Introduction
Prostate cancer (PCa), the second most common
malignancy in men, is the fifth leading cause of cancer-related
mortality among men globally (1).
The incidence rate of PCa in Iran is lower than that in the rest of
the world (2–4). Despite the high prevalence of PCa,
little is known about the mechanisms underlying the development and
progression of PCa. It has been proposed that genomic and
environmental factors contribute to the development and progression
of PCa (5–8). Twin studies have indicated that 42% of
the variation in PCa risk may be attributed to genetics (9). Single nucleotide polymorphisms (SNPs),
the most common type of genetic variation in the human genome, have
been demonstrated to be associated with the risk of developing PCa
(10–12).
MicroRNA (miR) are small, non-coding, endogenous,
single-stranded RNA molecules that are ~22 nucleotides in length
(13,14). They regulate gene expression by
directing sequence-specific degradation or inhibiting translation
of target mRNA (13,14). Mounting evidence has suggested that
mutation or SNPs in miR genes may affect target-binding activity,
expression, or processes of mature miR, thus affecting the
expression of their target genes (15,16).
Polymorphisms in mature and/or pre-miR sequences may affect miR
biogenesis and be associated with the development of various types
of cancer (17–21). Small insertions and deletion (indels)
polymorphisms are one of the most common genetic alterations in the
human genome that influence human traits and diseases (22,23).
There is limited information regarding the association between
pre-miR-3131 polymorphisms and cancer risk. Recently, Wang et
al (20) investigated the impact
of a 3-bp indel polymorphism (rs57408770) in pre-miR-3131 on
hepatocellular carcinoma (HCC) and observed that the insertion
(ins) allele significantly increased the risk of HCC in a Chinese
population. To the best of our knowledge, for the first time, the
present study aimed to determine the impact of a 3-bp indel
polymorphism (rs57408770) within pre-miR-3131 on PCa susceptibility
in a sample of an Iranian population.
Patients and methods
Patients
The present case-control study involved 177 patients
with PCa (mean age, 61.45±6.78 years) and 177 individuals with
benign prostatic hyperplasia (BPH) as controls (mean age,
62.43±7.68 years) admitted to hospital between February 2014 and
March 2015. All cases and controls were elected from the Department
of Urology, Shahid Labbafinejad Medical Center, Shahid Beheshti
University of Medical Sciences (Tehran, Iran). The study design and
enrolment procedure were described previously (17,18,24). The
project was approved by the local Ethics Committee of Zahedan
University of Medical Sciences (Zahedan, Iran) and written informed
consent was taken from all participants. Peripheral blood samples
were collected in tubes containing EDTA and genomic DNA was
extracted using the salting out method, as described previously
(25).
Genotyping
Mismatch polymerase chain reaction-restriction
fragment length polymorphism (PCR-RFLP) methods for genotyping were
designed. Mismatched C was introduced into the forward primers of
rs57408770 at −2 bp from the polymorphic site to create an
AluI (New England BioLabs, Inc., Ipswich, MA, USA)
restriction site. The forward and reverse primers were
5′-CTGTGCAGCTGACTCTGAGAAGACG-3′ and 5′-TATTGGCTCCTAGGAAGGCTGAGT-3′,
respectively.
PCR was performed using commercially available prime
Taq Premix (GeNet Bio, Nonsan, Korea), according to the
manufacturer's instructions. Each 0.20 ml PCR reaction tube
contained 1 µl genomic DNA (100 ng/ml), 1 µl each primer (10 µM), 7
µl 2X master mix and the appropriate amount of double-distilled
H2O. Amplification was performed with an initial
denaturation at 95°C for 6 min, followed by 30 cycles of 30 sec at
95°C, 30 sec at 65°C and 72°C for 30 sec, with a final extension
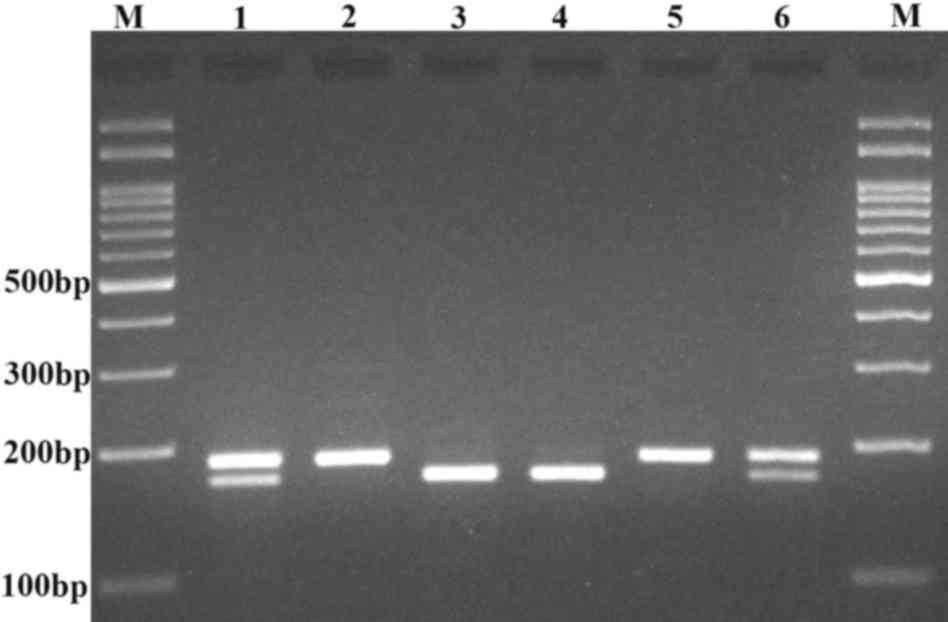
step of 72°C for 5 min. For genotyping, 10 µl PCR product was
digested by AluI and the digested products were separated by
2.5% agarose gel electrophoresis. The deletion (del) allele
produced a 188-bp fragment, while the ins allele produced 171- and
20-bp fragments (Fig. 1).
Statistical analysis
Statistical analysis was conducted using SPSS v. 22
software (IBM Corp., Armonk, NY, USA). Data were analyzed by
independent samples t-tests and χ2 tests. Unconditional
logistic regression analysis was used to examine the association
between the rs57408770 variant and PCa risk. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
The present study consisted of 177 patients with PCa
(mean age, 61.45±6.78 years) and 177 individuals with BPH (mean
age, 62.43±7.68 years). No significant difference was observed
between the age of the two groups (P=0.212; data not shown).
3-bp indel (rs57408770) polymorphism
and risk of PCa
The genotypes and allele frequencies of the 3-bp
indel (rs57408770) polymorphism within pre-miR-3131 in patients
with PCa and control subjects are demonstrated in Table I. The findings revealed that the
indel variant was significantly associated with increased risk of
PCa in codominant [odds ratio (OR)=2.23, 95% confidence interval
(CI)=1.13–4.37; P=0.021, ins/ins vs. reference del/del) and
recessive (OR=2.33, 95% CI=1.25–4.36; P=0.009, ins/ins vs.
reference del/del + del/ins). The ins allele was not significantly
associated with the risk of PCa (OR=1.33, 95% CI=0.98–1.81;
P=0.083).
 | Table I.Genotype and allele frequencies of a
3-bp indel (rs57408770) polymorphism of pre-microRNA-3131 in
patients with PCa and controls. |
Table I.
Genotype and allele frequencies of a
3-bp indel (rs57408770) polymorphism of pre-microRNA-3131 in
patients with PCa and controls.
|
| Group |
|
|
|---|
|
|
|
|
|
|---|
| Polymorphism | PCa, n (%) | Controls, n (%) | Odds ratio (95%
confidence interval) | P-value |
|---|
| Codominant |
|
|
|
|
|
del/dela | 62
(36.5) | 67
(39.4) | 1.00 | – |
|
del/ins | 73
(42.9) | 86
(50.6) | 0.92 (0.58–1.46) | 0.723 |
|
ins/ins | 35
(20.6) | 17
(10.0) | 2.23 (1.13–4.37) | 0.021 |
| Dominant |
|
|
|
|
|
del/dela | 62
(36.5) | 67
(39.4) | 1.00 | – |
| del/ins +
ins/ins | 108 (63.5) | 103 (60.6) | 1.3 (0.73–1.76) | 0.654 |
| Recessive |
|
|
|
|
| del/del +
del/insa | 135 (79.4) | 153 (90.0) | 1.00 | – |
|
ins/ins | 35
(20.6) | 17
(10.0) | 2.33
(1.25–4.36) | 0.009 |
| Allele |
|
|
|
|
|
dela | 197 (58.0) | 220 (64.7) | 1.00 | – |
|
ins | 143 (42.0) | 120 (35.3) | 1.33
(0.98–1.81) | 0.083 |
Association between 3-bp indel
polymorphism of pre-miR-3131 and clinicopathological
characteristics
The association between the 3-bp indel polymorphism
of pre-miR-3131 and clinicopathological characteristics, including
age, stage, prostate specific antigen levels, Gleason score
(18), perineural invasion and
surgical margin, are demonstrated in Table II. The findings revealed that the
3-bp indel polymorphism of pre-miR-3131 was only significantly
associated with perineural invasion (P=0.015). No significant
association was observed between the variant and other
clinicopathological characteristics in patients with PCa. The
Hardy-Weinberg equilibrium (HWE) was calculated and the results
indicated that the genotype distribution in cases and controls was
consistent with HWE (χ2=2.41, P=0.121 and
χ2=1.97, P=0.161, respectively; data not shown).
 | Table II.Association of 3-bp indel
(rs57408770) polymorphism of pre-microRNA-3131 with
clinicopathological characteristics of patients with prostate
cancer. |
Table II.
Association of 3-bp indel
(rs57408770) polymorphism of pre-microRNA-3131 with
clinicopathological characteristics of patients with prostate
cancer.
|
| rs3787016
C>T |
|
|---|
|
|
|
|
|---|
| Factors | del/del, n | del/ins, n | ins/ins, n | P-value |
|---|
| Age at diagnosis,
years |
|
|
| 0.876 |
|
≤65 | 43 | 52 | 26 |
|
|
>65 | 19 | 21 | 9 |
|
| Stage |
|
|
| 0.578 |
|
pT1 | 2 | 3 | 3 |
|
|
pT2a | 9 | 9 | 9 |
|
|
pT2b | 6 | 3 | 2 |
|
|
pT2c | 26 | 37 | 14 |
|
|
pT3a | 5 | 7 | 1 |
|
|
pT3b | 14 | 14 | 6 |
|
| Prostate specific
antigen level at diagnosis, ng/ml |
|
|
| 0.458 |
| ≤4 | 0 | 1 | 0 |
|
|
4–10 | 31 | 32 | 21 |
|
|
>10 | 30 | 40 | 14 |
|
| Gleason score |
|
|
| 0.538 |
| ≤7 | 44 | 58 | 28 |
|
|
>7 | 17 | 15 | 7 |
|
| Perineural
invasion |
|
|
| 0.015 |
|
Positive | 47 | 40 | 19 |
|
|
Negative | 14 | 33 | 16 |
|
| Surgical
margin |
|
|
| 0.180 |
|
Positive | 29 | 28 | 10 |
|
|
Negative | 32 | 45 | 25 |
|
Discussion
Recent expression profiling studies in PCa suggest
that miR may serve as potential biomarkers for PCa risk and disease
progression (26–30). Growing evidence has indicated that
mutation or polymorphisms in miR genes could affect target-binding
activity, expression or processes of mature miR, consequently
affecting the expression of their target genes (15,16).
Polymorphisms in miR have been demonstrated to be associated with
the development of PCa (17,18,31,32). In
present study, it was hypothesized that the 3-bp indel polymorphism
of pre-miR-3131 may be associated with the development of PCa. The
present results indicated that the ins/ins genotype of the
pre-miR-313 variant significantly increased the risk of PCa. To the
best of our knowledge, there has only been one report regarding the
impact of a 3-bp indel polymorphism of pre-miR-3131 on cancer risk
(20). Wang et al (20) demonstrated that the insertion allele
of a 3-bp indel polymorphism of pre-miR-3131 was significantly
associated with an increased risk for HCC. Furthermore, their
findings revealed that the 3-bp indel polymorphism could affect the
expression level of miR-3131 by influencing the binding of splicing
factor SRp20 with pre-miR-3131 (20). Hsa-miR-3131 is located on chromosome
2 in intron 2 of the Indian hedgehog gene and the 3-bp indel
variant (rs57408770) is located in the 3′ end of miR-3131 (20). A study by Shen et al (33) demonstrated that the miR-3131
expression level was upregulated by 92-fold in HepG2 cells treated
with Ganoderma lucidum polysaccharide, proposing that
miR-3131 may be involved in the proliferation and differentiation
of HCC cells.
Polymorphisms in miR genes, including pri-miR
(17,34,35),
pre-miR (36) and mature miR, may
affect the processing of miR as well as the regulatory function on
their target genes, and consequently may be implicated in the
development and prognosis of various types of cancer (37–39). In
conclusion, to the best of our knowledge, the present study
provided evidence for the first time that the 3-bp indel
polymorphism of pre-miR-3131 significantly increased the risk of
developing PCa in a sample of an Iranian population. Therefore, the
pre-miR-3131 3-bp indel variant may be a potential biomarker for
prostate cancer. Further studies with different ethnicities and
larger sample sizes are required to certify the present
findings.
Acknowledgements
The present study was supported by a grant from
Zahedan University of Medical Sciences (Zahedan, Iran; grant no.
7832).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Farahmand M, Khademolhosseini F and
Mehrabani D: Trend of prostate cancer in Fars Province, Southern
Iran, 2001-2007. J Res Med Sci. 15:295–297. 2010.PubMed/NCBI
|
|
3
|
Talaiezadeh A, Tabesh H, Sattari A and
Ebrahimi S: Cancer incidence in southwest of iran: First report
from khuzestan population-based cancer registry, 2002–2009. Asian
Pac J Cancer Prev. 14:7517–7522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baade PD, Youlden DR, Cramb SM, Dunn J and
Gardiner RA: Epidemiology of prostate cancer in the Asia-Pacific
region. Prostate Int. 1:47–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ntais C, Polycarpou A and Tsatsoulis A:
Molecular epidemiology of prostate cancer: Androgens and
polymorphisms in androgen-related genes. Eur J Endocrinol.
149:469–477. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chokkalingam AP, Stanczyk FZ, Reichardt JK
and Hsing AW: Molecular epidemiology of prostate cancer:
Hormone-related genetic loci. Front Biosci. 12:3436–3460. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou X, Wei L, Jiao G, Gao W, Ying M, Wang
N, Wang Y and Liu C: The association between the APE1 Asp148Glu
polymorphism and prostate cancer susceptibility: A meta-analysis
based on case-control studies. Mol Genet Genomics. 290:281–288.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bratt O: Hereditary prostate cancer:
Clinical aspects. J Urol. 168:906–913. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of
cancer-analyses of cohorts of twins from Sweden, Denmark, and
Finland. N Engl J Med. 343:78–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang Q, Whitington T, Gao P, Lindberg JF,
Yang Y, Sun J, Väisänen MR, Szulkin R, Annala M, Yan J, et al: A
prostate cancer susceptibility allele at 6q22 increases RFX6
expression by modulating HOXB13 chromatin binding. Nat Genet.
46:126–135. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hazelett DJ, Rhie SK, Gaddis M, Yan C,
Lakeland DL and Coetzee SG: Ellipse/GAME-ON consortium; Practical
consortium. Henderson BE, Noushmehr H, et al: Comprehensive
functional annotation of 77 prostate cancer risk loci. PLoS Genet.
10:e10041022014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eeles RA, Olama AA, Benlloch S, Saunders
EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C,
Dennis J, Jugurnauth-Little S, et al: Identification of 23 new
prostate cancer susceptibility loci using the iCOGS custom
genotyping array. Nat Genet. 45:385–91, 391e1-2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
16
|
Sibin MK, Harshitha SM, Narasingarao KV,
Dhananjaya IB, Dhaval PS and Chetan GK: Effect of rs11614913
polymorphism on mature miR196a2 Expression and its target gene
HOXC8 expression in human glioma. J Mol Neurosci. 61:144–151. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hashemi M, Danesh H, Bizhani F, Narouie B,
Sotoudeh M, Nouralizadeh A, Sharifiaghdas F, Bahari G and Taheri M:
Pri-miR-34b/c rs4938723 polymorphism increased the risk of prostate
cancer. Cancer Biomark. 18:155–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hashemi M, Moradi N, Ziaee SA, Narouie B,
Soltani MH, Rezaei M, Shahkar G and Taheri M: Association between
single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and
miR-149 and prostate cancer risk in a sample of Iranian population.
J Adv Res. 7:491–498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu F, Dear K, Huang L, Liu L, Shi Y, Nie
S, Liu Y, Lu Y and Xiang H: Association between microRNA-27a
rs895819 polymorphism and risk of colorectal cancer: A
meta-analysis. Cancer Genet. 209:388–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Li L, Yin Z, Zhang Q, Zhao H, Tao
R, Wang S, Hu S, He Y, Wang D, et al: An Indel Polymorphism within
pre-miR3131 confers risk for hepatocellular carcinoma.
Carcinogenesis. 38:168–176. 2017.PubMed/NCBI
|
|
21
|
Omrani M, Hashemi M, Eskandari-Nasab E,
Hasani SS, Mashhadi MA, Arbabi F and Taheri M: hsa-mir-499
rs3746444 gene polymorphism is associated with susceptibility to
breast cancer in an Iranian population. Biomark Med. 8:259–267.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mullaney JM, Mills RE, Pittard WS and
Devine SE: Small insertions and deletions (INDELs) in human
genomes. Hum Mol Genet. 19:R131–R136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eskandari-Nasab E, Hashemi M, Ebrahimi M
and Amininia S: The functional 4-bp insertion/deletion ATTG
polymorphism in the promoter region of NF-KB1 reduces the risk of
BC. Cancer Biomark. 16:109–115. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hashemi M, Shahkar G, Simforoosh N, Basiri
A, Ziaee SA, Narouie B and Taheri M: Association of polymorphisms
in PRKCI gene and risk of prostate cancer in a sample of Iranian
Population. Cell Mol Biol (Noisy-le-grand). 61:16–21.
2015.PubMed/NCBI
|
|
25
|
Hashemi M, Hanafi Bojd H, Eskandari Nasab
E, Bahari A, Hashemzehi NA, Shafieipour S, Narouie B, Taheri M and
Ghavami S: Association of Adiponectin rs1501299 and rs266729 gene
polymorphisms with nonalcoholic fatty liver disease. Hepat Mon.
13:e95272013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luu HN, Lin HY, Sørensen KD, Ogunwobi OO,
Kumar N, Chornokur G, Phelan C, Jones D, Kidd L, Batra J, et al:
miRNAs associated with prostate cancer risk and progression. BMC
Urol. 17:182017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei W, Leng J, Shao H and Wang W: MiR-1, a
potential predictive biomarker for recurrence in prostate cancer
after radical prostatectomy. Am J Med Sci. 353:315–319. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Valentino A, Reclusa P, Sirera R,
Giallombardo M, Camps C, Pauwels P, Crispi S and Rolfo C: Exosomal
microRNAs in liquid biopsies: Future biomarkers for prostate
cancer. Clin Transl Oncol. 19:651–657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shukla KK, Misra S, Pareek P, Mishra V,
Singhal B and Sharma P: Recent scenario of microRNA as diagnostic
and prognostic biomarkers of prostate cancer. Urol Oncol.
35:92–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qiu X and Dou Y: miR-1307 promotes the
proliferation of prostate cancer by targeting FOXO3A. Biomed
Pharmacother. 88:430–435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chu H, Zhong D, Tang J, Li J, Xue Y, Tong
N, Qin C, Yin C, Zhang Z and Wang M: A functional variant in
miR-143 promoter contributes to prostate cancer risk. Arch Toxicol.
90:403–414. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Porkka KP, Ogg EL, Saramaki OR, Saramäki
OR, Vessella RL, Pukkila H, Lähdesmäki H, van Weerden WM, Wolf M,
Kallioniemi OP, et al: The miR-15a-miR-16-1 locus is homozygously
deleted in a subset of prostate cancers. Genes Chromosomes Cancer.
50:499–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen J, Park HS, Xia YM, Kim GS and Cui
SW: The polysaccharides from fermented Ganoderma lucidum mycelia
induced miRNAs regulation in suppressed HepG2 cells. Carbohydr
Polym. 103:319–324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hashemi M, Bahari G, Naderi M,
Sadeghi-Bojd S and Taheri M: Pri-miR-34b/c rs4938723 polymorphism
is associated with the risk of childhood acute lymphoblastic
leukemia. Cancer Genet. 209:493–496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Okubo M, Tahara T, Shibata T, Yamashita H,
Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T and
Hirata I: Association between common genetic variants in
pre-microRNAs and gastric cancer risk in Japanese population.
Helicobacter. 15:524–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiu F, Yang L, Zhang L, Yang X, Yang R,
Fang W, Wu D, Chen J, Xie C, Huang D, et al: Polymorphism in mature
microRNA-608 sequence is associated with an increased risk of
nasopharyngeal carcinoma. Gene. 565:180–186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hashemi M, Sanaei S, Rezaei M, Bahari G,
Hashemi SM, Mashhadi MA, Taheri M and Ghavami S: miR-608 rs4919510
C>G polymorphism decreased the risk of breast cancer in an
Iranian subpopulation. Exp Oncol. 38:57–59. 2016.PubMed/NCBI
|