Introduction
Metastasis is one of the biological features of
malicious cancer, and is one the causes of mortality as a result of
colon cancer. The mechanism of metastasis is always the hot-spot of
molecular biology research. Previous evidence shows that during the
metastasis of malicious cancer, numerous genes are involved in the
control and regulation at various levels. To date, researchers have
identified that numerous genes can suppress cancer metastasis,
including nm23, multiple cancer inhibition suppressor gene-1 and
DLC-1 (1,2). The nm23/NDP kinase is known as a
non-metastasis gene, since Steeg et al (3) identified it from murine K-1735 melanoma
cell lines in 1988, Subsequently, Biggs et al (4) reported that the microtubule-associated
abnormal wing disc gene (Awd)/NDP kinase is critical in spindle
microtubule polymerization, and the product of awd of Drosophila is
78% identical to the product of the nm23 gene of mammals. In the
following decades, the nm23 gene was further investigated and it
was shown that suppression of metastatic mechanism of human nm23 in
various cancer types may be through a dynamin-mediated pathway
(5). The nm23 gene regulates cell
invasion, cell differentiation and motility via the regulation of
growth factor receptor signaling (6,7). In 2013,
the expression in chronic myelogenous leukemia cells was detected,
and revealed that the nm23-H1 gene may inhibit K562 cell
proliferation and migration (8). In
order to further assess the role of the nm23-H1 gene mutation in
colon cancer evolution, the present study used the polymerase chain
reaction-single strand conformation polymorphism (PCR-SSCP)-silver
staining method to detect changes in the nm23 gene in the colon
cancer tissue, and an immunohistochemistry assay to analyze the
protein expression of nm23 in patient samples.
Patients and methods
Patient samples
The present study obtained 63 specimens of human
colorectal cancer tissue from Shanghai Pudong Hospital Affiliated
to Fudan University (Shanghai, China), which were all diagnosed
pathologically. Of these, 20 (32%) exhibited lymph gland
metastasis. The pathological types classification were as follows:
3 (5%) cases of high differentiation gland cancer, 56 (89%) cases
of medium differentiation gland cancer and 4 (6%) cases of low
differentiation gland cancer. The present study was approved by the
Ethics Committee of Shanghai Pudong Hospital Affiliated to Fudan
University.
DNA-extraction
The genomic DNA extraction of fresh cancer tissue
was performed using standard methods. The DNAwas isolated by
digestion with Proteinase K (Takara Bio, Inc., Shiga, Japan)
followed by phenol:chloroform (1:1) (Takara Bio, Inc.) extraction.
Briefly, the cancer tissue was mashed up, and mixed with 10% sodium
dodecyl sulfate and protein enzyme K (10 g/l) (Takara Bio, Inc.)
for digestion at 55°C for 3 h. Precipitation was performed using
NaCl adn the samples were centrifuged at 81,600 × g for 20 mins.
The upper clear liquid layer was obtained and 700 ml/l cool alcohol
was added for 20°C for 30 mins. The samples were centrifuged at
142,800 × g for 10 mins. Phenol-chloroform and chloroform-isoamyl
alcohol (Takara Bio, Inc.) were added separately and the samples
were centrifuged at 132,600–153,000 × g for 5 mins. TE buffer
(Takara Bio, Inc.) was added to dissolve and preserve the DNA.
PCR-SSCP
Four pairs of primers were designed, corresponding
to the numbers 2–5 position exons of the nm23-H1 gene. PCR was
performed, according to the stated conditions (Table I) on a Mastercycler Gradient PCR
system (Eppendorf, Hamburg, Germany).
 | Table I.Primers used and PCR amplification
conditions. |
Table I.
Primers used and PCR amplification
conditions.
| Primer | Sequence (5′→3′) | Conditions | Size of amplified
products (bp) |
|---|
| Exons 2 |
TGTTTCATTCCTCTACCTGC | 95°C for 5 min | 171 |
|
|
GTTCTCCTATACTATGAAGG | (95°C for 45 sec,
52°C for 45 sec, 72°C for 60 sec) ×32 72/5 min |
|
| Exons 3 |
GTTATTCTCATTCTCTGTCC | 95°C for 5 min | 128 |
|
|
CATACTTGGAGTATCCCAC | (95°C for 45 sec,
52°C for 45 sec, 72°C for 60 sec) ×32 72°C for 5 min |
|
| Exons 4 |
GACCATATCTTCTTCTGTCC | 95°C for 5 min | 154 |
|
|
CCTTGTGGCAACTAAATCAG | (95°C for 45 sec,
53°C for 45 sec, 72°C for 60 sec) ×32 72°C for 5 min |
|
| Exons 5 |
GTCTTGGTCATGTGACTATC | 95°C for 5 min | 141 |
|
|
GTGAAAAGCAATGTGGTCTG | (95°C for 45 sec,
53°C for 45 sec, 72°C for 60 sec) ×32 72°C for 5 min |
|
Following PCR, 10 µl PCR product was added to the
sample-loading buffer (Takara Bio, Inc.), including 950 g/l
formamide. The samples was denatured for 5 min at 95°C, and was
immediately put on ice. The denatured samples were separated by
polyacrylamide gel electrophoresis in electrophoresis buffer (0.5 ×
TBE) at 70 V, 4°C, overnight. Following running, silver staining
(Wuhan Boster Company, Wuhan, China) was performed.
Immunohistochemistry
Rabbit polyclonal anti-nm23-H1 antibody and
biotinylated goat anti-rabbit immunoglobulin G (each from Wuhan
Boster Company) were used at concentrations of 1:50 and 1:500,
respectively. The human colon tissue samples were preserved in 10%
formaldehyde solution, and were dehydrated and embedded in
paraffin, according to routine methods. Briefly, the 4 µm-thick
paraffin sections were removed and blocked with 3%
peroxide-methanol at room temperature for 20 min for endogenous
peroxidase ablation. The sections were then incubated with the
rabbit anti-nm23 antibody diluted in phosphate-buffered saline
(PBS; 0.01 M; pH 7.4) for 2 h at 37°C. Following incubation with
primary antibody, the samples were incubated with goat anti-rabbit
immunoglobulin G for 30 min at 37°C. The samples were subsequently
stained with 3,3-diaminobenzidine (Wuhan Boster Company) followed
by washing in PBS. Nuclear counterstaining was performed using
Mayer's hematoxylin, followed by dehydration, clearing and mounted
with neutral gums.
Results
PCR-SSCP
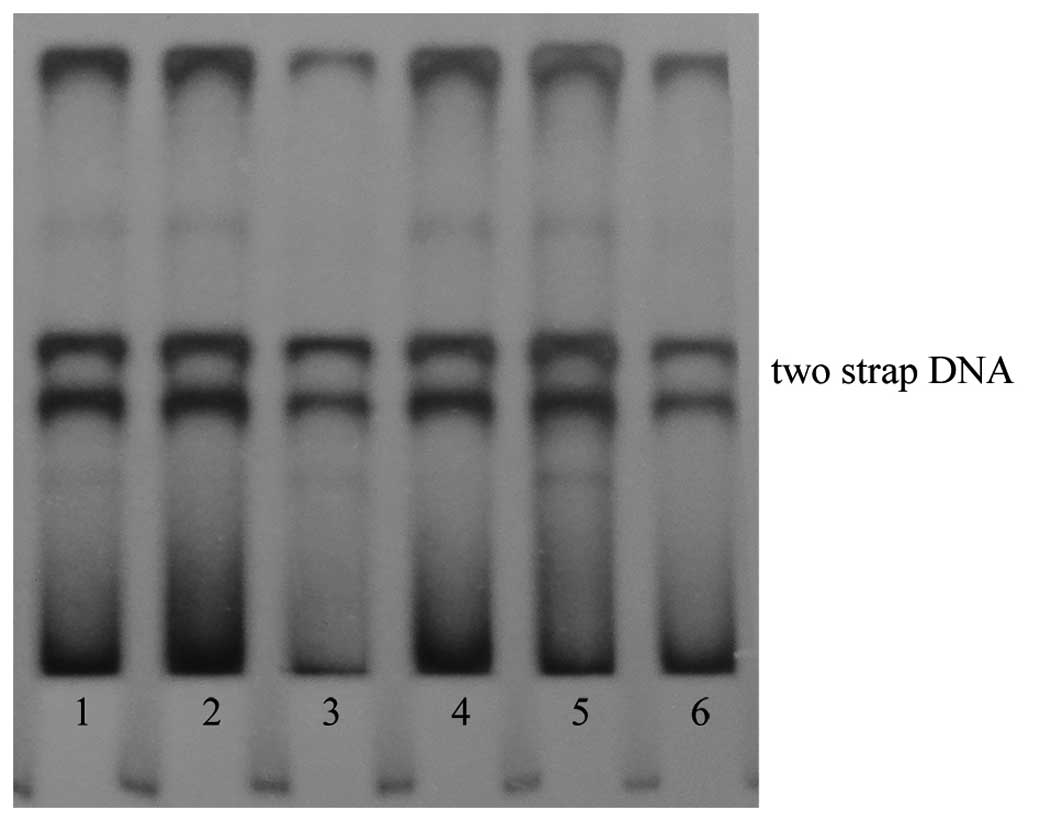
The present study performed PCR-SSCP detection on
the 63 cases of colon cancer tissue specimens to analyze any point
mutation of the nm23 gene on the 2nd-5th exon
position. Through repeated screening and operation, the results
from the DNA samples revealed that the
2nd-4th exons of nm23-H1 exhibit two straps
(Fig. 1), but the 5th exon
displays four straps for certain specimens (data not shown). The
coding protein was then analyzed and revealed that only the
5th exon sequence of the nm23-H1 gene has a
polymorphism. The 360th position base exhibits a T→C
replacement, but the coding amino acid AGT→AGC remains unchanged
(serine), therefore, it is a synonymous mutation.
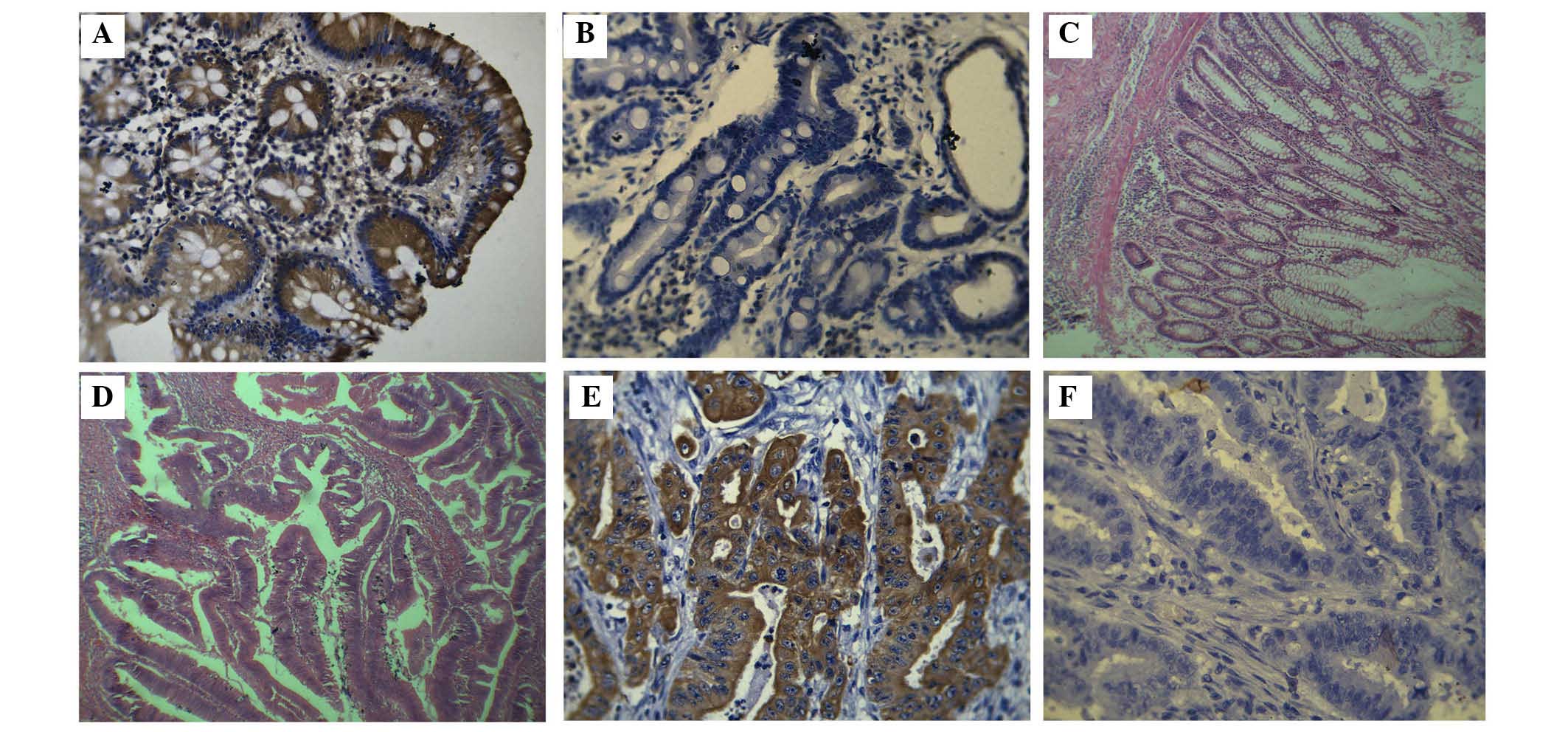
Immunohistochemistry
nm23-H1 is predominantly expressed in the cytoplasm
of the colorectal cancer gland. Among the 63 colorectal
adenocarcinoma specimens, 19 (30%) exhibited positive
immunostaining for the nm23-H1 protein, 44 (70%) exhibited negative
immunostaining, while the adjacent normal tissues were 72% positive
staining (Fig. 2).
Discussion
The nm23 gene has been regarded as a metastatic
suppressive gene in various tumor types (9). It is an important cancer metastasis
suppressor gene. The nm23-H1 gene cDNA has a total length of 534 bp
and 152 amino acids. The nm23 gene family has two homologous genes,
nm23-H1 and nm23-H2, and both are located at 17q22, encoding a
protein with a molecular weight of 18 and 17 kDa, respectively. The
nm23 gene encodes a nucleoside diphosphate kinase (NDPK). Both
nm23-H1 and nm23-H2 encode, the A and B subunits of NDPK,
respectively. They are 88% identical in their amino acid sequences.
NDPK activity is responsible for the synthesis of most cellular
nucleoside triphosphates, other activities include cell
proliferation, differentiation and development.
DR-nm23 cDNA was cloned in 1997. It is highly
homologous to the putative metastasis suppressor nm23-H1 gene and
the closely associated nm23-H2 gene (10,11). The
expression of DR-nm23 in colorectal cancer tissue was significantly
lower compared with that in adenoma and normal tissue. Therefore,
the expression status of DR-nm23 may act as a potential prognostic
factor in patients with colorectal cancer, and it may be involved
in the regulation of differentiation of colorectal cancer cells
(12). At the invasive front of colon
carcinoma, nm23-H1 levels were also reduced or lost, and its
downregulated expression was closely associated with the invasion
and metastasis of colorectal cancer (13). The protein expression of nm23 was
assessed in the colorectal carcinoma tissue by
immunohistochemistry. This revealed that the protein expression was
higher in carcinoma tissue compared with that in adjacent
non-neoplastic mucosa, similar to the results of a previous study
(14).
In gastric and colorectal carcinomas, the genomic
region 17q21 is frequently associated with microsatellite
instability and loss-of-heterozygosity. The nm23 gene is located in
this area. This region contains several putative tumor suppressor
genes, including the Spn gene. Spn downregulation is associated
with a poorer survival in patients with advanced stages of
colorectal carcinoma (15), and
downregulation of nm23-H1 contributes to tumorigenesis in lung
cancer (16). Also, Qin et al
(17) support the potential utility
of targeting Nm23-H1 as a therapeutic approach for the treatment of
Kaposi's sarcoma (17).
Data from a previous study has suggested that part
of the antimetastatic function of Nm23-H1 lies in the pathways that
it interrupts via binding and inactivation of proteins (18). In human hepatoma and colon carcinoma
cells, nm23-H1 was implicated as a metastasis suppressor, in
maintaining adherens junctions and limiting the invasive potential.
It appears that nm23 is crucial for inhibiting invasive migration,
cell-cell adhesion and cell migration (13). Enhanced expression of nm23-H1 protein
in Chinese patients with digestive system cancer inhibited cancer
metastasis (19). However, the
mechanistic basis of the metastasis suppressor function of nm23 and
its regulated expression remains to be elucidated.
Disorders of gene structure or gene expression can
cause abnormalities in cell growth and differentiation, thereby
causing cancer and having a malicious biological behavior.
Generally, these are mechanisms that lead to gene silencing,
mutation, genomic deletion and promoter methylation. Mutation
screening is always performed by PCR-SSCP. At present, this
technique has been used for detecting the known mutation hot-spots,
screening of unknown mutations and analyzing of the gene mutation
and cancer suppressing gene (20).
The present study assessed 63 specimens of Chinese human colon
cancer tissue via PCR-SSCP. The results indicated that the nm23
gene mutation does not serve the dominant role in the metastasis of
colon cancer.
A previous study demonstrated that treatment with
5-aza-2-deoxycytidine reduces the expression of Nm23-H1, which
causes hypermethylation in the nm23-H1 promoter (17,21).
Protein arginine methyltransferase 5 (PRMT5) methylates histones H3
at the nm23 promoter region and inhibits methylation of H3K9. PRMT5
regulates cell growth and proliferation by modulating the
expression of nm23 (22). As with the
previous studies, it is not possible to determine the mechanism of
nm23 responsible for the regulation of cell growth and
proliferation involved in tumor suppression. The mechanisms by
which nm23-H1 exerts its metastasis-suppressor functions involve
multiple pathways, which remain to be elucidated (23,24).
Although colon cancer metastasis may be due to other
genes or due to the methylation of the control portion of nm23
gene, the mechanism of the nm23 as a suppressor gene is more
complicated than previously hypothesized. Further research is
required to fully understand the mechanism and to identify its role
in the cancer process.
Acknowledgements
The present study was supported by Shanghai Pudong
Science and Technology Commission of China. (no. PKJ2014-Y23).
References
|
1
|
Jin Y, Tian X, Shang Y and Huang P:
Inhibition of DLC-1 gene expression by RNA interference in the
colon cancer LoVo cell line. Oncol Rep. 19:669–674. 2008.PubMed/NCBI
|
|
2
|
Dai Z and Jin Y: Promoter methylation of
the DLC-1 gene and its inhibitory effect on human colon cancer.
Oncol Rep. 30:1511–1517. 2013.PubMed/NCBI
|
|
3
|
Steeg PS, Bevilacqua G, Kopper L,
Thorgeirsson UP, Talmadge JE, Liotta LA and Sobel ME: Evidence for
a novel gene associated with low tumor metastatic potential. J Natl
Cancer Inst. 80:200–204. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Biggs J, Hersperger E, Steeg PS, Liotta LA
and Shearn A: A drosophila gene that is homologous to a mammalian
gene associated with tumor metastasis codes for a nucleoside
diphosphate kinase. Cell. 63:933–940. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dammai V, Adryan B, Lavenburg KR and Hsu
T: Drosophila awd, the homolog of human nm23, regulates FGF
receptor levels and functions synergistically with shi/dynamin
during tracheal development. Genes Dev. 17:2812–2824. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nallamothu G, Woolworth JA, Dammai V and
Hsu T: Awd, the homolog of metastasis suppressor gene Nm23,
regulates drosophila epithelial cell invasion. Mol Cell Biol.
28:1964–1973. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nallamothu G, Dammai V and Hsu T:
Developmental function of Nm23/awd: A mediator of endocytosis. Mol
Cell Biochem. 329:35–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dai Z, Xiao W and Jin Y: Inhibition of
nm23-H1 gene expression in chronic myelogenous leukemia cells.
Oncol Lett. 6:1093–1097. 2013.PubMed/NCBI
|
|
9
|
Mitre-Aguilar IB, Cabrera-Quintero AJ and
Zentella-Dehesa A: Genomic and non-genomic effects of
glucocorticoids: Implications for breast cancer. Int J Clin Exp
Pathol. 8:1–10. 2015.PubMed/NCBI
|
|
10
|
Martinez R, Venturelli D, Perrotti D,
Veronese ML, Kastury K, Druck T, Huebner K and Calabretta B: Gene
structure, promoter activity, and chromosomal location of the
DR-nm23 gene, a related member of the nm23 gene family. Cancer Res.
57:1180–1187. 1997.PubMed/NCBI
|
|
11
|
McDermott WG, Boissan M, Lacombe ML, Steeg
PS and Horak CE: Nm23-H1 homologs suppress tumor cell motility and
anchorage independent growth. Clin Exp Metastasis. 25:131–138.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu L, Liang L, Su J and Yang Z: Inhibitory
effect of upregulated DR-nm23 expression on invasion and metastasis
in colorectal cancer. Eur J Cancer Prev. 22:512–522. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boissan M, De Wever O, Lizarraga F, Wendum
D, Poincloux R, Chignard N, Desbois-Mouthon C, Dufour S,
Nawrocki-Raby B, Birembaut P, et al: Implication of metastasis
suppressor nm23-H1 in maintaining adherens junctions and limiting
the invasive potential of human cancer cells. Cancer Res.
70:7710–7722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oliveira LA, Artigiani-Neto R, Waisberg
DR, Fernandes LC, Lima Fde O and Waisberg J: nm23 protein
expression in colorectal carcinoma using TMA (tissue microarray):
Association with metastases and survival. Arq Gastroenterol.
47:361–367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Estevez-Garcia P, Lopez-Calderero I,
Molina-Pinelo S, Muñoz-Galvan S, Salinas A, Gomez-Izquierdo L,
Lucena-Cacace A, Felipe-Abrio B, Paz-Ares L, Garcia-Carbonero R and
Carnero A: Spinophilin loss correlates with poor patient prognosis
in advanced stages of colon carcinoma. Clin Cancer Res.
19:3925–3935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Molina-Pinelo S, Ferrer I, Blanco-Aparicio
C, Peregrino S, Pastor MD, Alvarez-Vega J, Suarez R, Verge M, Marin
JJ, Hernandez-Losa J, et al: Down-regulation of spinophilin in lung
tumours contributes to tumourigenesis. J Pathol. 225:73–82. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin Z, Dai L, Toole B, Robertson E and
Parsons C: Regulation of Nm23-H1 and cell invasiveness by Kaposi's
sarcoma-associated herpesvirus. J Virol. 85:3596–3606. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marino N, Marshall JC and Steeg PS:
Protein-protein interactions: A mechanism regulating the
anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch
Pharmacol. 384:351–362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang YQ, Wu L, Chen JX, Sun JZ, Li M, Li
DM, Lu HY, Su ZH, Lin XQ and Li JC: Relationship between nm23H1
genetic instability and clinical pathological characteristics in
Chinese digestive system cancer patients. World J Gastroenterol.
13:5549–5556; discussion 5555. 2008. View Article : Google Scholar
|
|
20
|
Piwkham D, Siriboonpiputtana T, Beuten J,
Pakakasama S, Gelfond JA, Paisooksantivatana K, Tomlinson GE and
Rerkamnuaychoke B: Mutation screening and association study of the
Folylpolyglutamate Synthetase (FPGS) gene with susceptibility to
childhood acute lymphoblastic leukemia. Asian Pac J Cancer Prev.
16:4727–4732. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hartsough MT, Clare SE, Mair M, Elkahloun
AG, Sgroi D, Osborne CK, Clark G and Steeg PS: Elevation of breast
carcinoma Nm23-H1 metastasis suppressor gene expression and reduced
motility by DNA methylation inhibition. Cancer Res. 61:2320–2327.
2001.PubMed/NCBI
|
|
22
|
Pal S, Vishwanath SN, Erdjument-Bromage H,
Tempst P and Sif S: Human SWI/SNF-associated PRMT5 methylates
histone H3 arginine 8 and negatively regulates expression of ST7
and nm23 tumor suppressor genes. Mol Cell Biol. 24:9630–9645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Marino N, Nakayama J, Collins JW and Steeg
PS: Insights into the biology and prevention of tumor metastasis
provided by the Nm23 metastasis suppressor gene. Cancer Metastasis
Rev. 31:593–603. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Esposito S, Russo MV, Airoldi I, Tupone
MG, Sorrentino C, Barbarito G, Di Meo S and Di Carlo E: SNAI2/Slug
gene is silenced in prostate cancer and regulates neuroendocrine
differentiation, metastasis-suppressor and pluripotency gene
expression. Oncotarget. 6:17121–17134. 2015. View Article : Google Scholar : PubMed/NCBI
|