Prostate cancer (PCa) is one of the most frequently
diagnosed cancers in men and the sixth leading cause of
cancer-related mortality among men worldwide (1). The prevalence of PCa differs
significantly among populations, indicating that the host's genetic
background may play an important role in susceptibility to PCa
(2). Single-nucleotide polymorphisms
(SNPs) are the most common type of genetic variations in human
genome, and they have been found to be associated with the risk of
PCa (3–6). Genome tiling arrays have indicated that
1% of the human genome is composed of protein-coding sequences and
~4–9% of the sequences of the human genome are transcribed to
non-coding RNAs (ncRNAs) (7).
NcRNAs are documented to be the main regulators of a
number of biological processes, such as transcription, splicing,
translation, epigenetic gene expression, cell cycle (8–12), stem
cell pluripotency and reprogramming (12,13),
embryogenesis (14), and regulation
of the immune response (15). They
are divided into small ncRNAs (<200 nt) and long ncRNAs
(lncRNAs; >200 nt) (16,17). LncRNAs are classified according to
the correlation between their location and the location of the
corresponding protein-coding gene, such as sense, antisense,
intergenic, intronic and bidirectional lncRNAs (18,19).
Aberrant expression of lncRNAs may contribute to the development
and progression of various cancers (20–25).
In total, 358 subjects participated in this
hospital-based case-control study, including 178 unrelated men with
histopathologically confirmed prostate cancer and 180 age-matched
unrelated men with benign prostatic hyperplasia (BPH), with no
history of any type of cancer, as the control group (36–39). All
cases and controls were selected from a university-affiliated
referral center (Shahid Labbafinejad Medical Center, Shahid
Beheshti University of Medical Sciences, Tehran, Iran). The local
Ethics Committee of Zahedan University of Medical Sciences approved
the project (IR.ZAUMS.REc.1395.102), and written informed consent
was obtained from all the participants. Genomic DNA was extracted
by the salting out method and stored at −20°C until use. Peripheral
blood samples were collected in tubes containing EDTA and genomic
DNA was extracted by the salting out method.
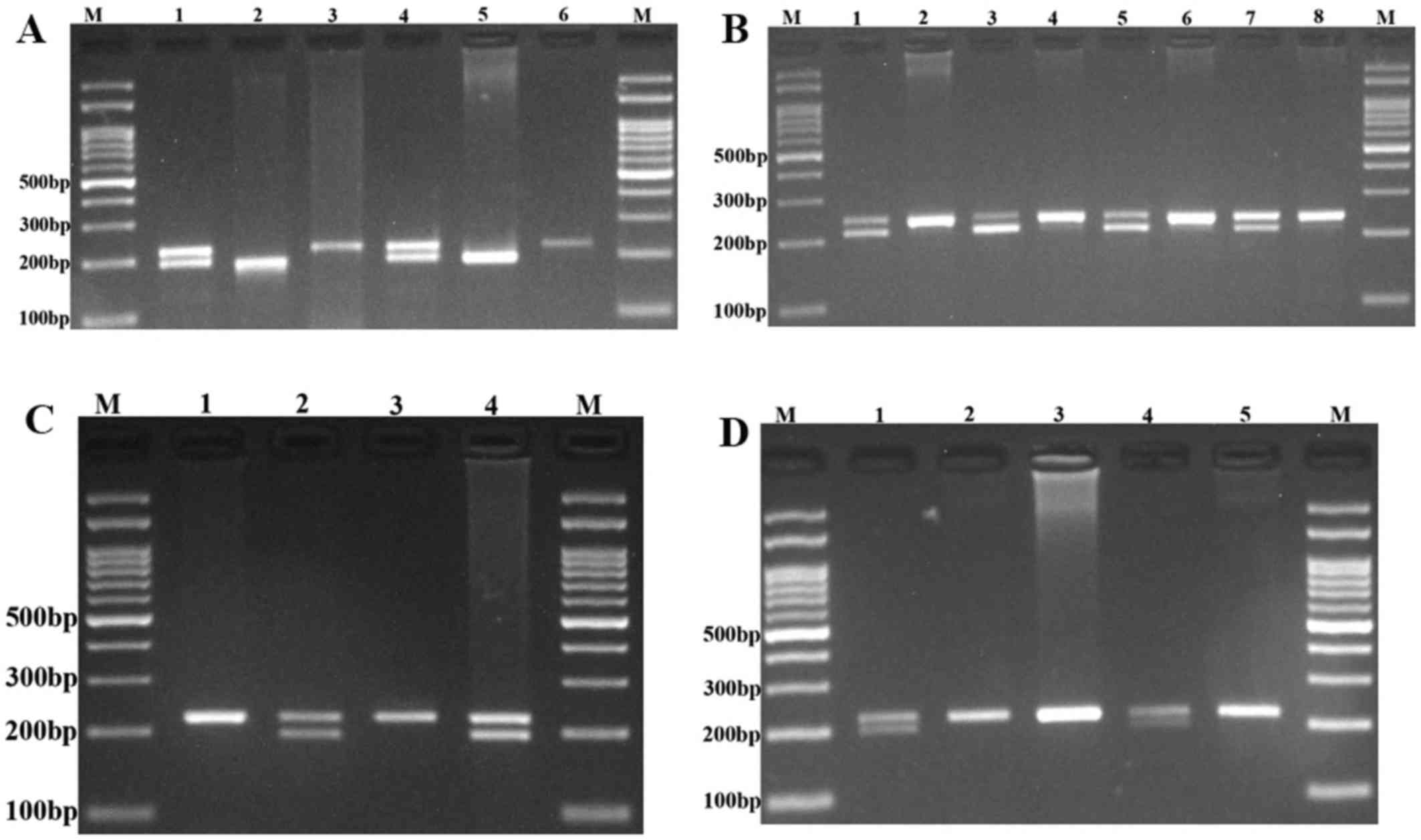
The polymerase chain reaction (PCR)-restriction
fragment length polymorphism assay was used for genotyping of the
PRNCR1 rs13252298, rs1456315, rs7841060, and rs7007694
polymorphisms. The primer sequences, restriction enzymes and the
length of the PCR products are listed in Table I. PCR was performed with the
commercially available prime Taq Premix (Genet Bio, Daejeon, Korea)
according to the manufacturer's recommended protocol. Into each
0.20-ml PCR reaction tube, 1 µl of genomic DNA (100 ng/ml), 1 µl of
each primer (10 µM), 7 µl of 2X master mix and 6 µl of
ddH2O were added. Amplification was performed with an
initial denaturation at 95°C for 30 sec, followed by 30 cycles of
30 sec at 95°C, 30 sec at 62°C for rs13252298, 60°C for rs1456315,
56°C for rs7841060, and 64°C for rs7007694, 72°C for 30 sec, with a
final extension step at 72°C for 10 min. Subsequently, 10 µl of the
PCR products were digested with the appropriate restriction enzymes
(Table I). The digested products
were separated by agarose gel electrophoresis, visualized by a UV
transilluminator and photographed (Fig.
1).
All data were analyzed using the statistical package
SPSS 22.0 software (IBM Corp., Armonk, NY, USA). The continuous and
categorical data were analyzed by the independent samples t-test
and χ2 test, respectively. The association among
polymorphisms and PCa was calculated by computing the odds ratio
(OR) and 95% confidence interval (95% CI) from unconditional
logistic regression analyses. Haplotype analysis was performed
using SNPStats software (40). The
level of statistical significance was set at P<0.05.
The present study included 178 PCa patients with a
mean age ± standard deviation of 61.53±6.91 years, and 180 patients
with BPH with a mean age of 62.40±7.64 years. No significant
difference was found between the groups in terms of age (P=0.258).
The genotypes and allele frequencies of PRNCR1 polymorphisms
in cases and controls are presented in Table II. As regards the rs13252298 A>G
variant, our findings demonstrated that this variant significantly
increased the risk of PCa in the recessive (OR=3.49, 95% CI:
1.79–6.81, P=0.0001, GG vs. AA+AG) inheritance model. As regards
the rs1456315 A>G polymorphism, the AG genotype as well as the G
allele significantly increased the risk of PCa (OR=5.16, 95% CI:
3.16–8.41, P<0.0001 and OR=2.20, 95% CI: 1.60–3.03, P<0.0001,
respectively). The TG genotype as well as the G allele of the
rs7841060 variant significantly increased the risk of PCa (OR=5.14,
95% CI: 3.15–8.37, P<0.0001 and OR=2.37, 95% CI: 1.71–3.26,
P<0.0001, respectively). Our findings demonstrated that the
rs7007694 T>C polymorphism was not significantly associated with
the risk of PCa. A haplotype analysis was performed, and the
findings indicated that GTGA and GTGG significantly increased the
risk of PCa compared with
rs1456315A/rs7007694T/rs7841060T/rs13252298G (ATTG) (Table III).
The associations between clinicopathological
characteristics, including age, stage, prostate-specific antigen
(PSA) levels, Gleason score, perineural invasion and surgical
margin, and PRNCR1 polymorphisms are shown in Table IV. The findings did not support an
association between PRNCR1 polymorphisms and the
clinicopathological characteristics of PCa patients.
The Hardy-Weinberg equilibrium (HWE) was calculated
and the findings revealed that the genotype distribution in
controls was not in HWE.
LncRNAs are involved in tumorigenesis through their
function as proto-oncogenes (41) or
tumor-suppressor genes (42).
Androgen receptor, a member of the nuclear receptor family, is a
ligand-activated transcription factor (43). It has been suggested that lncRNA
PRNCR1 promotes prostate carcinogenesis via activating AR
(26). SNPs, a class of genetic
variations, are commonly used in the prediction of cancer risk
(38,44,45),
prognosis (46) and clinical outcome
(47). Cumulative evidence indicates
that non-coding genes may be involved in gene expression complexity
in humans (48,49). Abnormal expression of lncRNAs has
been found to be associated with the development of numerous
cancers (50–52). Genome-wide association studies
suggested significant and consistent associations of multiple
genetic polymorphisms on chromosome 8q24 with PCa susceptibility
(53–58). To date, several studies investigated
the effect of PRNCR1 polymorphisms on the risk of PCa
(26,28–30).
However, to the best of our knowledge, no study investigating the
impact of PRNCR1 variants on cancer risk in an Iranian
population has been conducted to date. The present study aimed to
evaluate the possible association between rs13252298, rs1456315,
rs7841060 and rs7007694 polymorphisms of PRNCR1 and the risk
of PCa in a sample of Iranian population.
LncRNAs, a new class of functional ncRNAs, are
composed of >200 nucleotides and lack protein-coding ability
(19). LncRNAs potentially interact
with DNA, RNA, as well as protein molecules, to perform diverse
regulatory functions, including chromatin remodelling (59), RNA splicing and editing (60), translational inhibition (61), mRNA destruction (62) and epigenetic regulation of gene
expression (63–65). The most important function of lncRNAs
is involvement in the transcriptional or post-transcriptional
regulation of gene expression (66).
Abnormal expression of lncRNAs may facilitate tumor cell
proliferation, invasion and metastasis (67–70).
There were certain limitations to the present study,
including the number of SNPs that were investigated for the
PRNCR1 gene, as well as lack of the information regarding
survival outcomes and the patients' response to treatment. The
reason for the deviation from HWE in our population was not clear;
it may be attributed to genetic drift.
The authors would like to thank all the subjects who
willingly participated in the study. This project was funded by a
dissertation grant (MSc thesis of HS no. 7832) from Zahedan
University of Medical Sciences, Zahedan, Iran.
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hsing AW and Devesa SS: Trends and
patterns of prostate cancer: What do they suggest? Epidemiol Rev.
23:3–13. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang Q, Whitington T, Gao P, Lindberg JF,
Yang Y, Sun J, Väisänen MR, Szulkin R, Annala M, Yan J, et al: A
prostate cancer susceptibility allele at 6q22 increases RFX6
expression by modulating HOXB13 chromatin binding. Nat Genet.
46:126–135. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hazelett DJ, Rhie SK, Gaddis M, Yan C,
Lakeland DL and Coetzee SG: Ellipse/GAME-ON consortium; Practical
consortium, Henderson BE, Noushmehr H, et al: Comprehensive
functional annotation of 77 prostate cancer risk loci. PLoS Genet.
10:e10041022014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spisák S, Lawrenson K, Fu Y, Csabai I,
Cottman RT, Seo JH, Haiman C, Han Y, Lenci R, Li Q, et al: CAUSEL:
An epigenome- and genome-editing pipeline for establishing function
of noncoding GWAS variants. Nat Med. 21:1357–1363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen H, Yu H, Wang J, Zhang Z, Gao Z, Chen
Z, Lu Y, Liu W, Jiang D, Zheng SL, et al: Systematic enrichment
analysis of potentially functional regions for 103 prostate cancer
risk-associated loci. Prostate. 75:1264–1276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
ENCODE Project Consortium, . Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Groß S, Immel UD, Klintschar M and Bartel
F: Germline genetics of the p53 pathway affect longevity in a
gender specific manner. Curr Aging Sci. 7:91–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brannan CI, Dees EC, Ingram RS and
Tilghman SM: The product of the H19 gene may function as an RNA.
Mol Cell Biol. 10:28–36. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen B, Yu M, Chang Q, Lu Y, Thakur C, Ma
D, Yi Z and Chen F: Mdig de-represses H19 large intergenic
non-coding RNA (lincRNA) by down-regulating H3K9me3 and
heterochromatin. Oncotarget. 4:1427–1437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grote P and Herrmann BG: The long
non-coding RNA Fendrr links epigenetic control mechanisms to gene
regulatory networks in mammalian embryogenesis. RNA Biol.
10:1579–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z and Rana TM: Decoding the noncoding:
Prospective of lncRNA-mediated innate immune regulation. RNA Biol.
11:979–985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pei Z, Du X, Song Y, Fan L, Li F, Gao Y,
Wu R, Chen Y, Li W, Zhou H, et al: Down-regulation of lncRNA CASC2
promotes cell proliferation and metastasis of bladder cancer by
activation of the Wnt/β-catenin signaling pathway. Oncotarget.
8:18145–18153. 2017.PubMed/NCBI
|
|
22
|
Li T, Xu C, Cai B, Zhang M, Gao F and Gan
J: Expression and clinicopathological significance of the lncRNA
HOXA11-AS in colorectal cancer. Oncol Lett. 12:4155–4160.
2016.PubMed/NCBI
|
|
23
|
He A, Chen Z, Mei H and Liu Y: Decreased
expression of LncRNA MIR31HG in human bladder cancer. Cancer
Biomark. 17:231–236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pibouin L, Villaudy J, Ferbus D, Muleris
M, Prospéri MT, Remvikos Y and Goubin G: Cloning of the mRNA of
overexpression in colon carcinoma-1: A sequence overexpressed in a
subset of colon carcinomas. Cancer Genet Cytogenet. 133:55–60.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung S, Nakagawa H, Uemura M, Piao L,
Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono
T, et al: Association of a novel long non-coding RNA in 8q24 with
prostate cancer susceptibility. Cancer Sci. 102:245–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hui J, Xu Y, Yang K, Liu M, Wei D, Wei D,
Zhang Y, Shi XH, Yang F, Wang N, et al: Study of genetic variants
of 8q21 and 8q24 associated with prostate cancer in Jing-Jin
residents in northern China. Clin Lab. 60:645–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng SL, Hsing AW, Sun J, Chu LW, Yu K,
Li G, Gao Z, Kim ST, Isaacs WB, Shen MC, et al: Association of 17
prostate cancer susceptibility loci with prostate cancer risk in
Chinese men. Prostate. 70:425–432. 2010.PubMed/NCBI
|
|
30
|
Salinas CA, Kwon E, Carlson CS,
Koopmeiners JS, Feng Z, Karyadi DM, Ostrander EA and Stanford JL:
Multiple independent genetic variants in the 8q24 region are
associated with prostate cancer risk. Cancer Epidemiol Biomarkers
Prev. 17:1203–1213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Jia F, Bai P, Liang Y, Sun R, Yuan
F, Zhang L and Gao L: Association between polymorphisms in long
non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumour
Biol. 37:299–303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He BS, Sun HL, Xu T, Pan YQ, Lin K, Gao
TY, Zhang ZY and Wang SK: Association of genetic polymorphisms in
the LncRNAs with gastric cancer risk in a Chinese population. J
Cancer. 8:531–536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, Sun R, Liang Y, Pan X, Li Z, Bai P,
Zeng X, Zhang D, Zhang L and Gao L: Association between
polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of
colorectal cancer. J Exp Clin Cancer Res. 32:1042013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chu H, Chen Y, Yuan Q, Hua Q, Zhang X,
Wang M, Tong N, Zhang W, Chen J and Zhang Z: The HOTAIR, PRNCR1 and
POLR2E polymorphisms are associated with cancer risk: A
meta-analysis. Oncotarget. 8:43271–43283. 2017.PubMed/NCBI
|
|
35
|
Lv Z, Xu Q and Yuan Y: A systematic review
and meta-analysis of the association between long non-coding RNA
polymorphisms and cancer risk. Mutat Res. 771:1–14. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hashemi M, Shahkar G, Simforoosh N, Basiri
A, Ziaee SA, Narouie B and Taheri M: Association of polymorphisms
in PRKCI gene and risk of prostate cancer in a sample of Iranian
Population. Cell Mol Biol (Noisy-le-grand). 61:16–21.
2015.PubMed/NCBI
|
|
37
|
Hashemi M, Moradi N, Ziaee SA, Narouie B,
Soltani MH, Rezaei M, Shahkar G and Taheri M: Association between
single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and
miR-149 and prostate cancer risk in a sample of Iranian population.
J Adv Res. 7:491–498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hashemi M, Danesh H, Bizhani F, Narouie B,
Sotoudeh M, Nouralizadeh A, Sharifiaghdas F, Bahari G and Taheri M:
Pri-miR-34b/c rs4938723 polymorphism increased the risk of prostate
cancer. Cancer Biomark. 18:155–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hashemi M, Bahari G, Sattarifard H and
Narouie B: Evaluation of a 3-base pair indel polymorphism within
pre-microRNA-3131 in patients with prostate cancer using mismatch
polymerase chain reaction-restriction fragment length polymorphism.
Mol Clin Oncol. 7:696–700. 2017.PubMed/NCBI
|
|
40
|
Solé X, Guinó E, Valls J, Iniesta R and
Moreno V: SNPStats: A web tool for the analysis of association
studies. Bioinformatics. 22:1928–1929. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li L, Feng T, Lian Y, Zhang G, Garen A and
Song X: Role of human noncoding RNAs in the control of
tumorigenesis. Proc Natl Acad Sci USA. 106:pp. 12956–12961. 2009,
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang X, Rice K, Wang Y, Chen W, Zhong Y,
Nakayama Y, Zhou Y and Klibanski A: Maternally expressed gene 3
(MEG3) noncoding ribonucleic acid: Isoform structure, expression,
and functions. Endocrinology. 151:939–947. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heemers HV and Tindall DJ: Androgen
receptor (AR) coregulators: A diversity of functions converging on
and regulating the AR transcriptional complex. Endocr Rev.
28:778–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bahari G, Hashemi M, Naderi M and Taheri
M: IKZF1 gene polymorphisms increased the risk of childhood acute
lymphoblastic leukemia in an Iranian population. Tumour Biol.
37:9579–9586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hashemi M, Amininia S, Ebrahimi M,
Simforoosh N, Basiri A, Ziaee SAM, Narouie B, Sotoudeh M,
Mollakouchekian MJ, Rezghi Maleki E, et al: Association between
polymorphisms in TP53 and MDM2 genes and susceptibility to prostate
cancer. Oncol Lett. 13:2483–2489. 2017.PubMed/NCBI
|
|
46
|
Bao BY, Lin VC, Yu CC, Yin HL, Chang TY,
Lu TL, Lee HZ, Pao JB, Huang CY, Huang SP, et al: Genetic variants
in ultraconserved regions associate with prostate cancer recurrence
and survival. Sci Rep. 6:221242016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Murali A, Varghese BT, Kumar RR and Kannan
S: Combination of genetic variants in cyclin D1 and retinoblastoma
genes predict clinical outcome in oral cancer patients. Tumour
Biol. 37:3609–3617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kapranov P, Willingham AT and Gingeras TR:
Genome-wide transcription and the implications for genomic
organization. Nat Rev Genet. 8:413–423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu SW, Hao YP, Qiu JH, Zhang DB, Yu CG and
Li WH: High expression of long non-coding RNA CCAT2 indicates poor
prognosis of gastric cancer and promotes cell proliferation and
invasion. Minerva Med. 108:317–323. 2017.PubMed/NCBI
|
|
51
|
Guo J, Ma J, Zhao G, Li G, Fu Y, Luo Y and
Gui R: Long noncoding RNA LINC0086 functions as a tumor suppressor
in nasopharyngeal carcinoma by targeting miR-214. Oncol Res.
25:1189–1197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu JN and Shangguan YM: Long non-coding
RNA CARLo-5 upregulation associates with poor prognosis in patients
suffering gastric cancer. Eur Rev Med Pharmacol Sci. 21:530–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yeager M, Chatterjee N, Ciampa J, Jacobs
KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N,
Berndt S, et al: Identification of a new prostate cancer
susceptibility locus on chromosome 8q24. Nat Genet. 41:1055–1057.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
54
|
Amundadottir LT, Sulem P, Gudmundsson J,
Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir
KR, Cazier JB, Sainz J, et al: A common variant associated with
prostate cancer in European and African populations. Nat Genet.
38:652–658. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gudmundsson J, Sulem P, Manolescu A,
Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson
JT, Agnarsson BA, Baker A, et al: Genome-wide association study
identifies a second prostate cancer susceptibility variant at 8q24.
Nat Genet. 39:631–637. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haiman CA, Patterson N, Freedman ML, Myers
SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C,
McDonald GJ, et al: Multiple regions within 8q24 independently
affect risk for prostate cancer. Nat Genet. 39:638–644. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Eeles RA, Kote-Jarai Z, Giles GG, Olama
AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards
SM, Morrison J, et al: Multiple newly identified loci associated
with prostate cancer susceptibility. Nat Genet. 40:316–321. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Al Olama AA, Kote-Jarai Z, Giles GG, Guy
M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar
S, Saunders E, et al: Multiple loci on 8q24 associated with
prostate cancer susceptibility. Nat Genet. 41:1058–1060. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Keller C and Bühler M:
Chromatin-associated ncRNA activities. Chromosome Res. 21:627–641.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Luco RF and Misteli T: More than a
splicing code: Integrating the role of RNA, chromatin and
non-coding RNA in alternative splicing regulation. Curr Opin Genet
Dev. 21:366–372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pircher A, Gebetsberger J and Polacek N:
Ribosome-associated ncRNAs: An emerging class of translation
regulators. RNA Biol. 11:1335–1339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:pp. 11667–11672. 2009,
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mercer TR, Dinger ME, Sunkin SM, Mehler MF
and Mattick JS: Specific expression of long noncoding RNAs in the
mouse brain. Proc Natl Acad Sci USA. 105:pp. 716–721. 2008,
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Heemers H, Maes B, Foufelle F, Heyns W,
Verhoeven G and Swinnen JV: Androgens stimulate lipogenic gene
expression in prostate cancer cells by activation of the sterol
regulatory element-binding protein cleavage activating
protein/sterol regulatory element-binding protein pathway. Mol
Endocrinol. 15:1817–1828. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li
M, Xu L and Yin R: Upregulation of long non-coding RNA PRNCR1 in
colorectal cancer promotes cell proliferation and cell cycle
progression. Oncol Rep. 35:318–324. 2016. View Article : Google Scholar : PubMed/NCBI
|