Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide with an estimated one million new cases and
500,000 deaths annually (1). Early
detection of CRC is vital to reduce the mortality of this disease.
Several CRC screening tests, such as carcinoembryonic and
carbohydrate antigens 19-9, fecal occult-blood testing (FOBT) and
colonoscopy, have been used to detect CRC (2,3).
However, none of these methods has been established as a
well-accepted screening tool, due to the low sensitivity, high cost
or invasive nature. Thus, novel non-invasive biomarkers are
required to improve the detection of CRC.
Previous studies showed that microRNAs (miRNAs),
which are involved in tumorigenesis and in the development of
various types of cancer, are detectable in the plasma/serum
(4,5). Mitchell et al (4) clearly demonstrated that circulating
miRNAs originate from cancer tissues (4). Additional studies have demonstrated
the presence of circulating miRNAs and their potential use as novel
biomarkers of various types of cancer, such as pancreatic (6), colorectal (7), breast (8) and gastric cancers (9). These studies have provided new
opportunities for a non-invasive examination for the early
diagnosis of various types of cancer.
miR-200c and miR-18a are significantly upregulated
in CRC tumor tissues compared to their adjacent normal tissues
(10,11). miR-18a is significantly higher in
the plasma of pancreatic cancer patients and is a potential
screening biomarker for pancreatic cancer (12). Additional studies have demonstrated
that miR-200c was detectable at a higher level in the plasma of CRC
patients (7). In CRC tissues,
patients with overexpressed miR-200c or miR-18a exhibited a poorer
clinical prognosis (10,11). However, to the best of our
knowledge, this is the first study to report whether the
circulating miR-200c and miR-18a in plasma samples are potential
noninvasive markers for CRC.
In this study, we investigated whether the
circulating miR-200c and miR-18a in plasma samples might be used to
screen for CRC by comparing findings in CRC patients and volunteer
controls. Results clearly demonstrated their potential usefulness.
The findings of this study provided evidence that the plasma levels
of miR-200c and miR-18a may be used to distinguish CRC patients
from healthy individuals with a clinically satisfactory degree of
sensitivity and specificity.
Materials and methods
Patients and tissue samples
Plasma samples were collected from 164 patients,
including 78 CRC patients and 86 normal controls between January,
2010 and April, 2011 (Table I).
Exclusion criteria included inflammatory bowel disease, a family
history of familial adenomatous polyposis, hereditary non-polyposis
colon cancer or previous colonic surgery. Normal controls were
asymptomatic individuals recruited from a colonoscopy screening
programme at the First Department of General Surgery of the
Affiliated Hospital of the North Sichuan Medical College (Nanchong,
China). Plasma samples of CRC patients were collected prior to
surgical resection. The normal control samples were collected prior
to colonoscopy and at least 2 weeks following the initial
colonoscopy. Tumors were staged according to the 6th edition of the
International Union Against Cancer (UICC) Tumor-Nodes-Metastasis
(TNM) staging system. Patients who had undergone neoadjuvant
therapy prior to surgery were excluded. To investigate the changes
in plasma-based miRNA levels following removal of the lesion,
plasma samples were also collected at least 1 month after surgery.
Forty two pairs of tissue samples were collected from CRC patients.
The tumor and adjacent normal samples (≥4 cm apart from the tumor)
were biopsied during surgical resection. Samples were stored at
−80°C for subsequent analysis. The study was approved by the
Medical Ethics Committee of the North Sichuan Medical College, and
written informed consent was obtained from the patients.
 | Table IPatient characteristics for plasma
miRNA analysis. |
Table I
Patient characteristics for plasma
miRNA analysis.
| Characteristics | Patients
|
|---|
| Normal controls
(n=86) | CRC (n=78) |
|---|
| Age (years) | | |
| Mean ± SD | 60.3±11.8 | 61.4±13.6 |
| Gender | | |
| Male | 53 | 43 |
| Female | 33 | 35 |
| Histological
type | | |
| Well, moderate | | 44 |
| Poor, mucinous | | 34 |
| Location | | |
| Colon | | 31 |
| Rectum | | 47 |
| Lymph node
status | | |
| Positive | | 37 |
| Negative | | 41 |
| TNM stage | | |
| I, II | | 36 |
| III, IV | | 42 |
miRNA quantitation by real-time
quantitative reverse transcription-polymerase chain reaction
(qRT-PCR)
Total RNA was extracted from tissues using the
miRNeasy mini kit (Qiagen, Hilden, Germany) following the
manufacturer’s instructions. Total RNA containing small RNA was
extracted from 500 μl of plasma using TRIzol LS reagent
(Invitrogen, Carlsbad, CA, USA) and the miRNeasy Mini kit (Qiagen).
according to the manufacturer’s instructions. The final elution
volume was 30 μl. The RNA concentration was measured with a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific,
Wilmington, DE, USA). For miRNA qPCR, reverse transcription was
performed using the QuantiMir™ RT kit (System Biosciences, Mountain
View, CA, USA). The complementary DNA (cDNA) served as the template
for SYBR-Green real-time PCR using Power SYBR-Green PCR Master mix
(Applied Biosystems, Foster City, CA, USA). The reactions were
performed in triplicate on the iCycler iQ™ Multi-Color Real-Time
PCR Detection System (Bio-Rad, Hercules, CA, USA) using
miRNA-specific primers (Applied Biosystems). The amplification
profile was denatured at 95°C for 10 min, followed by 50 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 1 min. The comparative cycle threshold
(CT) method was applied to quantify the expression
levels of miRNAs. The relative amount of miR-200c and miR-18a to
small nuclear U6 RNA (RNU6B) was calculated using the equation
2−ΔCT, where ΔCT= (CT
miR-200c/miR-18a − CT RNU6B). The fold change of
the gene expression was calculated using the equation
2−ΔΔCT.
Statistical analysis
The difference in miRNA levels between paired tissue
samples was determined using the Wilcoxon matched-pairs test.
Expression levels of plasma miRNAs were compared using the
Mann-Whitney U or the Kruskall-Wallis test. Receiver-operating
characteristics (ROC) curves were established to evaluate the
diagnostic value of plasma miRNAs to distinguish between tumors and
controls. Statistical analyses were carried out using the SPSS 16.0
software (SPSS, Chicago, IL, USA). Two-sided P-values were
calculated, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
Patient characteristics are summarised in Table I. A total of 164 participants
including 78 CRC patients and 86 healthy individuals were included
in this study. No significant differences of age or gender were
found in CRC patients and normal controls (Student’s t-test,
P=0.564; χ2 test, P= 0.399).
Enhanced expression of miR-200c and
miR-18a in CRC tissue samples
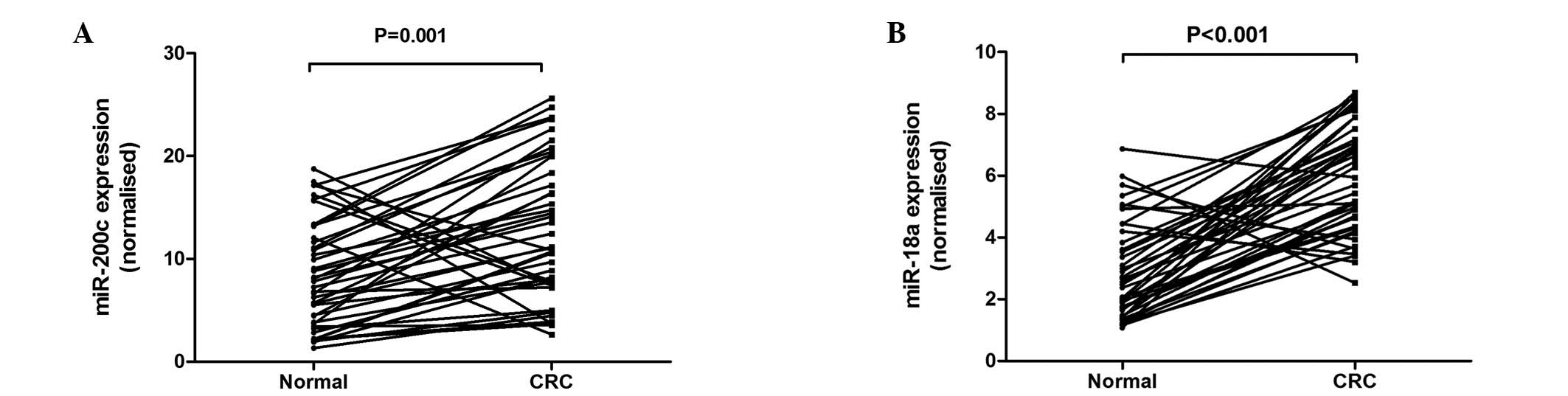
Of the 42 pairs of CRC tissue samples, 36 (85.7%)
demonstrated higher miR-200c expression in tumor tissue compared to
adjacent normal tissue (P=0.001; Fig.
1A), with an average increase of 2.02-fold. Thirty-seven
(88.1%) pairs of tissue samples demonstrated higher miR-18a
expression in the tumor tissue compared to adjacent normal tissue
(P<0.001; Fig. 1B), with an
average increase of 2.66-fold.
Increased levels of plasma miR-200c and
miR-18a in CRC patients
Concentration of miR-200c and miR-18a was
investigated in an independent group of 164 plasma samples that
included 78 CRC patients and 86 healthy controls (Table I). miR-200c and miR-18a
concentration significantly increased in the plasma of CRC patients
compared to normal controls (P<0.001, Mann-Whitney U test)
(Fig. 2A and B).
Sensitivity of plasma miR-200c and
miR-18a towards the detection of CRC
ROC curve analyses demonstrated that the plasma
levels of miR-200c and miR-18a were useful biomarkers for
distinguishing between CRC patients and controls with ROC curve
areas of 0.749 [95% confidence interval (CI), 0.675–0.822] and
0.804 (95% CI, 0.736–0.872), respectively (Fig. 2B and D). At the cut-off value of
7.6 for miR-200c (relative expression in comparison to RNU6B), the
sensitivity was 64.1% and specificity 73.3%. At the cut-off value
of 2.3 for miR-18a (relative expression in comparison to RNU6B),
the sensitivity and specificity were 73.1 and 79.1%, respectively.
miR-18a demonstrated a stronger discriminating ability compared to
miR-200c, with no statistical significance (P=0.243). The predicted
values of logistic regression showed a significant difference
between these two groups (P<0.001, Fig. 2E) and combination ROC analysis
demonstrated an increased AUC value to 0.839 (Fig. 2F, miR-18a, P= 0.013; miR-200c,
P=0.022) with a sensitivity and specificity of 84.6 and 75.6%,
respectively.
Correlation between plasma miR-200c and
miR-18a expression, and clinicopathological characteristics
The correlation between plasma-based
miR-200c/miR-18a and clinicopathological characteristics is shown
in Tables II and III. The expression levels of plasma
miR-200c and miR-18a were classified as low or high on the basis of
the median value. No significant correlation was found between the
two miRNAs and gender, age, histological type, tumor location,
lymph nodal status or Tumor-Nodes-Metastasis (TNM) classification
(P>0.05), while the levels of miR-18a demonstrated an elevation
trend in patients with more advanced TNM stage (P= 0.069).
 | Table IICorrelation between plasma miR-200c
expression and clinicopathological characteristics of CRC
patients. |
Table II
Correlation between plasma miR-200c
expression and clinicopathological characteristics of CRC
patients.
| Characteristics | miR-200c expression
| P-value |
|---|
| Low (n=39) | High (n=39) |
|---|
| Age (years) | | | |
| Mean ± SD | 63.1±14.5 | 59.7±12.6 | 0.281 |
| Gender | | | 0.255 |
| Male | 19 | 24 | |
| Female | 20 | 15 | |
| Histological
type | | | 0.361 |
| Well, moderate | 20 | 24 | |
| Poor, mucinous | 19 | 15 | |
| Tumor location | | | 0.247 |
| Colon | 18 | 13 | |
| Rectum | 21 | 26 | |
| Lymph node
status | | | 0.496 |
| Positive | 17 | 20 | |
| Negative | 22 | 19 | |
| TNM stage | | | 0.364 |
| I, II | 20 | 16 | |
| III, IV | 19 | 23 | |
 | Table IIICorrelation between plasma miR-18a
expression and clinicopathological characteristics of CRC
patients. |
Table III
Correlation between plasma miR-18a
expression and clinicopathological characteristics of CRC
patients.
|
Characteristics | miR-18a expression
| P-value |
|---|
| Low (n=39) | High (n=39) |
|---|
| Age (years) | | | |
| Mean ± SD | 62.1±13.7 | 60.7±13.6 | 0.655 |
| Gender | | | 0.495 |
| Male | 23 | 2O | |
| Female | 16 | 19 | |
| Histological
type | | | 0.171 |
| Well,
moderate | 25 | 19 | |
| Poor,
mucinous | 14 | 20 | |
| Tumor location | | | 0.488 |
| Colon | 14 | 17 | |
| Rectum | 25 | 22 | |
| Lymph node
status | | | 0.112 |
| Positive | 14 | 23 | |
| Negative | 25 | 16 | |
| TNM stage | | | 0.069 |
| I, II | 22 | 14 | |
| III, IV | 17 | 25 | |
Follow-up of plasma miR-200c and miR-18a
levels following removal of the lesion
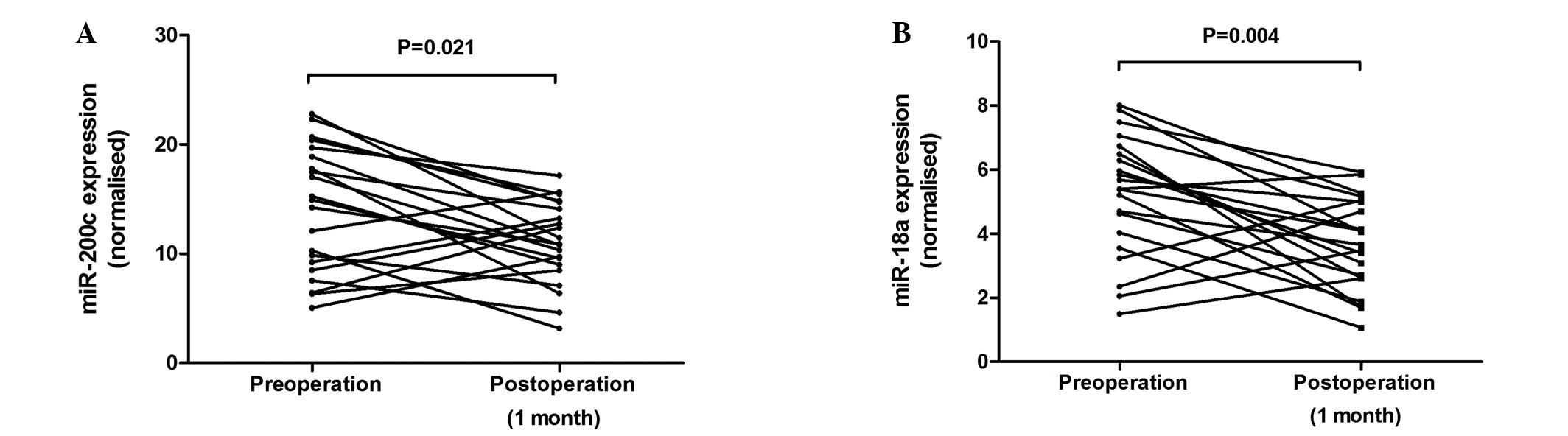
Plasma miR-200c and miR18a levels were measured in a
subgroup of CRC patients (n=21), following the resection of
lesions. A significant decrease in miR-200c (P= 0.021, Fig. 3A) and miR-18a (P= 0.004, Fig. 3B) levels following the resection
was noted.
Discussion
miRNAs have been reported to be involved in
tumorigenesis, acting as either oncogenes (13,14)
or tumor suppressors (15).
Several studies have described altered expression of miRNAs in
cancer tissues compared to normal tissues, suggesting miRNAs to be
potential novel diagnostic and prognostic markers (16,17).
Studies investigating plasma miRNAs comprise a
promising field for clinical application. Tumor-derived miRNA in
the plasma was first described by Mitchell et al (4). According to this study, high
stability was exhibited after prolonged incubation at room
temperature and/or multiple freezing-thawing processes (4). Additionally, the characteristics of
miRNAs, such as tissue-specific miRNA signatures and the
availability of numerous copies/cells, indicated potential
advantages as biomarkers compared to those of other nucleic acids,
such as circulating DNA and mRNA. An increasing number of studies
also suggest the potential use of miRNAs in the early detection of
patients with several malignancies, such as gastric, pancreatic,
colorectal and breast cancer (6–9).
Recent studies have focused ons the role of
circulating miRNAs in the plasma of CRC patients (7,18).
Huang et al (18) reported
that the combined analyses of miR-29a and miR-92a in the plasma
were able to distinguish CRC patients from healthy individuals.
However, the investigation of plasma miR-200c and miR-18a for the
detection of CRC has not been previously reported. In this study,
we have systematically established the feasibility of miR-200c and
miR-18a in the plasma, as non-invasive biomarkers for CRC
screening.
This study has shown that miR-200c and miR-18a were
overexpressed in colorectal tumor tissues compared to their
corresponding adjacent non-tumor tissues. In line with their
enhanced expression levels in primary tumor specimens, miR-200c and
miR-18a were also detected at high levels in the plasma samples of
CRC patients. Based on ROC curves, we selected cut-off values that
best differentiated CRC patients from healthy individuals. miR-200c
was found to have a sensitivity of 64.1% and a specificity of
73.3%, whereas miR-18a had a sensitivity of 73.1% and a specificity
of 79.1% for CRC. miR-18a demonstrated a higher discriminating
ability compared to miR-200c, while not statistically significant
(P=0.243). Combined ROC analyses using these two targets may yield
an increased AUC of 0.839, with 84.6% sensitivity and 75.6%
specificity in discriminating CRC patients from normal controls
(miR-18a, P= 0.013; miR-200c, P= 0.022), indicating the additive
effect in the diagnostic value of these two miRNAs.
miR-18a is part of the miR-17-92 gene cluster,
located on chromosome 13q13. As a known oncomir, miR-17-92 cluster
is able to promote cell proliferation, suppress apoptosis, induce
tumor angiogenesis and accelerate tumor progression (19,20).
miR-18a has been found to be significantly upregulated in gastric
cancer (21), hepatocellular,
pancreatic and colorectal carcinomas (11), suggesting its importance in
tumorigenesis. In CRC tissues, patients with overexpressed miR-18a
exhibited a poorer clinical prognosis. Recently, Morimura et
al (12) reported that plasma
miR-18a was a potential marker for pancreatic cancer that was not
associated with clinical factors. Our findings were comparable to
those of Morimura et al (12).
miR-200c is a member of the miR-200 family,
previously shown to inhibit the epithelial-to-mesenchymal
transition (EMT) by targeting the transcriptional repressors of
cadherin 1 (CDH1) and zinc finger E-box binding homeobox 1 (ZEB1),
suggesting that miR-200c is able to prevent tumor progression by
negatively regulating ZEB transcriptional repressors and preventing
EMT (22,23). A recent study demonstrated high
expression of serum miR-200c associated with poor prognosis in
patients with lung cancer (24).
miR-200c was also detectable at a higher level in the plasma of CRC
patients, and patients with overexpressed miR-200c exhibited a
poorer clinical prognosis (7,10).
These results indicate that miR-200c acts as an oncogene or a tumor
suppressor, depending on the circumstances. This study suggested
that similar to miR-18a, the expression of miR-200c, did not
correlate with gender, age, histological type, tumor location,
lymph nodal status or TNM classification (P>0.05), and yielded
low predictive value compared to miR-18a. However, miR-200c was
able to improve the differentiation power of miR-18a between CRC
patients and the controls, resulting in an increased sensitivity
and specificity.
In this study, we also measured circulating miRNAs
in paired-plasma samples obtained prior to and 1 month after
surgical removal of the tumors, to confirm the release of
circulating miRNAs. Results showed that both miR-18a and miR-200c
concentrations decreased significantly following removal of the
lesion. These findings suggest that the high levels of miR-18a and
miR-200c in the plasma of CRC patients are derived from neoplastic
cells.
Plasma miRNA assays possess several potential
clinical uses: screening patients at high risk for CRC and
monitoring disease recurrence during the follow-up period after CRC
resection. miRNA biomarkers might also be powerful and useful for
confirming the completeness of tumor resection and for evaluating
the efficacy of adjuvant therapies when the elimination clearance
of plasma miRNAs can be elucidated.
Although the results of this study are promising,
there are several limitations. First, further validations of the
usefulness of these markers in large cohorts are necessary, since
the sample size used in this study was small. Second, although
miR-200c and miR-18a elevation in plasma are likely to be derived
from CRC, it is uncertain whether this elevation is specific for
CRC, and whether it can be used to distinguish sporadic from
familial types of CRC. Thus, additional studies are required to
examine familial and sporadic cases. Third, despite the
significantly elevated levels of plasma miR-200c and miR-18a in
CRC, it is important to examine whether its plasma level changes in
patients with adenoma with various degree of dysplasia. The
determination of whether plasma miR-200c and miR-18a can be used
for the detection of pre-malignant lesions, such as adenoma, is
likely to add more value to the use of this marker for CRC
prevention.
In conclusion, this study has demonstrated the
feasibility of using plasma miR-18a and miR-200c as non-invasive
tools for the detection of CRC. These data serve as a basis for
further investigation, preferably in large-scale prospective
studies, before these two miRNAs can be incorporated into routine
clinical practice as non-invasive screening tools for colorectal
neoplasia.
Acknowledgements
This study was partly supported by the
Scientific Research Program of the Sichuan Provincial Health
Department of China (110319).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Levin B, Lieberman DA, McFarland B, et al:
Screening and surveillance for the early detection of colorectal
cancer and adenomatous polyps, 2008: a joint guideline from the
American Cancer Society, the US Multi-Society Task Force on
Colorectal Cancer, and the American College of Radiology. CA Cancer
J Clin. 58:130–160. 2008. View Article : Google Scholar
|
|
3
|
Mandel JS: Screening for colorectal
cancer. Gastroenterol Clin North Am. 37:97–115. 2008. View Article : Google Scholar
|
|
4
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Chen J, Chang P, et al: MicroRNAs
in plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res (Phila).
2:807–813. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ng EK, Chong WW, Jin H, et al:
Differential expression of microRNAs in plasma of patients with
colorectal cancer: a potential marker for colorectal cancer
screening. Gut. 58:1375–1381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heneghan HM, Miller N, Lowery AJ, et al:
Circulating microRNAs as novel minimally invasive biomarkers for
breast cancer. Ann Surg. 251:499–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsujiura M, Ichikawa D, Komatsu S, et al:
Circulating microRNAs in plasma of patients with gastric cancers.
Br J Cancer. 102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xi Y, Formentini A, Chien M, et al:
Prognostic values of microRNAs in colorectal cancer. Biomark
Insights. 2:113–121. 2006.PubMed/NCBI
|
|
11
|
Motoyama K, Inoue H, Takatsuno Y, et al:
Over- and under-expressed microRNAs in human colorectal cancer. Int
J Oncol. 34:1069–1075. 2009.PubMed/NCBI
|
|
12
|
Morimura R, Komatsu S, Ichikawa D, et al:
Novel diagnostic value of circulating miR-18a in plasma of patients
with pancreatic cancer. Br J Cancer. 105:1733–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Voorhoeve PM, le Sage C, Schrier M, et al:
A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in
testicular germ cell tumors. Cell. 124:1169–1181. 2006. View Article : Google Scholar
|
|
15
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mattie MD, Benz CC, Bowers J, et al:
Optimized high-throughput microRNA expression profiling provides
novel biomarker assessment of clinical prostate and breast cancer
biopsies. Mol Cancer. 5:242006. View Article : Google Scholar
|
|
17
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang Z, Huang D, Ni S, et al: Plasma
microRNAs are promising novel biomarkers for early detection of
colorectal cancer. Int J Cancer. 127:118–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Diosdado B, van de Wiel MA, Terhaar Sive,
Droste JS, et al: MiR-17-92 cluster is associated with 13q gain and
c-myc expression during colorectal adenoma to adenocarcinoma
progression. Br J Cancer. 101:707–714. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Connolly E, Melegari M, Landgraf P, et al:
Elevated expression of the miR-17-92 polycistron and miR-21 in
hepadnavirus-associated hepatocellular carcinoma contributes to the
malignant phenotype. Am J Pathol. 173:856–864. 2008. View Article : Google Scholar
|
|
21
|
Yao Y, Suo AL, Li ZF, et al: MicroRNA
profiling of human gastric cancer. Mol Med Rep. 2:963–970.
2009.PubMed/NCBI
|
|
22
|
Park SM, Gaur AB, Lengyel E, et al: The
miR-200 family determines the epithelial phenotype of cancer cells
by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Develop. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Korpal M, Lee ES, Hu G, et al: The miR-200
family inhibits epithelial-mesenchymal transition and cancer cell
migration by direct targeting of E-cadherin transcriptional
repressors ZEB1 and ZEB2. J Biol Chem. 283:14910–14914. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu XG, Zhu WY, Huang YY, et al: High
expression of serum miR-21 and tumor miR-200c associated with poor
prognosis in patients with lung cancer. Med Oncol. 29:618–626.
2012. View Article : Google Scholar : PubMed/NCBI
|