovery of urine biomarkers for bladder cancer via global
metabolomics. Biomarkers. 21:578–588. 2016. View Article : Google Scholar : PubMed/NCBI
|