Introduction
Gliomas are the most common tumors of the central
nervous system (CNS). The 2014 International Congress of
Neuropathology added molecular findings to the diagnostic
guidelines for brain tumors, whereas in 2016, the World Health
Organization (WHO) classification standard introduced molecular
features in the reclassification of CNS tumors (1). This included diffuse astrocytomas,
oligodendrogliomas, ependymomas, choroid plexus tumors, neuronal
and mixed neuronal-glial tumors, pineal region tumors and embryonal
tumors (1). The 2021 edition of
the classification standard highlighted the role of molecular
diagnostics in the classification of CNS tumors (2). It integrated histological
classification with molecular phenotypes, which included IDH-1,
MGMT, 1p/19q, BRAF and ATRX. Glioblastomas are the most common
malignant tumors of the CNS, accounting for 46% of all CNS tumors.
Patients with glioblastomas have a relatively poor prognosis, with
a five-year survival rate of <5% (3). Identifying specific molecular
phenotypes is crucial for individualized treatment. One such
molecular phenotype involves detecting methylation of the
O6-methylguanine DNA methyltransferase (MGMT) promoter,
which is associated with the glioblastoma prognosis (4-11).
MGMT is a DNA repair protein that reverses DNA damage caused by
alkylating agents by removing the alkyl group from
O6-alkyl-guanine. When the MGMT promoter
is hypermethylated, MGMT expression is silenced, leading to the
absence of MGMT-mediated DNA repair. The alkylating agent
temozolomide capitalizes on this lack of repair to improve the
survival rates in patients with glioblastomas (12-15).
However, the invasiveness of pathological biopsy and the high cost
of detecting the methylation status of MGMT promoter, poses
challenges for implementing individualized treatment in patients
with glioblastomas. Glioblastomas with different genotypes exhibit
vast heterogeneity on both genetic and histopathological levels,
including intratumoral spatial variation in cellularity,
angiogenesis, extravascular extracellular matrix and areas of
necrosis (16-18).
This high intratumoral heterogeneity of glioblastoma is an
indicator of tumor malignancy, as it reflects areas of high cell
density, necrosis, hemorrhage and mucoid degeneration. Intratumoral
heterogeneity is also an important factor affecting prognosis and
is related to tumor grade (19-21).
While glioblastoma heterogeneity cannot be discerned
with the naked eye, MRI texture feature analysis is a valuable
tool. In the present study, the 3D Slicer software was employed to
analyze the magnetic resonance imaging (MRI) texture features of
glioblastoma heterogeneity. The primary aim was to facilitate the
discrimination of glioblastoma molecular phenotypes and improve the
accuracy of molecular imaging diagnosis. This approach aims to
provide patients with individualized treatment, predicting and
improving their prognosis, and enhancing their quality of life.
Materials and methods
General information
A total of 128 patients who were pathologically
diagnosed with glioblastoma at Tangshan Gongren Hospital
(Tangshan, China) between June 2018 and September 2020 were
enrolled in the present retrospective study. General
clinical data included the age, sex, preoperative Karnofsky
performance status (KPS) score, treatment method and overall
survival (OS) of patients. Inclusion criteria were as
follows: i) Patients with complete and reliable
clinical, pathological and imaging data; ii) patients who
did not undergo radiotherapy, chemotherapy, or other neoadjuvant
therapies before surgery; and iii) patients pathologically
diagnosed with glioblastoma after surgery. The following
exclusion criteria were applied: i) Patients with a
history of other malignant tumors; ii) patients with
postoperative complications, such as intracranial hematoma or
intracranial infection; and iii) patients who succumbed
due to other causes. All biopsied specimens were tested for
MGMT promoter methylation. In the study population, 79 patients had
glioblastomas with methylated MGMT promoters, and 49 patients had
glioblastomas with unmethylated MGMT promoters. All patients
underwent conventional and contrast-enhanced MRIs. The
Institutional Review Board of the Ethics committee at the Tangshan
Gongren Hospital (Tangshan, China) granted an exemption to obtain
informed consent due to the use of anonymous data (approval no.
GRYY-LL-2020-44). The Ethics Committee exempted informed consent
because of the retrospective nature of the present study.
Detection of MGMT promoter methylation
status
Nested methylation-specific polymerase chain
reaction was performed to test the methylation status of the MGMT
promoter. Assessment criteria were as follows: Samples with the
corresponding fragments amplified using only the MGMT-U primer were
unmethylated; samples with the corresponding fragments amplified
using only the MGMT-M primer or both the MGMT-U and MGMT-M primers
were methylated (22).
Testing methods
The Philips 3.0-T MRI scanner and 8-Channel Sense
Head Coil were used on each patient to perform conventional and
contrast-enhanced MRI including transverse T1-weighted (T1WI),
transverse and sagittal T2-weighted (T2WI), transverse T2-weighted
fluid-attenuated inversion recovery (T2-FLAIR), and
contrast-enhanced T1WI (T1WI + C) imaging. Scan parameters were as
follows. Transverse T1WI: repetition time (TR) 2,270 msec, echo
time (TE) 20 msec, field of view (FOV) 196x196 mm, matrix 288x190,
number of excitations 2, slice thickness 6 mm, interslice gap 1 mm.
Transverse and sagittal T2WI: TR 2,500 msec, TE 90 msec, FOV
230x230 mm, matrix 420x306, number of excitations 2, slice
thickness 6 mm, interslice gap 1 mm. Transverse T2-FLAIR: TR 8,000
ms, TE 120 msec, FOV 230x230 mm, matrix 304x216, number of
excitations 2, slice thickness 6 mm, interslice gap 1 mm. T1WI + C:
TR 200 msec, TE 2 msec, FOV 230x230 mm, matrix 256x256, number of
excitations 2, slice thickness 6 mm, interslice gap 1 mm. For
contrast-enhanced MRI, gadopentetate dimeglumine was injected
intravenously at a dose of 0.1 ml/kg body weight and at a flow rate
of 3 ml/sec.
Image processing
All patient MRI images were exported from the PACS
workstation in DICOM format and imported into the 3D Slicer
open-source software (version 4.4.0; available at: https://slicer.org/). First, three experienced
neuroimaging experts extracted MRI texture features from the
transverse T1WI + C images using the Editor module in 3D Slicer.
The regions of interest were delineated manually on each slice to
ensure that the tumor boundaries were delineated as accurately as
possible, and the three experts reached a consensus for the
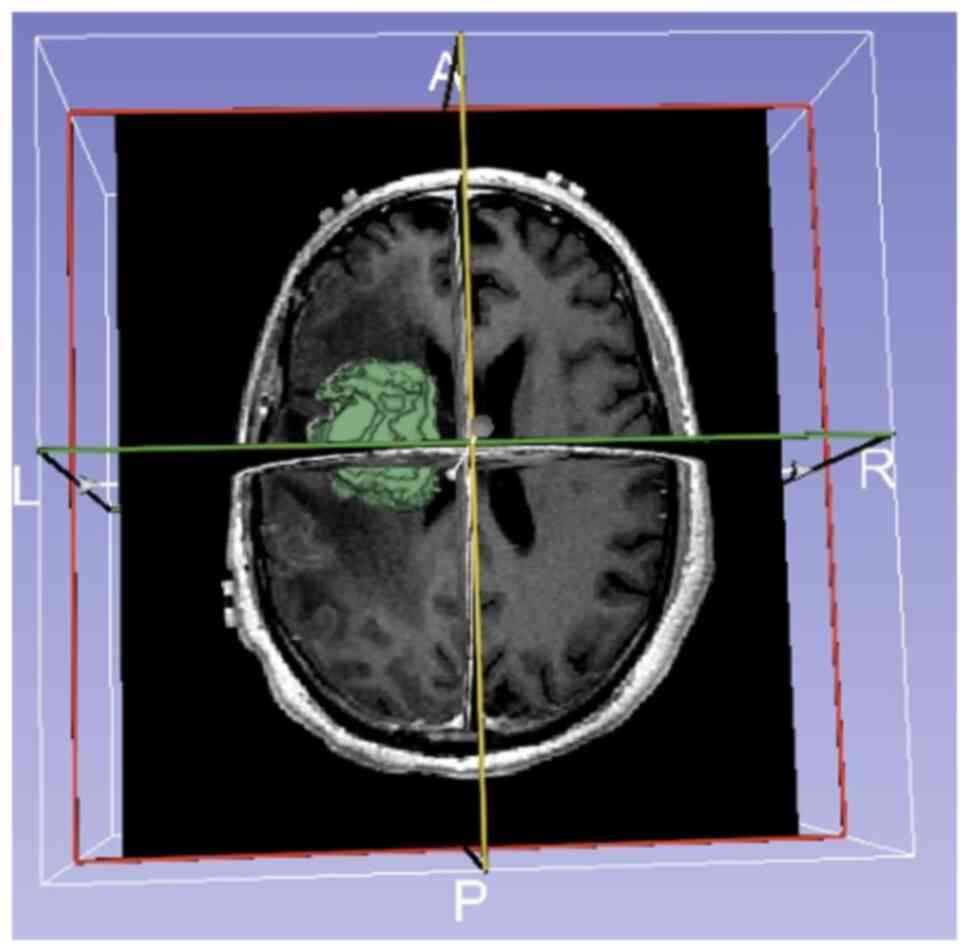
reconstruction of the three-dimensional tumor model (Fig. 1). Second, the three experts used
the Heterogeneity CAD extension module to extract the MRI texture
features of the tumors. Subsequent analyses were performed only
when the intraclass correlation coefficient of the region of
interest delineated by the three experts was >0.75. Finally,
first-order statistics on the following four aspects of MRI texture
features were calculated: Morphology, shape, texture: gray level
co-occurrence matrix (GLCM), and texture: Gray level run-length
matrix.
Follow-up
Follow-up was mainly conducted through medical
records reviews and telephone interviews. The follow-up queries
included the following: Inquiring about postoperative survival
status (death or survival) of the patient; for patients who had
survived, inquiring about their general condition following
postoperative radiotherapy and chemotherapy; for patients who had
succumbed, inquiring about the specific cause of death; other
information was determined from the medical records of patient. OS
was defined as the period from the date of surgery to the date of
the last follow-up or death.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
(IBM Corp.). Among the variables obtained through the 3D Slicer
program, those that were normally distributed and demonstrating
homogenous variance were subjected to the independent samples
t-test. By contrast, those with non-normal distributions or
non-homogenous variance were subjected to the Mann-Whitney test, to
analyze the association between MGMT promoter methylation status
and glioblastoma MRI texture analysis. The Chi square test was
performed on the relationship between glioblastomas with the
methylated MGMT promoter and MRI features and location. P<0.05
was considered to indicate a statistically significant difference.
Binomial logistic regression analysis was then performed to further
analyze the predictive power of MRI features and texture analysis
heterogeneity for MGMT promoter methylation. Receiver operating
characteristic curves were generated to compare the diagnostic
performance between these two methods. In addition, Kaplan-Meier
univariate survival analysis was performed to analyze the factors
affecting glioblastoma prognosis. The corresponding survival curves
were plotted, and the log-rank test was performed. Finally,
multivariate analysis was conducted using the Cox proportional
hazards model to further analyze the relationship of the relevant
factors with prognosis.
Results
Relationship between MGMT promoter
methylation status and glioblastoma MRI texture features
Comparisons were made between glioblastomas with
methylated and unmethylated MGMT promoters using 31 texture
features extracted with 3D Slicer. Among those, seven texture
features showed statistically significant differences between the
two groups (P<0.05): Energy, Entropy, Uniformity,
Autocorrelation, Variance (GLCM), Gray Level Non-Uniformity (GLN),
and Cluster Shade (Table IA and
B).
 | Table IMRI texture analysis of MGMT
promoter methylation status and glioblastoma heterogeneity. |
Table I
MRI texture analysis of MGMT
promoter methylation status and glioblastoma heterogeneity.
| A, MRI texture
analysis of MGMT promoter methylation status and
glioblastoma heterogeneity. |
|---|
| | MRI texture
features | Methylated
MGMT promoter (n=79) | Unmethylated
MGMT promoter (n=49) | P-value |
|---|
| F1 | Sphericity | 0.372±0.106 | 0.391±0.116 | 0.505 |
| F2 | SRE | 0.268±0.103 | 0.285±0.097 | 0.479 |
| F3 | RP | 0.158±0.0470 | 0.173±0.054 | 0.227 |
| F4 | SRLGLE | 0.268±0.103 | 0.285±0.097 | 0.479 |
| F5 | Surface Area
mm^2 |
13969.933±9232.480 |
12279.785±7976.843 | 0.430 |
| F6 | Surface:Volume
Ratio | 0.497±0.208 | 0.538±0.266 | 0.474 |
| F7 | Compactness 1 | 30.676±13.806 | 29.745±17.756 | 0.807 |
| F8 | Maximum 3D
Diameter | 70.464±32.460 | 70.699±36.383 | 0.978 |
| Data are presented
as the mean ± standard deviation and were compared using the
independent samples t-test. MGMT,
O6-methylguanine DNA methyltransferase; MRI,
magnetic resonance imaging. |
| B, MRI texture
analysis of MGMT promoter methylation status and
glioblastoma heterogeneity. |
| | MRI texture
features | Methylated
MGMT promoter (n=79) | Unmethylated
MGMT promoter (n=49) | P-value |
| F9 | Energy | 39.38 | 28.97 | 0.033 |
| F10 | Entropy | 39.37 | 28.95 | 0.032 |
| F11 | Uniformity | 39.30 | 28.86 | 0.030 |
| F12 | Volume mm^3 | 36.17 | 33.84 | 0.585 |
| F13 | Volume cc | 36.13 | 33.45 | 0.584 |
| F14 | Compactness 2 | 33.70 | 36.80 | 0.527 |
| F15 | Spherical
Disproportion | 33.98 | 36.41 | 0.618 |
| F16 |
Autocorrelation | 39.54 | 29.80 | 0.035 |
| F17 | Cluster
Prominence | 39.40 | 28.86 | 0.300 |
| F18 | Cluster
Tendency | 39.43 | 28.90 | 0.310 |
| F19 | Difference
Entropy | 39.45 | 28.86 | 0.370 |
| F20 | Energy (GLCM) | 39.48 | 28.83 | 0.340 |
| F21 | Entropy (GLCM) | 30.55 | 41.14 | 0.390 |
| F22 | Homogeneity 1 | 39.42 | 28.98 | 0.330 |
| F23 | Sum Average | 39.46 | 28.80 | 0.350 |
| F24 | Sum Entropy | 30.55 | 41.14 | 0.360 |
| F25 | Sum Variance | 39.51 | 28.94 | 0.390 |
| F26 | Variance
(GLCM) | 39.49 | 28.50 | 0.031 |
| F27 | LRE | 37.18 | 32.00 | 0.290 |
| F28 | GLN | 39.50 | 28.79 | 0.029 |
| F29 | RLN | 37.78 | 31.17 | 0.177 |
| F30 | LRLGLE | 37.58 | 32.13 | 0.280 |
| F31 | Cluster Shade | 30.58 | 41.10 | 0.032 |
Relationship between MGMT promoter
methylation and tumor location
The analysis revealed that glioblastomas with
methylated MGMT promoters were predominantly located in the
temporal lobe (Table II), with
significant differences in the tumor location between the two
groups (P=0.039).
 | Table IIRelationship between MGMT
promoter methylation and tumor location. |
Table II
Relationship between MGMT
promoter methylation and tumor location.
| Molecular
phenotype | Temporal lobe
(%) | Frontal lobe
(%) | Other lobes
(%) | χ2 | P-value |
|---|
| MGMT | | | | 6.649 | 0.039 |
| Methylated
MGMT promoter | 41(52) | 18(23) | 20(25) | | |
| Unmethylated
MGMT promoter | 17(35) | 22(45) | 10(20) | - | - |
Binomial logistic regression model and
receiver operating characteristic (ROC) curve analysis
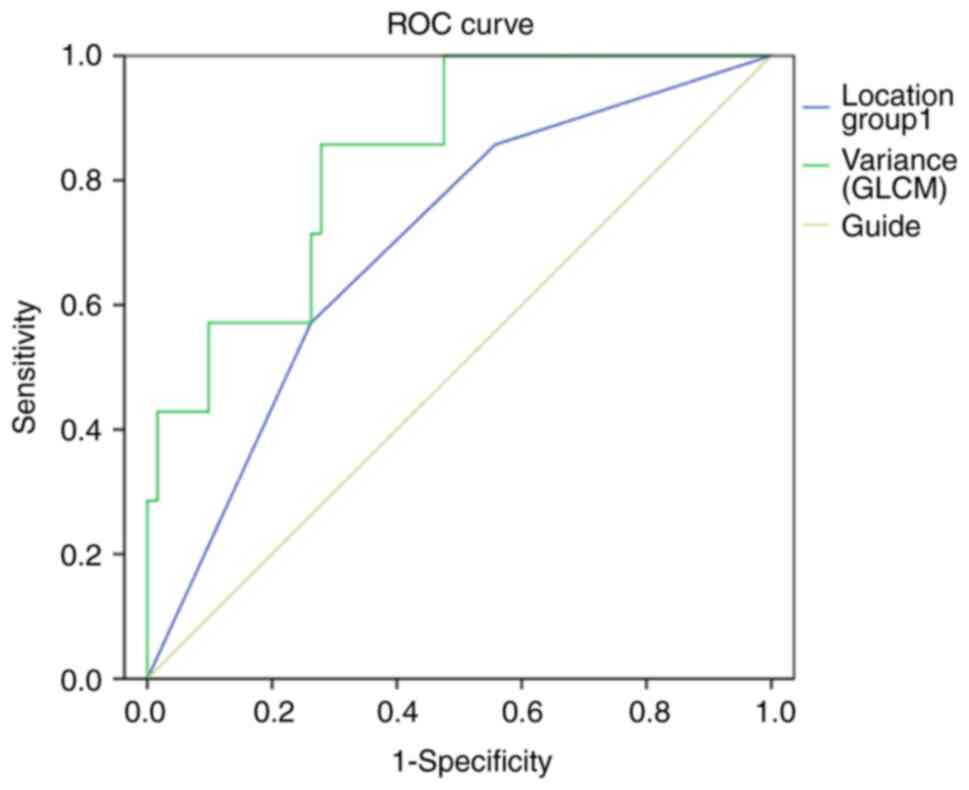
In the logistic regression model, glioblastomas with
methylated MGMT promoters were primarily located in the temporal
lobes [Model I, odds ratio (OR): 0.277, 95% confidence interval
(95% CI): 0.100-0.766, P=0.013], which showed an area under the
curve (AUC) of 0.679 (95% CI: 0.503-0.893). MRI texture analysis
showed a significant difference in Variance (GLCM) (Model II, OR:
1.68, 95% CI, 1.030-1.582, P=0.005), with an AUC of 0.838 (95% CI,
0.701-0.976). Consequently, Model II demonstrated significantly
improved diagnostic performance than that of Model I (Fig. 2, Table III).
 | Table IIIBinomial logistic regression model of
the MRI features and texture features of MGMT promoter
methylation. |
Table III
Binomial logistic regression model of
the MRI features and texture features of MGMT promoter
methylation.
| Model | Feature | OR (95% CI) | P-value | AUC (95% CI) |
|---|
| Model I: MRI
features | Tumor location
(frontal lobe/temporal lobe) | 0.277
(0.1000-0.766) | 0.013 | 0.679
(0.503-0.893) |
| Model II: MRI
texture features | F26: Variance
(GLCM) | 1.68
(1.030-1.582) | 0.005 | 0.838
(0.701-0.976) |
Factors affecting glioblastoma
prognosis
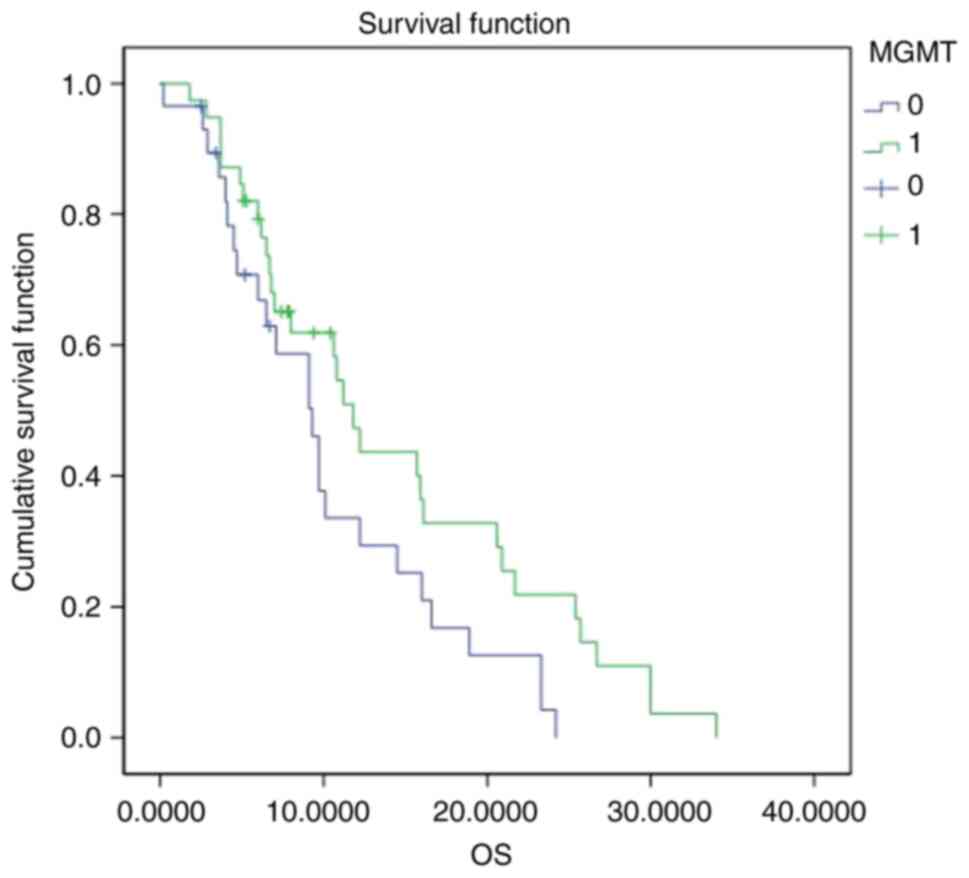
Kaplan-Meier univariate survival analysis results
are shown in Table IV. Briefly, a
significant difference in OS between glioblastomas with methylated
and unmethylated MGMT promoters was noticed (P=0.045; Fig. 3). The median survival for patients
with methylated MGMT promoters was 11.8 months, whereas for
patients with the unmethylated MGMT promoters the median survival
time was 9.3 months. Thus, glioblastoma with methylated MGMT
promoters had a significantly longer OS. When comparing patients
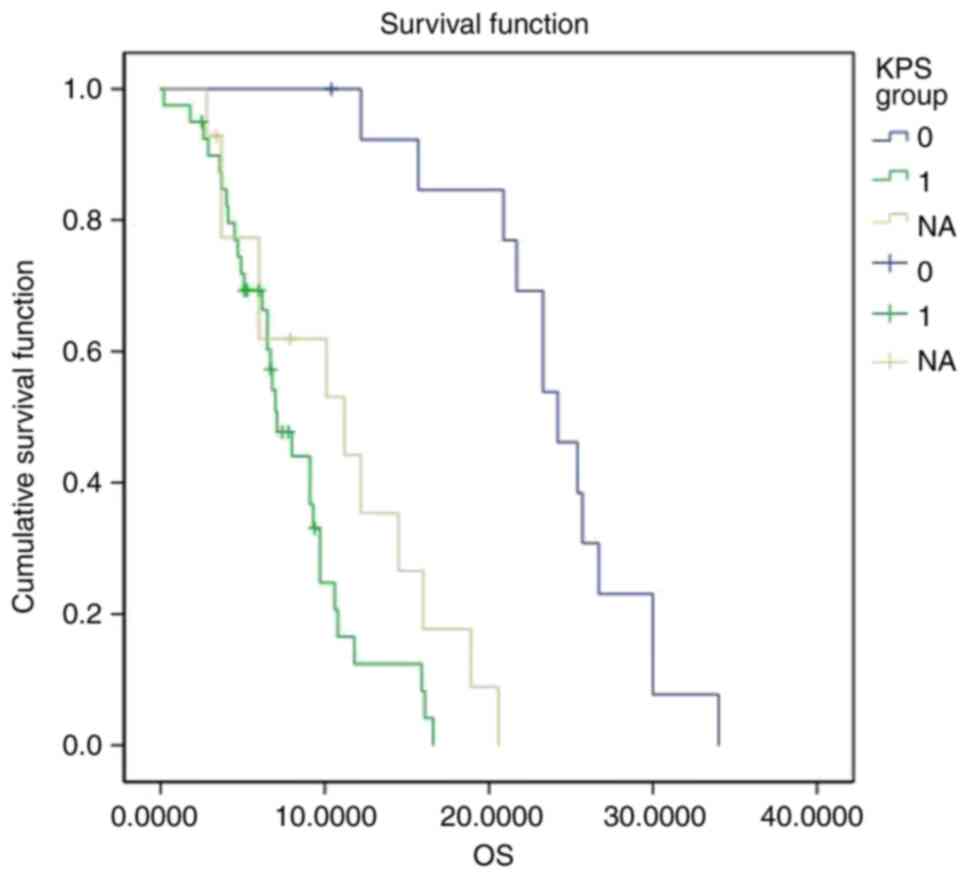
with preoperative KPS scores ≥80 to those with scores <80, the
median survival was 24.2 months for the former and 7.1 months for
the latter, with a statistically significant difference
(P<0.001). Thus, patients with preoperative KPS scores ≥80 had a
longer OS (Fig. 4). Among the
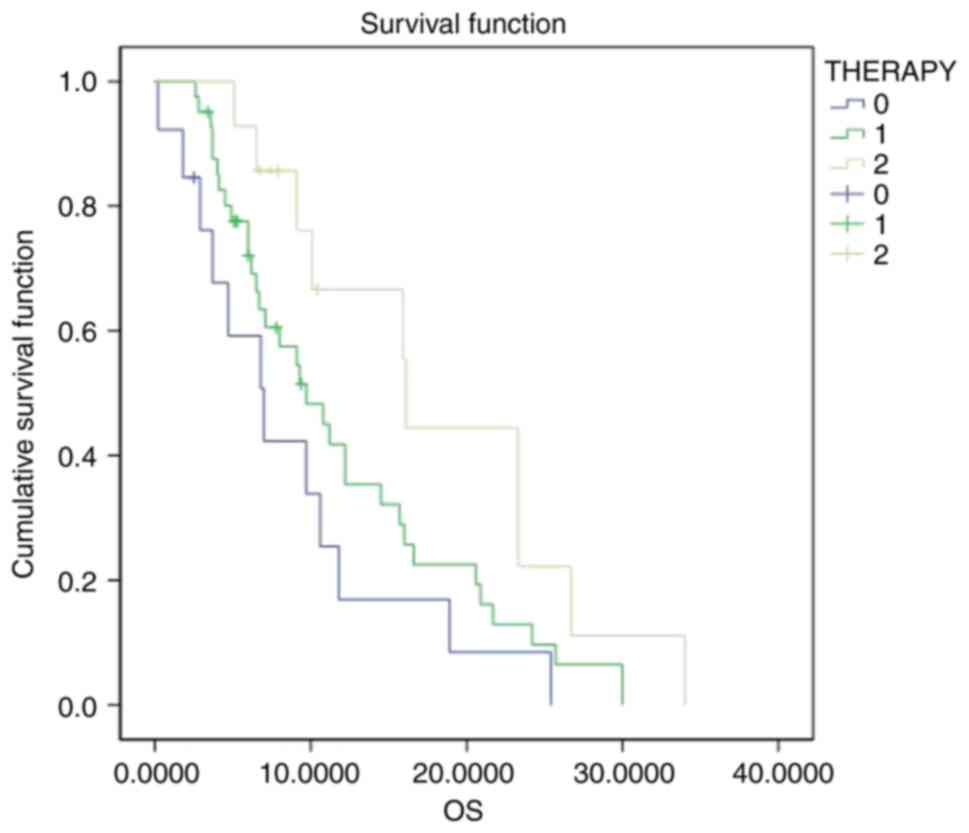
treatment groups, 33 patients received radiotherapy, 26 received
chemotherapy, and 69 received concurrent chemoradiotherapy. The
differences in OS among the three groups were statistically
significant (P=0.035), with a median survival of 7.0 months for
radiotherapy, 9.7 months for chemotherapy and 16.1 months for
concurrent chemoradiotherapy. Thus, patients who received
concurrent chemoradiotherapy had a significantly longer OS than
those who received either radiotherapy or chemotherapy alone
(Fig. 5). No significant
differences in OS were found based on age, sex, or glioblastoma
subtype.
 | Table IVFactors affecting glioblastoma
prognosis. |
Table IV
Factors affecting glioblastoma
prognosis.
| Factors | Number of
patients | Median
survival | SE | 95% CI | χ2 | P-value |
|---|
| MGMT
promoter | | | | | | |
|
Methylated | 79 | 11.8 | 1.656 | 11.131-17.264 | | |
|
Unmethylated | 49 | 9.3 | 1.367 | 7.732-13.090 | 4.019 | 0.045 |
| Age, years | | | | | | |
|
<60 | 52 | 9.1 | 2.063 | 8.557-16.644 | | |
|
≥60 | 76 | 10.8 | 1.334 | 10.287-15.515 | 0.231 | 0.631 |
| Sex | | | | | | |
|
Female | 58 | 9.7 | 1.324 | 8.954-14.145 | | |
|
Male | 70 | 12.2 | 1.920 | 10.383-17.910 | 1.103 | 0.294 |
| Preoperative KPS
score | | | | | | |
|
≥80 | 54 | 24.2 | 1.617 | 20.914-27.255 | | |
|
<80 | 68 | 7.1 | 0.727 | 6.583-9.433 | 34.163 | <0.001 |
|
NA | 6 | | | | | |
| Treatment
method | | | | | | |
|
Radiotherapy | 33 | 7.0 | 2.120 | 4.587-12.897 | | |
|
Chemotherapy | 26 | 9.7 | 1.369 | 9.446-14.814 | | |
|
Concurrent
chemoradiotherapy | 69 | 16.1 | 2.85 | 12.550-23.720 | 6.706 | 0.035 |
Multivariate survival analysis
Multivariate survival analysis using the Cox
proportional hazards model (Table
V) showed the following results: The difference in OS between
patients with preoperative KPS scores ≥80 and <80 was
significant (P=0.032), with a hazard ratio (HR) of 2.315 and a 95%
CI of 1.075-4.987. The OS among patients receiving different
postoperative adjuvant therapies (radiotherapy, chemotherapy, or
concurrent chemoradiotherapy) also showed a significant difference
(P=0.033), with an HR of 2.817 and a 95% CI of 1.086-7.306.
Patients receiving concurrent chemoradiotherapy had a significantly
longer OS than that of patients receiving only radiotherapy or
chemotherapy. No significant differences in glioblastoma prognosis
were observed based on MGMT promoter methylation, age and sex.
 | Table VFactors affecting glioblastoma
prognosis. |
Table V
Factors affecting glioblastoma
prognosis.
| Factors | n (%) | HR (95% CI) | P-value |
|---|
| MGMT
promoter | | | |
|
Methylated | 79 | References | |
|
Unmethylated | 49 | 1.630
(0.474,5.608) | 0.439 |
| Age, years | | | |
|
<60 | 52 | References | |
|
≥60 | 76 | 1.901 (0.991,
3.648) | 0.053 |
| Sex | | | |
|
Male | 58 | References | |
|
Female | 70 | 0.698
(0.377,1.292) | 0.252 |
| Preoperative KPS
score | | | |
|
<80 | 54 | References | |
|
≥80 | 68 | 2.315
(1.075,4.987) | 0.032 |
|
NA | 6 | - | - |
| Treatment
method | | | |
|
Radiotherapy | 33 | References | |
|
Chemotherapy | 26 | 2.010
(0.930,4.344) | 0.076 |
|
Concurrent
chemoradiotherapy | 69 | 2.817
(1.086,7.306) | 0.033 |
Discussion
Owing to the extremely high malignancy and high
invasiveness of glioblastoma, it has been difficult to achieve
satisfactory outcomes using radiotherapy, chemotherapy, or
concurrent chemoradiotherapy. Using the 3D Slicer, a visualization
and data analysis tool used in MRI studies, the MRI texture
features of glioblastoma with methylated and unmethylated MGMT
promoters were compared. The present results identified seven MRI
texture features [Energy, Entropy, Uniformity, Autocorrelation,
Variance (GLCM), GLN and Cluster Shade] that showed significant
differences between the two groups.
MGMT has been shown to be closely linked to
glioblastoma (4-11).
Different molecular phenotypes of glioblastoma can reflect the
different biological states and processes of the body (23). Current literature suggests that the
location of intracranial gliomas is related to the origin of the
tumor cell genetic phenotype, and hence there is some correlation
between tumor location and molecular phenotype (24). The current findings revealed a
significant difference in tumor location between glioblastomas with
methylated and unmethylated MGMT promoters, with the former
mostly located in the temporal lobe. This observation is consistent
with the results obtained by Li et al (25) and may be related to the origin and
genetic alterations of mutations in MGMT promoter
methylation. It mainly refers to texture features on T2-weighted
images assessed by the space-frequency analysis, which were
significantly different between methylated and unmethylated cases
by Drabycz et al (26). The
present retrospective study analyzed the correlation of MGMT
promoter methylation with glioblastoma MRI texture features and
prognosis. Although there are similarities between these two
studies, there are still some differences.
Previously, Kickingereder et al (27) demonstrated the correlation between
MRI texture features and molecular characteristics. However, the
reliability and accuracy of software analysis used in such studies
still require further improvement. In fact, only a few MRI studies
have analyzed glioblastoma heterogeneity, and currently, there is
no uniform standard among the software used. Currently available
software includes OsiriX, MaZda and 3D Slicer. Among these, OsiriX
is proprietary software dedicated to the Apple system, with special
requirements for the imported data type and device model. Although
MaZda is open-source software, it can only extract tumor
information on a two-dimensional level and delineate tumor
information layer by layer on images stored in the BMP format,
which inevitably leads to loss of tumor information. 3D Slicer is
one such open-source, freely available software used in the
analysis and interpretation of medical imaging data. Over the past
20 years, the US National Institutes of Health (NIH) has allocated
multiple grants for the creation of 3D Slicer, with the aim of
achieving powerful medical image processing capabilities. This
visualization and data analysis tool is extensible and has powerful
plug-in capabilities, enabling the addition of algorithms and
application programs. 3D Slicer is able to reconstruct the tumor
volume in three-dimensional space, thereby capturing all available
information about the tumor and minimizing the loss of tumor
information. Egger et al (28) found that using the 3D Slicer
semi-quantitative segmentation tool can improve the reliability of
glioblastoma segmentation, overcoming the effects of human factors,
and enabling the quantification of texture parameters while also
yielding more intuitive and accurate values. By utilizing the
Heterogeneity CAD extension module of 3D Slicer, 31 texture
features of tumor heterogeneity were extracted. A total of seven
MRI texture features with significant differences were identified
between glioblastomas with methylated and unmethylated MGMT
promoters. These features included Energy, Entropy, Uniformity,
Autocorrelation, Variance (GLCM), GLN and Cluster Shade.
MGMT promoters with variance showed particularly strong
diagnostic performance. Glioblastomas with methylated MGMT
promoters were frequently found in the temporal lobe. The present
study also found that patients with higher preoperative KPS scores
and those receiving concurrent chemotherapy have improved OS. Such
findings raise the hope that texture features extracted using 3D
Slicer can facilitate determining the MGMT promoter
methylation status in patients with glioblastomas. The main texture
features commonly used in clinic are energy, entropy and
autocorrelation. Among them, energy is mainly related to the
differences in the heterogeneity of tumor cells, which mainly
reflects the arrangement of tumor cells. Entropy is a widely used
texture feature in clinical practice and plays an important role in
the diagnosis and treatment of various malignant tumors. Texture
analysis based on T1WI-enhancement is expected to provide some help
in identifying the methylation status of MGMT promoter in
patients with GBM. By understanding the texture characteristics of
magnetic resonance, the limitations of human eyes in evaluating
tumors are overcome. At the same time, the results of texture
analysis were quantified by quantitative extraction of parameters
related to tumor heterogeneity.
Overall, the findings of the present study suggested
that MRI texture analysis is a valuable, non-invasive,
less-expensive method for detecting MGMT promoter
methylation and personalizing treatment, potentially improving
patient outcome. Despite providing such valuable information, this
was a single-center study with a small sample size. Therefore,
selection bias cannot be ruled out. Future studies should involve
multicenter cooperation to verify the results of the present
study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the 2021 Hebei
Medical Science Research Project Program (grant no. 20210447).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RW collected and analyzed data, post-processed
imaging data, performed literature review and follow-up, and wrote
the manuscript. ZS and HW confirm the authenticity of all the raw
data, designed the project and revised the manuscript. JS and MM
analyzed imaging data and performed statistical analysis. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The Institutional Review Board of the Ethics
committee at the Tangshan Gongren Hospital (Tangshan, China)
granted an exemption to obtain informed consent due to the use of
anonymous data (approval no. GRYY-LL-2020-44). The Ethics Committee
exempted informed consent because of the retrospective nature of
the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO classification of tumors of the
central nervous system: A summary. Neuro-Oncology. 23:1231–1251.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ostrom QT, Gittleman H, Fulop J, Liu M,
Blanda R, Kromer C, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 17
(Suppl):iv1–iv62. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Della Monica R, Cuomo M, Buonaiuto M,
Costabile D, Franca RA, Del Basso De Caro M, Catapano G, Chiariotti
L and Visconti R: MGMT and whole-genome DNA methylation impacts on
diagnosis, prognosis and therapy of glioblastoma multiforme. Int J
Mol Sci. 23(7148)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Teske N, Karschnia P, Weller J, Siller S,
Dorostkar MM, Herms J, von Baumgarten L, Tonn JC and Thon N:
Extent, pattern, and prognostic value of MGMT promotor methylation:
Does it differ between glioblastoma and IDH-wildtype/TERT-mutated
astrocytoma? J Neurooncol. 156:317–327. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Min TL, Allen JW, Velazquez Vega JE, Neill
SG and Weinberg BD: MRI imaging characteristics of glioblastoma
with concurrent gain of chromosomes 19 and 20. Tomography.
7:228–237. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pease M, Gersey ZC, Ak M, Elakkad A,
Kotrotsou A, Zenkin S, Elshafeey N, Mamindla P, Kumar VA, Kumar AJ,
et al: Pre-operative MRI radiomics model non-invasively predicts
key genomic markers and survival in glioblastoma patients. J
Neurooncol. 160:253–263. 2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang M, Chen HZ, Cui YY, Zhang ZZ and Ma
XD: The associations between preoperative conventional MRI features
and genetic biomarker status in newly diagnosed GBMs: A clinical
summary and prognostic analysis. Turk Neurosurg. 31:880–887.
2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Do DT, Yang MR, Lam LHT, Le NQK and Wu YW:
Improving MGMT methylation status prediction of glioblastoma
through optimizing radiomics features using genetic algorithm-based
machine learning approach. Sci Rep. 12(13412)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Verduin M, Primakov S, Compter I, Woodruff
HC, van Kuijk SMJ, Ramaekers BLT, te Dorsthorst M, Revenich EGM,
ter Laan M, Pegge SAH, et al: Prognostic and predictive value of
integrated qualitative and quantitative magnetic resonance imaging
analysis in glioblastoma. Cancers (Basel). 13(722)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Safaei R, Mojtahedi H, Hanaei S, Razavi A,
Esmaeili M, Sadr M, Rezaei A, Edalatfar M, Kashani HK,
Sadeghi-Naini M, et al: MGMT gene rs1625649 polymorphism in Iranian
patients with brain glioblastoma: A case control study. Avicenna J
Med Biotechnol. 15:48–52. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kitange GJ, Mladek AC, Carlson BL,
Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta
SK, et al: Inhibition of histone deacetylation potentiates the
evolution of acquired temozolomide resistance linked to MGMT
upregulation in glioblastoma xenografts. Clin Cancer Res.
18:4070–4079. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Blakstad H, Brekke J, Rahman MA, Arnesen
VS, Miletic H, Brandal P, Lie SA, Chekenya M and Goplen D: Survival
in a consecutive series of 467 glioblastoma patients: Association
with prognostic factors and treatment at recurrence at two
independent institutions. PLoS One. 18(e0281166)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kurdi M, Shafique Butt N, Baeesa S,
Alghamdi B, Maghrabi Y, Bardeesi A, Saeedi R, Al-Sinani T, Alghanmi
N, Bari MO, et al: The impact of IDH1 mutation and MGMT promoter
methylation on recurrence-free interval in glioblastoma patients
treated with radiotherapy and chemotherapeutic agents. Pathol Oncol
Res. 27(1609778)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Haque W, Thong E, Andrabi S, Verma V,
Butler BE and the BS: Prognostic and predictive impact of MGMT
promoter methylation in grade 3 gliomas. J Clin Neurosci.
85:115–121. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Davnall F, Yip CS, Ljungqvist G, Selmi M,
Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ and Goh V:
Assessment of tumor heterogeneity: An emerging imaging tool for
clinical practice? Insights Imaging. 3:573–589. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ladenhauf VK, Galijasevic M, Kerschbaumer
J, Freyschlag CF, Nowosielski M, Birkl-Toeglhofer AM, Haybaeck J,
Gizewski ER, Mangesius S and Grams AE: Peritumoral ADC values
correlate with the MGMT methylation status in patients with
glioblastoma. Cancers (Basel). 15(1384)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Qureshi SA, Hussain L, Ibrar U,
Alabdulkreem E, Nour MK, Alqahtani MS, Nafie FM, Mohamed A,
Mohammed GP and Duong TQ: Radiogenomic classification for MGMT
promoter methylation status using multi-omics fused feature space
for least invasive diagnosis through mpMRI scans. Sci Rep.
13(3291)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Eccles SA and Welch DR: Metastasis: Recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Saxena S, Jena B, Mohapatra B, Gupta N,
Kalra M, Scartozzi M, Saba L and Suri JS: Fused deep learning
paradigm for the prediction of O6-methylguanine-DNA
methyltransferase genotype in glioblastoma patients: A
neuro-oncological investigation. Comput Biol Med.
153(106492)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Choi HJ, Choi SH, You SH, Yoo RE, Kang KM,
Yun TJ, Kim JH, Sohn CH, Park CK and Park SH: MGMT promoter
methylation status in initial and recurrent glioblastoma:
Correlation study with DWI and DSC PWI features. AJNR Am J
Neuroradiol. 42:853–860. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yaltirik CK, Yilmaz SG, Ozdogan S, Bilgin
EY, Barut Z, Ture U and Isbir T: Determination of IDH1, IDH2, MGMT,
TERT and ATRX gene mutations in glial tumors. In Vivo.
36:1694–1702. 2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ius T, Pignotti F, Della Pepa GM, Bagatto
D, Isola M, Battistella C, Gaudino S, Pegolo E, Chiesa S, Arcicasa
M, et al: Glioblastoma: From volumetric analysis to molecular
predictors. J Neurosurg Sci. 66:173–186. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sanai N, Alvarez-Buylla A and Berger MS:
Neural stem cells and the origin of gliomas. N Engl J Med.
353:811–822. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li HY, Sun CR, He M, Yin LC, Du HG and
Zhang JM: Correlation between tumor location and clinical
properties of glioblastomas in frontal and temporal lobes. World
Neurosurg. 112:e407–e414. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Drabycz S, Roldán G, Robles P, Adler D,
Mcintyre J, Magliocco A, Cairncross JG and Mitchell JR: An analysis
of image texture, tumor location, and MGMT promoter methylation in
glioblastoma using magnetic resonance imaging. Neuroimage.
49:1398–1405. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kickingereder P, Bonekamp D, Nowosielski
M, Kratz A, Sill M, Burth S, Wick A, Eidel O, Schlemmer HP,
Radbruch A, et al: Radiogenomics of glioblastoma: Machine
learning-based classification of molecular characteristics by using
multiparametric and multiregional MR imaging features. Radiology.
281:907–918. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Egger J, Kapur T, Fedorov A, Pieper S,
Miller JV, Veeraraghavan H, Freisleben B, Golby AJ, Nimsky C and
Kikinis R: GBM volumetry using the 3D slicer medical image
computing platform. Sci Rep. 3(1364)2013.PubMed/NCBI View Article : Google Scholar
|