Introduction
Melanoma represents a minority of skin cancers but
it is associated with an increased risk of death compared with
other skin malignancies (1-4),
being responsible for ~90% of mortalities reported in cutaneous
tumors (5). The worldwide
incidence has risen rapidly over the last five decades (1-4).
Among all newly diagnosed cancers, melanoma was the 17th most
common type according to World Cancer Research Fund International
in 2019(6). Melanoma is a
heterogeneous tumor with alterations in specific genes involved in
pathways controlling cell proliferation, differentiation and
survival (7). The major pathways
involved are the cyclin-dependent kinase inhibitor 2A
(CDKN2A)-dependent pathway and MAPK- and PI3K-dependent pathways
(8,9).
As ~50% of advanced melanomas [unresectable Stage
III or metastatic Stage IV, according to the American Joint
Committee on Cancer Staging Manual, 8th edition (3)] harbor a mutation in the BRAF
gene and since the Federal Drug Administration and the European
Institutions approved targeted therapy with BRAF and MAPK kinase
inhibitors, molecular testing for codon V600 of the
BRAF oncogene became an integral part of the management of
such cases. Several approaches are available, from single gene
testing for the BRAF V600 mutation, such as PCR or
immunohistochemistry (IHC) using anti-BRAF (mutated V600E) antibody
(AB) [VE1 clone (BRAF VE1)], to whole-genome sequencing. The
targeted next-generation sequencing (NGS) technique allows the
detection of abnormalities across multiples genes. The
identification of multiple molecular aberrations provides several
treatment targets. Despite the valuable provided data, NGS remains
expensive and not widely distributed and requires sophisticated
bioinformatics systems, fast data processing and large data storage
capabilities. On the other hand, IHC has a shorter turnaround time
(TAT), is cheaper and widely available, but it detects only
specific antigens and false-positive and -negative results may be
obtained. IHC using BRAF VE1 has emerged as a powerful tool in
assessing the BRAF V600E mutation status; validation studies
performed to date reported high specificity (95.4-100%) and
sensitivity (94.4-100%) (10-14).
The tumor (suppressor) protein 53 gene
(TP53), ‘the guardian of the genome’, is inactivated in ~90%
of melanomas and only 10-20% of cases are carrying disabling point
mutations (15), but its role
remains controversial in melanoma and melanoma progression. IHC is
used routinely as a surrogate for TP53 mutation analysis,
particularly in ovarian cancer (16), Barrett's esophagus (17) and other cancer types (18-22).
Its application in melanoma requires further study (23-29).
In light of the importance of identifying the
molecular profile for therapeutic purposes, the main objective of
the present study was to evaluate whether BRAF VE1 and p53
immunoexpression are reliable surrogates for mutation detection.
For this purpose, BRAF and TP53 status were analyzed
by IHC in a series of melanoma cases for which molecular data were
previously obtained by NGS. Furthermore, the inter-observer
concordance for the IHC assessment was assessed and a
cost-effectiveness analysis for the BRAF VE1 AB was performed.
Materials and methods
Samples
Formalin-fixed paraffin-embedded (FFPE) samples of
37 melanomas from 37 patients were retrospectively analyzed,
including 8 (21.6%) primary tumors and 29 (78.4%) metastatic
tumors. The samples were provided by the Pathology Department of
Erasme University Hospital (Brussels, Belgium). All tumor sample
sites are summarized in Table
SI.
The selection criteria were as follows: Patients
with melanoma for whom molecular testing was required and performed
between January 2014 and February 2019. The sample types were
either biopsies (n=6), cell blocks (n=3) or surgical resections
(n=28). The samples were retrieved for the NGS results and
subsequently, the FFPE blocks were checked to assess if there is
sufficient residual tissue to perform IHC. Thus, the exclusion
criterion was an insufficient amount of residual tumor tissue as
determined by the pathologist and the cases were further excluded
from the statistical analyses.
NGS DNA extraction
DNA extraction from FFPE samples was performed as
previously described (30,31), using the QIAamp FFPE tissue kit
(Qiagen GmbH), according to the manufacturer's protocol. The
obtained DNA was quantified using the Qubit® fluorometer
in combination with the Qubit® dsDNA HS assay kit
(Thermo Fisher Scientific, Inc.).
Library preparation, cluster
amplification and sequencing
NGS was performed as previously described (30,31).
DNA (10 ng) was amplified using the Cancer Panel (Ampliseq™; Thermo
Fisher Scientific, Inc.). An amplicon library was generated for
sequencing 2,850 mutations in 50 genes, including the following:
ABL1, AKT1, ALK, APC, ATM,
BRAF, CDH1, CDKN2A, CSF1R,
CTNNB1, EGFR, ERBB2, ERBB4, EZH2,
FBXW7, FGFR1, FGFR2, FGFR3,
FLT3, GNA11, GNAQ, GNAS, HNF1A,
HRAS, IDH1, IDH2, JAK2, JAK3,
KDR, KIT, KRAS, MET, MLH1,
MPL, NOTCH1, NPM1, NRAS, PDGFRA,
PIK3CA, PTEN, PTPN11, RB1, RET,
SMAD4, SMARCB1, SMO, SRC, STK11,
TP53 and VHL. Library construction was performed
using the Ion AmpliSeq™ Library kit 2.0 and Ion Xpress™ barcode
adapters kit (Thermo Fisher Scientific, Inc.), while the
quantification was performed with the Qubit® fluorometer
and the Qubit® dsDNA HS assay kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
library was then quantified using the Qubit® fluorometer
and the Qubit® dsDNA HS assay kit (Thermo Fisher
Scientific, Inc.). Libraries were multiplexed and clonally
amplified by emulsion PCR using the Ion One Touch 2 instrument with
the Ion PGM™ template OT2 200 kit (Thermo Fisher Scientific, Inc.)
and sequenced using a PGM™ sequencer with the Ion PGM™ sequencing
200 kit v2. Quality control was performed using the Ionsphere™
quality control kit (Thermo Fisher Scientific, Inc.). All steps
were performed according to the manufacturer's protocols.
Data analysis
The raw data analysis was performed as previously
described (30,31), using Torrent Suite software
v4.0.2-v5.10.0 (Thermo Fisher Scientific, Inc.). The coverage
analysis was performed using the Coverage Analysis plugin v4.0-
v5.10. Cases for which the average base coverage was <500x were
considered non-informative. Detection of mutations was performed
using the Variant Caller plugins v4.0-v5.10 (Thermo Fisher
Scientific, Inc.). Each mutation was verified in the Integrative
Genome Viewer from the Broad Institute (http://www.broadinstitute.org/). Only mutations
reported in the COSMIC database (http://cancer.sanger.ac.uk/) were taken into account,
while silent or intronic mutations were not reported. Mutations
were then classified into three categories based on data obtained
from the literature and from the COSMIC database: Mutations with
known clinical impact (with a targeted therapy on the market, e.g.,
BRAF V600 mutation); mutations with potential clinical
impact (clinical trials are ongoing, e.g., NRAS,
EZH2, BRAF nonV600E); and mutations with unknown
clinical impact (e.g., TP53).
The cases with poor-quality sequencing and/or
insufficient material for IHC staining were considered
non-contributory.
IHC IHC procedure
Sections (4 µm thick) of the original FFPE block
used for molecular analysis were subjected to anti-BRAF (mutated
V600E) IHC staining (cat. no. ab228461; clone VE1; dilution, 1/100;
Abcam) and anti-p53 IHC staining (cat. no. M7001; clone DO-7;
dilution, 1/200; Agilent Technologies, Inc.) on a Dako Omnis
(Agilent Technologies, Inc.). Heat-induced epitope retrieval was
performed using Dako Target Retrieval Solution pH 9 (cat. no.
GV804; Agilent Technologies, Inc.) 30 min at 97˚C, followed by
primary antibody incubation 20 min at 32˚C and detection with Dako
Envision Flex detection system (cat. no. GV800; Agilent
Technologies, Inc.) according to the manufacturer's protocol. The
sections were counterstained with hematoxylin (cat. no. GC808;
Agilent Technologies, Inc.). Control tissues (tonsil for p53 IHC
and BRAF V600E mutated tumor for BRAF IHC) and universal negative
control antibody (negative control Mouse IgG1; cat. no. X0931;
dilution, 1/200; Agilent Technologies, Inc.) were processed in
parallel with tissues exposed to the primary as described
above.
Pathology scoring
Immunostained slides were evaluated by two trainee
pathologists (one and two years of experience) blinded to the
molecular data. For discordant cases, a third pathologist (>10
years of experience) blinded to previous results re-evaluated the
p53 and/or BRAF V600E IHC staining.
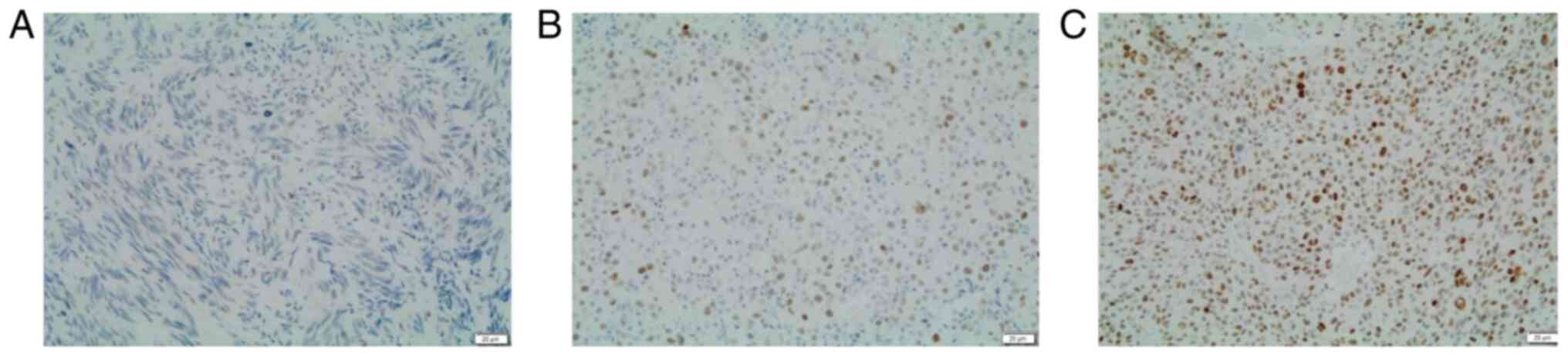
A semi-quantitative assessment of p53 IHC (Fig. 1) expression was performed by using
a three-tier score as previously described (32): 0-no staining (loss of expression);
1-≤25% of cells with variable, heterogeneous cytoplasmic and
nuclear staining (wild-type); 2->25% of cells with high
homogeneous nuclear staining (overexpression). This
semi-quantitative evaluation is similar to the ones used by
Kastelein et al (17) in
Barrett's esophagus or Guedes et al (20) in prostate cancer.
A qualitative assessment of BRAF VE1 (Fig. 2) was used as previously described
(10-13):
Negative-no cytoplasmic staining; positive-presence of cytoplasmic
staining.
Pathological data
The pathological data collected were sample type
(biopsy/cell block/surgical resection), tumor sample site
(skin/conjunctiva/brain/subcutaneous tissue/liver/lymph
node/adrenal glands/digestive tract/lung/undetermined), IHC
analysis (sufficient/insufficient material), IHC BRAF V600E
(positive/negative), IHC p53 (no
staining/wild-type/overexpression/non-contributory), NGS analysis
(contributory/non-contributory), results of NGS (gene mutation,
exon, coverage, % of mutated DNA, clinical impact).
Non-contributory IHC means lack of positive internal control, while
non-contributory NGS means that the sequencing quality was too poor
to allow for analysis. Clinicopathological data, age and sex
distributions were not available for the present study in line with
the Ethics Committee consent.
Cost analysis
Information regarding the costs associated with IHC
using BRAF VE1 AB Idylla™ (PCR-based method) and NGS were
retrieved. A comparison of the costs was performed between these
three methods.
Statistical analysis
The results from IHC and NGS testing were
cross-tabulated and, using the online calculator provided by
Medcalc (https://www.medcalc.org/calc/diagnostic_test.php),
the sensitivity, specificity, accuracy, positive predictive value
and negative predictive value were calculated. The association
between NGS and IHC was performed using Fisher's exact test with
Statistica® (StatSoft). P<0.05 was considered to
indicate statistical significance.
To assess the inter-observer coefficient, Cohen's
unweighted κ coefficient was calculated by running an online test,
provided by Vassarstats (http://vassarstats.net/kappa.html). The scale used for
the interpretation of the κ coefficient was the following: 0-0.2,
slight agreement; 0.21-0.40, fair agreement; 0.41-0.60, moderate
agreement; 0.61-0.80, substantial agreement; and 0.81-1, almost
perfect agreement.
Results
IHC results and inter-observer
concordance
The IHC staining was performed on 34 of 37 cases
(three cases did not have sufficient material for immunostaining).
A total of 34 cases were further considered for statistical
analyses. The IHC profile assessment for p53 and BRAF V600E is
summarized in Table I.
 | Table IResults of immunohistochemical
assessment of p53 and BRAF V600E by pathologists. |
Table I
Results of immunohistochemical
assessment of p53 and BRAF V600E by pathologists.
| Protein/result | 1st
Pathologist | 2nd
Pathologist | Final result |
|---|
| p53 | | | |
|
0 (loss of
expression) | 0 (0) | 1(3) | 1(3) |
|
1
(wild-type) | 23 (67.6) | 25 (73.5) | 22 (64.6) |
|
2
(overexpression) | 10 (29.4) | 7 (20.5) | 10 (29.4) |
|
Non-contributorya | 1(3) | 1(3) | 1(3) |
| BRAF V600E | | | |
|
Absence of
staining | 30 (88.2) | 30 (88.2) | 30 (88.2) |
|
Presence of
staining | 4b (11.8) | 4b (11.8) | 4 (11.8) |
Regarding p53, six of the 34 cases were discordant
(17.6%) and were reviewed by a third pathologist, who agreed three
times with the first pathologist and three times with the second.
One case was not assessable due to the lack of internal positive
control. The κ coefficient for evaluating the inter-observer
concordance was 0.554, suggesting a moderate agreement.
Concerning BRAF V600E, two of 34 analyzed
cases were discordant (5.90%) and were reviewed by a third
pathologist, who agreed with the first pathologist in both cases.
The Cohen unweighted κ coefficient was 0.717, which represents a
substantial agreement.
NGS results and NGS-IHC
correlation
Sequencing was optimal in 36 out of 37 cases
(97.3%). It failed in one sample (2.7%) due to poor quality of
sequencing (non-contributory). No mutation in the gene panel was
detected for 5 of the 36 cases (13.9%), 15 (41.7%) harbored a
BRAF mutation and 16 (44.4%) had other mutations than BRAF
(potentially clinically actionable mutations or mutations with
unknown impact). Of the 15 cases harboring a BRAF mutation,
eight were V600E (two cases had insufficient residual tissue
for IHC staining, leading to six cases included for NGS-IHC
correlation analysis), followed by three V600 non-E (two
V600K and one V600R), two D594N, one
K601E and one K601N. The most frequent mutations
among the non-BRAF mutations were NRAS (19.4%, 7/36) and
TP53 (13.9%, 5/36). BRAF V600E and TP53
mutations were not mutually exclusive and one case harbored BRAF
V600E and TP53 mutations.
When performing the statistical analysis, loss of
expression and overexpression of the p53 protein on IHC analysis
was considered as a positive result (aberrant IHC result-expression
of mutation in the tumor suppressor gene TP53) and the
non-contributory cases were eliminated from the analysis. NGS was
the reference technique for the analysis of both ABs. NGS and IHC
results were available in 32 cases for the evaluation of
TP53 protein expression and in 33 cases regarding BRAF
V600E protein expression. The cross-tabulated results of the
NGS and IHC assessment are summarized in Table II.
 | Table IICross-tabulated results for tumor
suppressor protein 53 gene and BRAF V600E mutation
status determined by IHC and NGS. |
Table II
Cross-tabulated results for tumor
suppressor protein 53 gene and BRAF V600E mutation
status determined by IHC and NGS.
| | NGS result |
|---|
| IHC result | Non-mutated | Mutated |
|---|
| p53 | | |
|
Wild-type | 19 | 2 |
|
Aberrant
expression | 8 | 3 |
| BRAF V600E | | |
|
No
staining | 27 | 2 |
|
Presence of
staining | 0 | 4 |
Considering the NGS technique to be the reference
for the TP53 mutation status, IHC had an overall diagnostic
accuracy of 68.8% for the TP53 mutation with 31.2% of cases
being misclassified (10/32). The overall sensitivity of IHC was 60%
and two cases were false-negative. The overall specificity of IHC
was 70.4% and eight cases were false-positive. The positive
predictive value of IHC for p53 was 27.3% (3/11), while the
negative predictive value was 90.5% (19/21). No statistically
significant association between the two diagnostic methods was
obtained (P=0.3098).
Considering the NGS technique to be the reference
for the BRAF V600E mutation status, IHC had an overall
diagnostic accuracy of 93.9% for the BRAF V600E mutation
with 6.1% of cases being misclassified (2/33). These two
misclassified cases were cases harboring a BRAF V600E
mutation and negative BRAF VE1 IHC staining. The overall
sensitivity of IHC was 66.7% and two cases were false-negative. The
overall specificity for IHC was 100% with no false-positive results
obtained. The positive predictive value of BRAF VE1 was 100%, while
the negative predictive value was 93.1% (27/29). A statistically
significant association between the two diagnostic methods was
determined (P=0.0004). All related results are summarized in
Table III.
 | Table IIINGS-IHC correlation results for both
genes tested. |
Table III
NGS-IHC correlation results for both
genes tested.
| | TP53 | BRAF
V600E |
|---|
| NGS-IHC
correlation | Value (%) | 95% CI | Value (%) | 95% CI |
|---|
| Diagnostic
accuracy | 68.8 | 50.0-83.9 | 93.9 | 79.8-99.3 |
| Overall
sensitivity | 60.0 | 14.7-94.7 | 66.7 | 22.3-95.7 |
| Overall
specificity | 70.4 | 49.8-86.3 | 100.0 | 87.2-100.0 |
| Positive predictive
value | 27.3 | 13.0-48.5 | 100.0 | 51.0-100.0 |
| Negative predictive
value | 90.5 | 76.0-96.6 | 93.1 | 81.3-97.7 |
| P-value | 0.3098 | 0.0004 |
Cost-effectiveness analysis for BRAF
status testing
In our laboratory, the BRAF V600E status may
be tested by IHC, Idylla™ (PCR-based method) or NGS. It was decided
to perform the cost analysis only based on the reagents' cost to
ensure harmonized results. Amortization costs, maintenance or human
resources were not considered, as these elements vary among
laboratories. It should be noted that the fact that reagents' costs
may also vary depending on the test volume, supplier or country, so
that the present cost-effectiveness analysis is only indicative.
All of the costs per test for each technique are summarized in
Tables SII and SIII. The cost per patient of an NGS
analysis considering only the reagents is ~280 €, that of Idylla™
120 € and BRAF VE1 IHC costs ~22 €. For the present case series,
several simulations were performed and it was concluded that
performing BRAF V600E IHC staining for all cases first followed by
NGS for negative BRAF V600E IHC cases saves ~5% of costs compared
to performing only NGS. Performing Idylla™ DNA-testing only was
revealed to cost less than performing BRAF V600E IHC staining for
all cases first followed by Idylla™. However, in larger case
series, IHC staining followed by a DNA-based test for negative
cases would cost less than only DNA-based testing.
Discussion
Melanoma is the most aggressive form of skin cancer
with ~200,000 new cases diagnosed each year worldwide (33). The identification of BRAF
mutations through DNA sequencing in the early 2000s led to a
revolution in the treatment of melanoma, allowing the development
of therapies targeting the BRAF oncogene and applications of other
kinase inhibitors (34). The
BRAF mutation status is critical for advanced-stage melanoma
and molecular testing should be performed routinely for stage III
and IV (unresectable and metastatic) (35) to promptly start the therapy,
particularly in melanomas with aggressive behavior. Several
BRAF mutation tests are available: DNA-based tests
(including PCR and NGS) and one AB-based test for mutant BRAF V600E
protein using the mouse monoclonal VE1 clone to detect the protein
expression by IHC.
BRAF mutations are observed to occur in
numerous different types of cancer, including thyroid, lung, colon
cancers, glioblastomas and certain hematological malignancies
(16-22,34).
BRAF mutations are identified in 40-60% of melanomas
(14,34,36).
BRAF V600E is the most common mutation and it accounts for
60-90% of all BRAF mutations, followed by V600K
mutation and V600D/R accounting for 10-30 and 3%,
respectively (37,38). The present results overlap with
those described in the literature, as BRAF mutations were
identified in ~40% of the present cases and among the mutated
cases, V600E mutation accounts for 53.3% of cases, followed
by V600K mutation with 13.3%. The present study demonstrated
a sensitivity of 66.7% and a specificity of 100% for the BRAF VE1
IHC as the method of detection for BRAF V600E compared to
NGS analysis (considered the gold standard) in FFPE tumor tissue
samples. Specimens with the V600 non-E mutation were not
immunoreactive with the VE1 clone, as already described (10,11).
The present results are comparable to previous validation studies
in which a qualitative assessment scale for VE1 staining was
employed regarding the specificity (95.4-100%), but the sensitivity
was inferior to previously reported results (94.4-100%) (10-14).
Different scoring systems have been used for the
evaluation of BRAF by IHC (two or more than two categories,
based on the intensity of the IHC staining or based on the pattern
of staining) in previous studies, leading to major difficulties in
comparing the results. For instance, Lo et al (10), Long et al (11), Colomba et al (12) and Boursault et al (13) used the same evaluation method as
that employed in the present study, while Marin et al
(39) and Pearlstein et al
(40) scored BRAF IHC expression
in more than two categories. The establishment of an objective and
widely accepted consensus scoring system will lead to the
uniformization of results and a decrease in interpretation
discrepancies and bias.
The inferior sensitivity may be explained by the
small number of cases harboring BRAF V600E included in the
present study. Immunostaining with BRAF VE1 AB did not produce any
false-positive final result. At the first assessment, one
pathologist misclassified a negative case as positive on IHC. This
false-positive result may be explained by the lack of experience of
the trainee pathologist and may be remedied by providing training
programs to correctly evaluate IHC profiles. In the present study,
two false-negative results occurred, characterized by the absence
of IHC staining but BRAF V600E mutation on NGS. In one of
the cases, the interpretation of immunostaining was difficult due
to the presence of a high quantity of melanin. In order to increase
the specificity of the test, the brown chromogen may be replaced
with red chromogen (40).
Discordance between pathologists in assessing the BRAF VE1 IHC was
determined in two of the 34 cases where a third pathologist's
opinion was required. The Cohen's κ coefficient obtained (0.717)
indicates a substantial agreement between the two pathologists.
Although molecular testing is considered the gold standard for the
detection of BRAF mutations, the monoclonal VE1 AB is
emerging as a reliable option (10-14,40-42).
In the present study, a statistically significant association was
demonstrated between the IHC and NGS analysis (P=0.0004), which
confirmed the hypothesis that IHC may be used as a surrogate in the
evaluation of the BRAF V600E mutation status. The advantages
of IHC are as follows: Shorter TAT, lower cost, small quantity of
tumor tissue required for detection and the service may be offered
by more histopathology departments than the genomics analysis.
Relying on the results obtained, the algorithm incorporating both
IHC and DNA-based analysis, as previously proposed by Pearlstein
et al (40) or the Anglian
Cancer Network Protocol (10), may
be considered to be used in current clinical practice (Fig. 3). In this algorithm, BRAF VE1 IHC
analysis is performed in all cases of unresectable or metastatic
melanoma, providing a quick and inexpensive way to detect BRAF
V600E mutations. Furthermore, it is the only modality of
BRAF mutation detection in cases with limited tumor tissue
that may otherwise not be tested by DNA-based analysis due to
insufficient material. Afterwards, cases with negative BRAF VE1
staining should be tested by a DNA-based test in order to identify
a possible false-negative result or a BRAF mutation other
than V600E. The use of the proposed algorithm would offer a
cost-saving of ~5% compared to the NGS test in all cases. However,
testing only by Idylla™ in the present case series compared to
prior BRAF VE1 testing followed by Idylla™ was more economical, but
this is highly dependent on the number of cases. Concerning
DNA-based testing, the NGS technique provides information beyond
BRAF, revealing more ‘actionable’ mutations leading to other
therapeutic options. However, NGS is not widely available, its
interpretation requires highly trained staff and it has a longer
TAT. The Idylla™ test is a PCR-based test and an alternative to IHC
or NGS for BRAF V600 mutation, with the following
advantages: It is less laborious, faster and allows the detection
of BRAF V600 non-E mutations; however, it has a higher cost
and it is not as widely available as IHC (43).
p53 is a transcription factor with a powerful tumor
suppressor function. Wild-type p53 is a potent inducer of
apoptosis, of cell cycle arrest and of cellular senescence
(14). A review published by Lu
et al (44) in 2013
highlights the potential role of targeting p53 in melanoma
treatment, stating that reactivation of p53 by simultaneously
blocking the p53 E3 ubiquitin ligase, MDM2 and inhibitor of
apoptosis-stimulating protein of p53, together with BRAF V600E
inhibition, induces apoptosis and suppresses melanoma growth in
cell lines and animal models. Furthermore, a novel hypothesis
regarding the importance of the p53 status emerges, stating that
TP53 mutation is a potential negative predictor of
metastatic melanoma treated by CTLA-4 blockade (45,46).
Thus, it may be worthwhile to routinely evaluate the TP53 status.
The frequency of TP53 mutation in melanoma reported in the
literature is ~20% (9,47). A slightly inferior rate (14%) was
obtained in the present cohort. The association between TP53
mutations and p53 nuclear accumulation remains to be fully
elucidated. IHC is used routinely as a surrogate for TP53
mutation analysis in several types of cancer (17-22,48),
but its interpretation is at times difficult (48). In the present study, the κ
coefficient obtained was 0.554, suggesting a moderate
inter-observer concordance, underlining the difficulty in
interpreting p53 protein expression. The IHC-NGS association was
not statistically significant, indicating that the use of p53 IHC
to replace a DNA-based test should not be considered for routine
use. In the present study, false-negative (two identified in the
present cohort) and false-positive (eight identified) results were
obtained. The false-negative results harbored different TP53
mutations. Furthermore, not all the mutations are leading to
corresponding transcriptional or translational changes in p53
protein products (28). The
false-positive results may be explained by the fact that p53 IHC
expression may result from malfunction of other components of the
p53 pathway other than the gene mutation or the fact that targeted
NGS focuses on genomic regions of particular interest, with various
regions of the TP53 gene remaining unsequenced. Besides
these, the tumors also exhibit genetic heterogeneity (29). Other studies (23-27)
also reported no correlation between the protein expression and the
TP53 mutational status, suggesting that a direct link
between aberrant protein expression of p53 and the presence of
TP53 mutation was not able to be established.
As at the present time, determination of the
TP53 status is not a standard of care for patients with
melanoma in contrast to the BRAF status, expanded molecular
testing is not required for all patients. Considering that IHC
analysis of p53 is not highly sensitive or specific, when
determination of the TP53 mutation will be warranted, it
should be assessed by a molecular method.
In conclusion, molecular profiling has become a key
piece of information in the management of advanced-stage melanoma
with the introduction of ‘personalized medicine’. The results
obtained demonstrated that IHC staining with BRAF VE1 AB is a
reliable surrogate for NGS in identifying the BRAF V600E
mutation, becoming an efficient screening tool prior to DNA-based
analysis. Loss of p53 expression/overexpression determined by IHC
is at times associated with TP53 gene mutations, but it was
not possible to establish a root-cause relationship. Aside from
that, BRAF VE1 allows the detection of a specific hotspot mutation,
while p53 expression on IHC is related to protein expression that
may be wild-type but may also mirror a large spectrum of
TP53 mutations.
Supplementary Material
Tumor sample site according to tumor
type (primary/metastatic).
Information regarding the reagent
costs of BRAF testing by different methods per patient.
Estimation of the reagent costs for
the present case series (n=34).
Acknowledgements
Not applicable.
Funding
This work was supported by the Fonds Yvonne Boël (Brussels,
Belgium).
Availability of data and material
The original targeted NGS data were submitted to
the European Nucleotide Archive. They are available as ‘Rusu et
al 2021: The use of immunohistochemistry as an accurate tool in
the assessment of BRAF V600E and TP53 mutations in primary and
metastatic melanoma’ with the following study ID: PRJEB42810
(ERP126721) (https://www.ebi.ac.uk/ena/browser/view/PRJEB42810?show=related-records).
Authors' contributions
NDH, IS, CD and OSC contributed to the conception
and design of the study. Material preparation, data collection and
IHC analysis were performed by SRu, CV and NDH. SDC, CVC, OB and
NDN performed IHC and molecular analyses. ALT, CM and SRo performed
the pathological diagnoses. SRU, CV, ND and CD analyzed the data.
SRU, CV, NDH and CVC approved the authenticity of the raw data. The
first draft of the manuscript was written by SRU and all authors
commented on previous versions of the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Approval was granted by the Ethics Committee of the
Erasme-Université Libre de Bruxelles (Brussels, Belgium) in May
2019 (no. P2019/310). The Ethics Committee waived the requirement
of informed consent under the condition that the researchers check
for the absence of any objection of the patient to use their
material for research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Glazer AM, Rigel DS, Winkelmann RR and
Farberg AS: Clinical diagnosis of skin cancer: Enhancing inspection
and early recognition. Dermatol Clin. 35:409–416. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
De Vries E, Elder DE, Bray F, Thompson JF,
Coebergh JW, Barnhill RL, Cerroni L, van Muijen GNP, Ruiter DJ,
Scolyer RA, et al: Chapter 2: Melanocytic tumours. Malignant
melanoma: Introduction. In: World Health Organization
Classification of Tumours. Pathology and Genetics of Skin Tumours.
LeBoit PE, Burg G, Weedon D and Sarasin A (eds). IARC Press, Lyon,
pp52-65, 2006.
|
|
3
|
Gershenwald GE, Scolyer RA, Hess KR,
Thompson JF, Long GV, Ross MI, Lazar AJ, Atkins MB, Balch CM,
Bamhill RL, et al: Part X. Skin-47. Melanoma of the Skin. In: AJCC
Cancer Staging Manual, 8th edition. Amin MB, Edge S, Greene F, Byrd
DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess
KR, Sullivan DC, et al (eds). Springer, Chicago, IL,
pp563-585, 2017.
|
|
4
|
Lazar A and Bastian B: Melanoma. In:
McKee's Pathology of the Skin. 4th edition. Calonje JE, Brenn T,
Lazar A and McKee P (eds). Elsevier/Saunders, Edinburgh,
pp1221-1267, 2012.
|
|
5
|
Matthews N, Li W, Qureshi A, Weinstock M
and Cho E: Epidemiology of melanoma. In: Cutaneous Melanoma:
Etiology and Therapy. Ward WH and Farma JM (eds) Codon
Publications, Brisbane, pp3-23, 2017.
|
|
6
|
World Cancer Research Fund, American
Institute for Cancer Research: Worldwide Cancer Data, Global Cancer
Statistics for the most common cancers. WCRF International, London,
2019. https://www.wcrf.org/dietandcancer/cancer-trends/worldwide-cancer-data.
Accessed May 22, 2019.
|
|
7
|
Curtin JA, Fridlyand J, Kageshita T, Patel
HN, Busam KJ, Kutzner H, Cho KW, Aiba S, Brocker EB, LeBoit PE, et
al: Distinct sets of genetic alterations in melanoma. N Engl J Med.
353:2135–2147. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Palmieri G, Ombra M, Colombino M, Casula
M, Sini M, Manca A, Paliogiannis P, Ascierto PA and Cossu A:
Multiple molecular pathways in melanomagenesis: Characterization of
therapeutic targets. Front Oncol. 5(183)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Giehl K: Oncogenic Ras in tumor
progression and metastasis. Biol Chem. 386:193–205. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lo MC, Paterson A, Maraka J, Clark R,
Goodwill J, Nobes J, Garioch J, Moncrieff M, Rytina E and Igali L:
A UK feasibility and validation study of the VE1 monoclonal
antibody immunohistochemistry stain for BRAF-V600E mutations in
metastatic melanoma. Br J Cancer. 115:223–227. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Long GV, Wilmott JS, Capper D, Preusser M,
Zhang YX, Thompson JF, Kefford RF, von Deimling A and Scolyer RA:
Immunohistochemistry is highly sensitive and specific for the
detection of V600E BRAF mutation in melanoma. Am J Surg Pathol.
37:61–65. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Colomba E, Helias-Rodzewicz Z, Von
Deimling A, Marin C, Terrones N, Pechaud D, Surel S, Côté JF,
Peschaud F, Capper D, et al: Detection of BRAF p.V600E mutations in
melanomas: Comparison of four methods argues for sequential use of
immunohistochemistry and pyrosequencing. J Mol Diagn. 15:94–100.
2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Boursault L, Haddad V, Vergier B,
Cappellen D, Bellocq JP, Jouary T and Merlio JP: Homogénéité et
conservation du statut BRAF entre mélanome primitif et métastases
déterminées par immunohistochimie et biologie moléculaire. Ann
Dermatol Venereol. 140(S396)2013.
|
|
14
|
Tetzlaff MT, Pattanaprichakul P, Wargo J,
Fox PS, Patel KP, Estrella JS, Broaddus RR, Williams MD, Davies MA,
Routbort MJ, et al: Utility of BRAF V600E immunohistochemistry
expression pattern as a surrogate of BRAF mutation status in 154
patients with advanced melanoma. Hum Pathol. 46:1101–1110.
2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Box NF, Vukmer TO and Terzian T: Targeting
p53 in melanoma. Pigment Cell Melanoma Res. 27:8–10.
2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Köbel M, Piskorz AM, Lee S, Lui S, LePage
C, Marass F, Rosenfeld N, Mes Masson AM and Brenton JD: Optimized
p53 immunohistochemistry is an accurate predictor of TP53 mutation
in ovarian carcinoma. J Pathol Clin Res. 2:247–258. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kastelein F, Biermann K, Steyerberg EW,
Verheij J, Kalisvaart M, Looijenga LH, Stoop HA, Walter L, Kuipers
EJ, Spaander MC, et al: Aberrant p53 protein expression is
associated with an increased risk of neoplastic progression in
patients with Barrett's oesophagus. Gut. 62:1676–1683.
2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Rodrigues NR, Rowan A, Smith ME, Kerr IB,
Bodmer WF, Gannon JV and Lane DP: p53 mutations in colorectal
cancer. Proc Natl Acad Sci USA. 87:7555–7559. 1990.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Iggo R, Gatter K, Bartek J, Lane D and
Harris AL: Increased expression of mutant forms of p53 oncogene in
primary lung cancer. Lancet. 335:675–679. 1990.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guedes LB, Almutairi F, Haffner MC,
Rajoria G, Liu Z, Klimel S, Zoino R, Yousefi K, Sharma R, De Marzo
AM, et al: Analytic, preanalytic, and clinical validation of p53
IHC for detection of TP53 missense mutation in prostate cancer.
Clin Cancer Res. 23:4693–4703. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lepelley P, Preudhomme C, Vanrumbeke M,
Quesnel B, Cosson A and Fenaux P: Detection of p53 mutations in
hematological malignancies: Comparison between immunocytochemistry
and DNA analysis. Leukemia. 8:1342–1349. 1994.PubMed/NCBI
|
|
22
|
Dobes P, Podhorec J, Coufal O, Jureckova
A, Petrakova K, Vojtesek B and Hrstka R: Influence of mutation type
on prognostic and predictive values of TP53 status in primary
breast cancer patients. Oncol Rep. 32:1695–1702. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Albino AP, Vidal MJ, McNutt NS, Shea CR,
Prieto VG, Nanus DM, Palmer JM and Hayward NK: Mutation and
expression of the p53 gene in human malignant melanoma. Melanoma
Res. 4:35–45. 1994.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Akslen LA, Monstad SE, Larsen B, Straume O
and Ogreid D: Frequent mutations of the p53 gene in cutaneous
melanoma of the nodular type. Int J Cancer. 79:91–95.
1998.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sparrow LE, English DR, Heenan PJ, Dawkins
HJ and Taran J: Prognostic significance of p53 over-expression in
thin melanomas. Melanoma Res. 5:387–392. 1995.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Florenes VA, Oyjord T, Holm R, Skrede M,
Borresen AL, Nesland JM and Fodstad O: TP53 allele loss, mutations
and expression in malignant melanoma. Br J Cancer. 69:253–259.
1994.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Weiss J, Heine M, Arden KC, Körner B,
Pilch H, Herbst RA and Jung EG: Mutation and expression of TP53 in
malignant melanomas. Recent Results Cancer Res. 139:137–154.
1995.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Soussi T, Legros Y, Lubin R, Ory K and
Schlichtholz B: Multifactorial analysis of p53 alterations in human
cancer: A review. Int J Cancer. 57:1–9. 1994.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hussein MR: The TP53 tumor supressor gene
and melanoma tumorigenesis: Is there a relationship? Tumor Biol.
25:200–207. 2004.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Le Mercier M, D'Haene N, De Nève N,
Blanchard O, Degand C, Rorive S and Salmon I: Next-generation
sequencing improves the diagnosis of thyroid FNA specimens with
indeterminate cytology. Histopathology. 66:215–224. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
D'Haene N, Le Mercier M, De Nève N,
Blanchard O, Delaunoy M, El Housni H, Dessars B, Heimann P,
Remmelink M, Demetter P, et al: Clinical validation of targeted
next generation sequencing for colon and lung cancers. PLoS One.
10(e0138245)2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lebrun L, Milowich D, Le Mercier M, Allard
J, Van Eycke YR, Roumeguere T, Decaestecker C, Salmon I and Rorive
S: UCA1 overexpression is associated with less aggressive subtypes
of bladder cancer. Oncol Rep. 40:2497–2506. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Miraflor AP, de Abreu FB, Peterson JD,
Turner SA, Amos CI, Tsongalis GJ and Yan S: Somatic mutation
analysis in melanoma using targeted next generation sequencing. Exp
Mol Pathol. 103:172–177. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Holderfield M, Deuker MM, McCormick F and
McMahon M: Targeting RAF kinases for cancer therapy: BRAF-mutated
melanoma and beyond. Nat Rev Cancer. 4:455–467. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Coit DG, Thompson JA, Albertini MR, Barker
C, Carson WE, Contreras C, Daniels GA, DiMaio D, Fields RC, Fleming
MD, et al: Cutaneous Melanoma, Version 2.2019, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
17:367–402. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lee JH, Choi JW and Kim YS: Frequencies of
BRAF and NRAS mutations are different in histological types and
sites of origin of cutaneous melanoma: A meta-analysis. Br J
Dermatol. 164:776–784. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kudchadkar R, Paraiso KH and Smalley KS:
Targeting mutant BRAF in melanoma: Current status and future
development of combination therapy strategies. Cancer J.
18:124–131. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Thiel A, Moza M, Kytola S, Orpana A,
Jahkola T, Hernberg M, Virolainen S and Ristimaki A: Prospective
immunohistochemical analysis of BRAF V600E mutation in melanoma.
Hum Pathol. 46:169–175. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Marin C, Beauchet A, Capper D, Zimmermann
U, Julie C, Ilie M, Saiag P, von Deimling A, Hofman P and Emile JF:
Detection of BRAF p.V600E mutations in melanoma by
immunohistochemistry has a good interobserver reproducibility. Arch
Pathol Lab Med. 138:71–75. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Pearlstein MV, Zedek DC, Ollila DW, Treece
A, Gulley ML, Groben PA and Thomas NE: Validation of the VE1
Immunostain for the BRAF V600E mutation in melanoma. J Cutan
Pathol. 41:724–732. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Schirosi L, Strippoli S, Gaudio F,
Graziano G, Popescu O, Guida M, Simone G and Mangia A: Is
immunohistochemistry of BRAF V600E useful as a screening tool and
during progression disease of melanoma patients? BMC Cancer.
16(905)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cheng L, Lopez-Beltran A, Massari F,
MacLennan GT and Montironi R: Molecular testing for BRAF mutations
to inform melanoma treatment decisions: A move toward precision
medicine. Mod Pathol. 31:24–38. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bisschop C, Ter Elst A, Bosman LJ,
Platteel I, Jalving M, van den Berg A, Diepstra A, van Hemel B,
Diercks GFH, Hospers GAP and Schuuring E: Rapid BRAF mutation tests
in patients with advanced melanoma: Comparison of
immunohistochemistry, Droplet Digital PCR, and the Idylla Mutation
Platform. Melanoma Res. 28:96–104. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lu M, Miller P and Lu X: Restoring the
tumour suppressive function of p53 as a parallel strategy in
melanoma therapy. FEBS Lett. 588:2616–2621. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Onyshchenko M: The puzzle of predicting
response to immune checkpoint blockade. EBioMedicine. 33:18–19.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xiao W, Du N, Huang T, Guo J, Mo X, Yuan
T, Chen Y, Ye T, Xu C, Wang W, et al: TP53 mutation as potential
negative predictor for response of Anti-CTLA-4 therapy in
metastatic melanoma. EBioMedicine. 32:119–124. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Siroy AE, Boland GM, Milton DR, Roszik J,
Frankian S, Malke J, Haydu L, Prieto VG, Tetzlaff M, Ivan D, et al:
Beyond BRAF(V600): Clinical mutation panel testing by
next-generation sequencing in advanced melanoma. J Invest Dermatol.
135:508–515. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Murniak B and Hortobagyi T:
Immunohistochemical correlates of TP53 somatic mutations in cancer.
Oncotarget. 7:64910–64920. 2016.PubMed/NCBI View Article : Google Scholar
|