Introduction
As one class of newly identified regulatory
molecules, miRNAs are important in numerous critical cell
processes, including cell proliferation, differentiation and
apoptosis (1–3). It has been found that ~30% of the
mammalian genome is regulated by these short endogenous RNAs. As
for their functions, it is widely recognized that microRNAs
(miRNAs) are able to integrate with specific target mRNAs, forming
close complementary structures and resulting in either mRNA
degradation or inhibition of protein translation (4). In addition, miRNAs alter gene
expression mainly through their effects on the methylation of genes
or by the targeting of transcription factors significant to cell
physiology (5,6).
Over the last several years, studies have
demonstrated that these tiny regulators are able to tune organ
development (7). However, few of
those involved the physiological or pathological mechanisms of the
lungs, particularly in mammalian fetal lung development, have been
identified. The lung has a specific stable miRNA expression
profile, which is conserved across mammalian species (8,9).
Therefore, in theory it is comparable among different species.
However, over the past several years, the number of expression
profile studies on fetal development in mammals has been relatively
small and these studies have been mostly confined to a few miRNAs,
including the Let-7 family (10,11)
and the miRNA-17-92 cluster (12,13).
However, with the improvement of chip technology and the deepening
understanding of the developmental mechanism, we are able to
identify more of the significant miRNAs involved in fetal lung
development using the latest microarrays. Therefore, in the present
study, we used the miRCURY LNA™ microRNA Array (v.16.0) (containing
>1891 probes) to determine expression profiles for fetal lung
development at three key time points to identify new miRNAs. We
then selected certain miRNAs for further study in order to attempt
to clarify the possible mechanism involved.
Materials and methods
Rats
Healthy adult Sprague-Dawley rats (12 female and 12
male) were maintained in a specific-pathogen-free animal facility
at the Animal Center of Nanjing Medical University. In this study,
3 time points (gestational days 16, 19 and 21) representing 3
stages of fetal lung development were selected. The 3 time points
were designated as groups S1 (E21), S2 (E19) and S3 (E16). For each
group, 4 pregnant rats were selected according to the random
contrast rule.
The pregnant rats were sacrificed with
CO2 and whole fetal lungs were immediately isolated from
the fetuses. The superfluous extraneous tissues were then removed
as thoroughly as possible. The procedures in this study followed
the protocols approved by the Nanjing Medical University Animal
Care and Use Committee.
Following isolation, a randomly selected fetal lung
was retained for histological observation. Subsequently, the total
RNA of the remaining lungs was isolated using TRIzol (Invitrogen
Life Technologies, Carlsbad, CA, USA) and the miRNeasy mini kit
(Qiagen, Hilden, Germany) according to the manufacturer’s
instructions. RNA quality and quantity were measured using a
NanoDrop spectrophotometer (ND-1000, Nanodrop Technologies,
Wilmington, DE, USA) and RNA integrity was evaluated by gel
electrophoresis.
Histology
The lung tissues of the 3 groups were fixed with 4%
paraformaldehyde solution, embedded in paraffin and cut into 4 μm
continuous sections. The sections were stained with hematoxylin and
eosin (H&E) for subsequent morphological observation. H&E
sections were then viewed by optical microscopy at ×40
magnification to observe the structural changes of the fetal lungs
in the 3 groups (S1, S2 and S3).
miRNA microarray
Following the isolation of the RNA from the fetal
lungs, the miRCURY Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek,
Denmark) was used according to the manufacturer’s guidelines for
miRNA labeling. Each sample (1 μg) was 3′-end-labeled with a Hy3™
fluorescent label using T4 RNA ligase by the following steps. RNA
in 2.0 μl water was combined with 1.0 μl CIP buffer and CIP
(Exiqon). The mixture was incubated for 30 min at 37°C and then the
reaction was terminated by incubation at 95°C for 5 min. Then 3.0
μl labeling buffer, 1.5 μl fluorescent label (Hy3™), 2.0 μl DMSO
and 2.0 μl labeling enzyme were added to the mixture. The labeling
reaction was incubated for 1 h at 16°C and the reaction was
terminated by incubation at 65°C for 15 min.
The Hy3™-labeled miRNA samples were hybridized using
the miRCURY LNA™ microRNA array system (v.16.0) (Exiqon) according
to the array manual. The total 25 μl mixture of Hy3™-labeled
samples in 25 μl hybridization buffer was denatured for 2 min at
95°C, incubated on ice for 2 min and then hybridized to the
microarray for 16–20 h at 56°C in the 12-Bay hybridization system
(Nimblegen Systems, Inc., Madison, WI, USA), which provides an
active mixing action and constant incubation temperature to improve
hybridization uniformity and enhance the signals. Following
hybridization, the slides were washed several times using a wash
buffer kit (Exiqon) and dried by centrifugation for 5 min at 400
rpm. The slides were scanned using the Axon GenePix 4000B
microarray scanner (Axon Instruments, Foster City, CA, USA).
The scanned images were imported into GenePix Pro
6.0 software (Axon Instruments) for grid alignment and data
extraction. Replicated miRNAs were averaged and miRNAs that had
intensities >50 in all samples were selected for calculation of
the normalization factor. The expressed data were normalized by
median normalization. Following normalization, the differentially
expressed miRNAs were identified through fold-change filtering
(fold change ≥1.0).
Scatter-plots and correlation coefficient matrices
which were visualizations used to assess the variation (or
reproducibility) between chips were prepared (Fig. 1 and Table I). The axes of the scatter-plot are
the normalized signal values of the samples (ratio scale).
 | Table ICorrelation coefficient matrix among 3
microarrays (groups S1, S2 and S3). |
Table I
Correlation coefficient matrix among 3
microarrays (groups S1, S2 and S3).
| Group | S1 | S2 | S3 |
|---|
| S1 | 1.0000 | 0.7084 | 0.5440 |
| S2 | 0.7084 | 1.0000 | 0.8600 |
| S3 | 0.5440 | 0.8600 | 1.0000 |
Quantitative real-time PCR
Total RNA was isolated from fetal lungs using the
TRIzol reagent. Single-strand cDNA was synthesized using a reverse
transcription mixture that contained 1 μg total RNA, 0.3 μl
rno-miRNA reverse primer (1 μM) (Table II), 0.1 μl MMLV revertase (200
U/μl; Epicentre, Madison, WI, USA), 2 μl 10× RT buffer, 2 μl dNTP
mix (2.5 mM each) and 0.3 μl ribonuclease inhibitor (40 U/μl) in a
20 μl total volume. The reaction was performed at 16°C for 30 min
and at 42°C for 40 min, followed by heat inactivation at 85°C for 5
min. For real-time PCR, 1 μl cDNA was added to 24 μl master mix
containing 2.5 μl dNTP (2.5 mM each), 2.5 μl 10× PCR buffer
(Promega, Madison, WI, USA), 1 unit Taq polymerase (Promega), final
concentration 0.25× SYBR-Green I (Invitrogen) and 2 μl reverse and
forward primers. cDNA was then amplified for 35 cycles using the
Applied Rotor-Gene 3000 (Corbett Research, Sydney, Australia)
real-time PCR system. The primer sequences used are listed in
Table III. RT and PCR for U6
snRNA were performed in each plate as an endogenous control. The
amount of PCR product was calculated from the threshold cycle (Ct).
In addition, the comparative CT method was used, and the
relative amount of miRNA to U6 snRNA was calculated with the
equation 2−(Ct microRNA - Ct U6).
 | Table IIRT primer sequences. |
Table II
RT primer sequences.
| Gene | RT primers
(5′-3′) |
|---|
| U6 |
CGCTTCACGAATTTGCGTGTCAT |
| rno-miR-126* |
GTCGTATCCAGTGCGTGTCGTGGA
GTCGGCAATTGCACTGGATACGAC CGCGTA |
 | Table IIIPrimers for real-time RT-PCR. |
Table III
Primers for real-time RT-PCR.
| Gene | Primers
(5′-3′) | T (°C) |
|---|
| U6 | F:
GCTTCGGCAGCAC | |
| ATATACTAAAAT | |
| R:
CGCTTCACGAAT | 60 |
| TTGCGTGTCAT | |
| rno-miR-126* | GSP: GGGCATTAT | |
| TACTTTTGG | |
| R: TGCGTGTC | 60 |
| GTGGAGTC | |
Statistical analysis
Data were analyzed using the SPSS 13.0 statistical
package and EXCEL 2003. The real-time PCR data were evaluated by
one-way ANOVA. P<0.05 was considered to indicate a statistically
significant result.
Results
Histology
In group S3 (E16), the epithelial cells had
differentiated into original acinar-like structures. The structure
of the interstitial tissue was extremely dense (Fig. 2A), while in group S2 (E19), the
acinus cavities had expanded rapidly. The interstitium was arranged
in cords and appeared to be thinner than in E16 (Fig. 2B); in group S1 (E21), more mature
alveolars emerged and were arranged around the bronchioles. The
interstitium was sparser than in the previous groups (Fig. 2C).
miRNA expression profile
The 6th generation miRCURY LNA™ microRNA array
(v.16.0) that we used covers all mouse, rat, human and virus
genomes. As a result, 202 differentially expressed miRNAs from the
3 groups (S1, S2 and S3) passed the fold-change filtering (fold
change ≥1.0), including 4 different types of expression patterns
(S2/S3 ↑or↓ and S1/S2 ↑or↓) (Table
IV).
 | Table IVAll differentially expressed miRNAs
among the 3 groups (S1, S2 and S3), including 4 expression patterns
(S2/S3 ↑or↓ and S1/S2 ↑or↓). |
Table IV
All differentially expressed miRNAs
among the 3 groups (S1, S2 and S3), including 4 expression patterns
(S2/S3 ↑or↓ and S1/S2 ↑or↓).
| S3-S2↑, S2-S1↑
(S2/S3>1 and S1/S2>1)a | S3-S2↓, S2-S1↓
(S2/S3<1 and S1/S2<1)a | S3-S2↑, S2-S1↓
(S2/S3>1 and S1/S2>1)a | S3-S2↓, S2-S1↑
(S2/S3<1 and S1/S2<1)a |
|---|
|
|
|
|
|---|
| Name | Fold changes | Name | Fold changes | Name | Fold changes | Name | Fold changes |
|---|
| rno-miR-3560 | 8.4211415 | 4.888905 | rno-miR-186* | 0.980325 | 0.68844749 | rno-miR-195 | 13.549629 | 0.9854888 | rno-miR-let-7b | 0.99745790 | 1.470588872 |
| rno-miR-126* | 7.5239524 | 1.511816 | rno-miR-466c* | 0.977220 | 0.87722704 | rno-miR-34a | 12.426133 | 0.6040662 |
rno-miR-466b-1* | 0.99315310 | 1.732802323 |
| rno-miR-672* | 5.2601791 | 1.175347 | rno-miR-19a | 0.971818 | 0.62082771 | rno-miR-347 | 10.659673 | 0.6544392 | rno-miR-107 | 0.95708310 | 1.453658569 |
| rno-miR-146b | 5.0163166 | 3.094662 | rno-miR-25 | 0.963684 | 0.57179299 | rno-miR-503* | 7.3622807 | 0.2163902 |
rno-miR-291a-5p | 0.94483460 | 1.724463405 |
| rno-miR-34c | 4.5061344 | 1.957254 | rno-miR-144 | 0.951005 | 0.64339048 | rno-miR-495 | 6.0282288 | 0.3506531 | rno-miR-3572 | 0.93592810 | 1.020299247 |
| rno-miR-653* | 3.4649380 | 1.764355 | rno-miR-329 | 0.923679 | 0.93020848 | rno-miR-10a-3p | 4.0970221 | 0.0661559 | rno-miR-883* | 0.92618030 | 2.217677800 |
| rno-miR-411* | 3.3178232 | 1.590243 | rno-miR-331 | 0.903534 | 0.02509753 | rno-miR-30c-1* | 3.8809911 | 0.5900630 |
rno-miR-466b-2* | 0.92215430 | 1.223710712 |
| rno-miR-let-7a | 3.1785206 | 2.289511 | rno-miR-19b | 0.900471 | 0.72076563 | rno-miR-24-2* | 3.8646984 | 0.9495285 | rno-miR-375 | 0.91535550 | 1.059154957 |
| rno-miR-let-7d | 2.9136439 | 1.452478 | rno-miR-151* | 0.892133 | 0.39417882 | rno-miR-339-5p | 3.1913082 | 0.3121446 | rno-miR-466c | 0.89813410 | 1.398870274 |
| rno-miR-26b | 1.9370148 | 1.723152 | rno-miR-451 | 0.878429 | 0.25339903 | rno-miR-379 | 3.1248495 | 0.4766527 | rno-miR-770* | 0.89243770 | 1.651569420 |
| rno-miR-874 | 1.8857111 | 2.413461 | rno-miR-21* | 0.874866 | 0.65841255 | rno-miR-672 | 3.0738620 | 0.9356720 | rno-miR-200c | 0.87946140 | 1.146764503 |
|
rno-miR-1188-3p | 1.7935323 | 2.298813 | rno-miR-7a-2* | 0.852774 | 0.38291656 | rno-miR-376-3p | 3.0653762 | 0.3206239 | rno-miR-300-5p | 0.87661250 | 1.733430021 |
| rno-miR-23a | 1.7799642 | 1.206440 | rno-miR-652 | 0.846386 | 0.71646338 | rno-miR-136* | 2.8064011 | 0.7382026 | rno-miR-325-3p | 0.86575560 | 1.913369777 |
| rno-miR-329* | 1.7775597 | 1.262534 | rno-miR-186 | 0.822947 | 0.05419442 | rno-miR-127* | 2.7881470 | 0.8166521 |
rno-miR-3573-3p | 0.85555160 | 2.199396790 |
| rno-miR-30d | 1.7484931 | 3.044324 | rno-miR-106b | 0.817602 | 0.50922622 | rno-miR-322 | 2.7181080 | 0.5153748 | rno-miR-210 | 0.85130860 | 1.444849465 |
| rno-miR-138-2* | 1.7377907 | 1.740641 | rno-miR-204* | 0.797339 | 0.5891234 | rno-miR-409-5p | 2.7004505 | 0.2778741 | rno-miR-30a | 0.83405280 | 4.448185340 |
| rno-miR-30b-5p | 1.7229757 | 1.254918 | rno-miR-20a | 0.790606 | 0.33531233 | rno-miR-16 | 2.4124438 | 0.5187975 | rno-miR-30e* | 0.82078630 | 1.911407722 |
| rno-miR-26a | 1.7208078 | 3.452369 | rno-miR-99a | 0.785670 | 0.28761068 | rno-miR-194* | 2.3146718 | 0.0961872 | rno-miR-433* | 0.78441850 | 1.462433489 |
| rno-miR-let-7c | 1.6864176 | 1.272732 | rno-miR-345-5p | 0.732098 | 0.79662712 | rno-miR-15b | 2.2900781 | 0.2570223 | rno-miR-30c | 0.78415490 | 1.641928548 |
| rno-miR-743b | 1.6569246 | 1.746423 | rno-miR-106b* | 0.721485 | 0.86241946 | rno-miR-365 | 2.1816797 | 0.2143063 | rno-miR-200a | 0.77613210 | 1.728426011 |
| rno-miR-352 | 1.6529537 | 2.337057 | rno-miR-23b | 0.712766 | 0.97392883 | rno-miR-185 | 2.1641823 | 0.1573393 |
rno-miR-344b-2-3p | 0.77200260 | 1.214345429 |
| rno-miR-539* | 1.6424152 | 2.621551 | rno-miR-674-5p | 0.695417 | 0.80928285 | rno-miR-98 | 2.1485470 | 0.8302507 | rno-miR-3596c | 0.73287290 | 1.961526568 |
| rno-miR-136 | 1.6394235 | 2.305536 | rno-miR-431 | 0.690826 | 0.29360741 | rno-miR-328a | 2.0544097 | 0.0597691 | rno-miR-551b* | 0.70730060 | 4.873483536 |
| rno-miR-138-1* | 1.4810756 | 1.463674 | rno-miR-152 | 0.682139 | 0.75124564 | rno-miR-448* | 2.0526945 | 0.4748063 | rno-miR-500 | 0.69860000 | 1.352811809 |
| rno-miR-376a | 1.4443813 | 1.631507 | rno-miR-758* | 0.677330 | 0.72947047 | rno-miR-335 | 1.9220501 | 0.6855958 | rno-miR-450a* | 0.69203410 | 3.284467450 |
| rno-miR-664 | 1.4361529 | 1.661272 |
rno-miR-344b-1-3p | 0.660900 | 0.51092973 | rno-miR-466b | 1.7737696 | 0.5701351 | rno-miR-300-3p | 0.67563510 | 1.421903839 |
| rno-miR-127 | 1.4246079 | 1.449653 | rno-miR-664-1* | 0.656291 | 0.33467595 | rno-miR-200b | 1.7686778 | 0.8490812 | rno-miR-675 | 0.66297150 | 1.881596970 |
| rno-miR-145 | 1.4116241 | 1.584110 | rno-miR-17-5p | 0.651909 | 0.71146565 |
rno-miR-199a-3p | 1.7519083 | 0.3819046 | rno-miR-667 | 0.66158250 | 1.851774707 |
| rno-miR-207 | 1.3967711 | 1.344485 | rno-miR-183 | 0.645298 | 0.06785863 | rno-miR-429 | 1.7345297 | 0.6532936 | rno-miR-101a | 0.63908846 | 1.599289011 |
| rno-miR-331* | 1.3332774 | 3.647136 | rno-miR-298 | 0.622817 | 0.32752771 | rno-miR-206 | 1.6451149 | 0.5619425 | rno-miR-668 | 0.62312700 | 1.546920336 |
| rno-miR-323 | 1.3300142 | 1.247038 |
rno-miR-199a-5p | 0.619426 | 0.42597925 | rno-miR-665 | 1.5267099 | 0.2914475 | rno-miR-183* | 0.61654370 | 2.938326520 |
| rno-miR-294 | 1.2375557 | 1.770187 | rno-miR-181d | 0.618991 | 0.30448551 |
rno-miR-17-1-3p | 1.4533188 | 0.2164874 | rno-miR-181a | 0.60586140 | 1.027205203 |
| rno-miR-21 | 1.2271042 | 1.679205 | rno-miR-140* | 0.612127 | 0.17284846 | rno-miR-99b* | 1.4381097 | 0.9020220 | rno-miR-22* | 0.59792190 | 1.458050402 |
| rno-miR-32* | 1.2216227 | 1.257030 | rno-miR-218 | 0.610725 | 0.02906206 | rno-miR-30e | 1.4249814 | 0.9009271 | rno-miR-3571 | 0.59185380 | 1.660526533 |
| rno-miR-378 | 1.2187415 | 1.174652 | rno-miR-140 | 0.602168 | 0.17782928 | rno-miR-664-2* | 1.3842126 | 0.0870265 | rno-miR-24-1* | 0.58317060 | 2.889240932 |
|
rno-miR-let-7d* | 1.1706845 | 1.128949 | rno-miR-505* | 0.546060 | 0.7634096 | rno-miR-142-3p | 1.3621145 | 0.6892047 | rno-miR-145* | 0.55848840 | 1.061437866 |
| rno-miR-134* | 1.1520524 | 2.038411 | rno-miR-320 | 0.535097 | 0.53630321 | rno-miR-423 | 1.3421092 | 0.7849327 | rno-miR-99b | 0.52918460 | 1.484742511 |
| rno-miR-742 | 1.1370318 | 2.165993 | rno-miR-361 | 0.521556 | 0.06827078 | rno-miR-let-7i | 1.3092522 | 0.4607812 | rno-miR-872* | 0.50684120 | 3.472207096 |
| rno-miR-465 | 1.1331676 | 1.424808 | rno-miR-483 | 0.518141 | 0.32997545 | rno-miR-410 | 1.3015181 | 0.7325175 | rno-miR-30b-3p | 0.50448300 | 1.170676650 |
| rno-miR-340-5p | 1.1306514 | 1.900459 | rno-miR-191 | 0.507004 | 0.14680991 | rno-miR-490 | 1.2756753 | 0.7856162 | rno-miR-877 | 0.48686730 | 1.339451220 |
| rno-miR-34b* | 1.0914578 | 2.423233 | rno-miR-434 | 0.504917 | 0.86745761 | rno-miR-466d | 1.2712970 | 0.9663230 | rno-miR-423* | 0.44084070 | 1.653788675 |
| rno-mir-141 | 1.0728281 | 1.002083 | rno-miR-485* | 0.498218 | 0.56491937 | rno-miR-193 | 1.2541245 | 0.7348141 | rno-miR-541 | 0.42540900 | 1.207167473 |
| rno-mir-181b | 1.0632784 | 1.197243 | rno-miR-214 | 0.451194 | 0.59062997 | rno-miR-22 | 1.1550101 | 0.7615345 | rno-miR-124 | 0.39048060 | 2.030618140 |
| rno-mir-27b | 1.0548732 | 1.005943 | rno-miR-185* | 0.385377 | 0.74238926 | rno-miR-374 | 1.1515834 | 0.6490015 | rno-miR-351 | 0.38371410 | 3.656196612 |
| rno-mir-465* | 1.0445459 | 2.298723 | rno-miR-1949 | 0.381056 | 0.42356006 | rno-miR-2985 | 1.1071099 | 0.9055459 | rno-miR-369-3p | 0.37871630 | 2.411181539 |
| rno-mir-382 | 1.0444827 | 1.091136 | rno-miR-130a | 0.374817 | 0.84478535 | rno-miR-301a | 1.0911573 | 0.4289909 |
rno-miR-125a-5p | 0.34660480 | 1.113517147 |
| rno-mir-503 | 1.0266687 | 1.160400 | rno-miR-93 | 0.359659 | 0.24274816 | rno-miR-142-5p | 1.0873938 | 0.9302832 | rno-miR-103-1* | 0.31836890 | 1.464945944 |
| | | rno-miR-130b | 0.353169 | 0.84478535 | rno-miR-205 | 1.0665678 | 0.0960427 |
rno-miR-1193-3p | 0.29953480 | 5.522615564 |
| | |
rno-miR-125b-5p | 0.350006 | 0.27382858 | rno-miR-92b | 1.0280013 | 0.3149766 | rno-miR-33 | 0.29346730 | 1.899358020 |
| | | rno-miR-9* | 0.304562 | 0.78517235 | rno-miR-126 | 1.0129183 | 0.9994663 | rno-miR-341 | 0.19430920 | 1.452073739 |
| | | rno-miR-296 | 0.248001 | 0.31706143 | rno-miR-let-7e | 1.0102800 | 0.9453700 | rno-miR-542-3p | 0.16812910 | 2.681449923 |
| | | rno-miR-101b | 0.221090 | 0.94937991 | | | |
rno-miR-148b-3p | 0.09409170 | 2.009611745 |
In addition, in view of their classic status in lung
development, we also selected some representative members of the
rno-Let-7 family and rno-miRNA-17-92 cluster from the
differentially expressed miRNAs (Table IV) to generate line charts
reflecting their expression trends during fetal lung development
(Fig. 3). Compared with these
intensely investigated miRNAs, certain other miRNAs, including
miRNA-126* and miRNA-126, have been scarcely reported on and
studied in lung development. However, both have been revealed to be
highly enriched in mouse embryonic stem cells (15). Additionally, more than one study
has reported their significance in the vascular endothelial cells
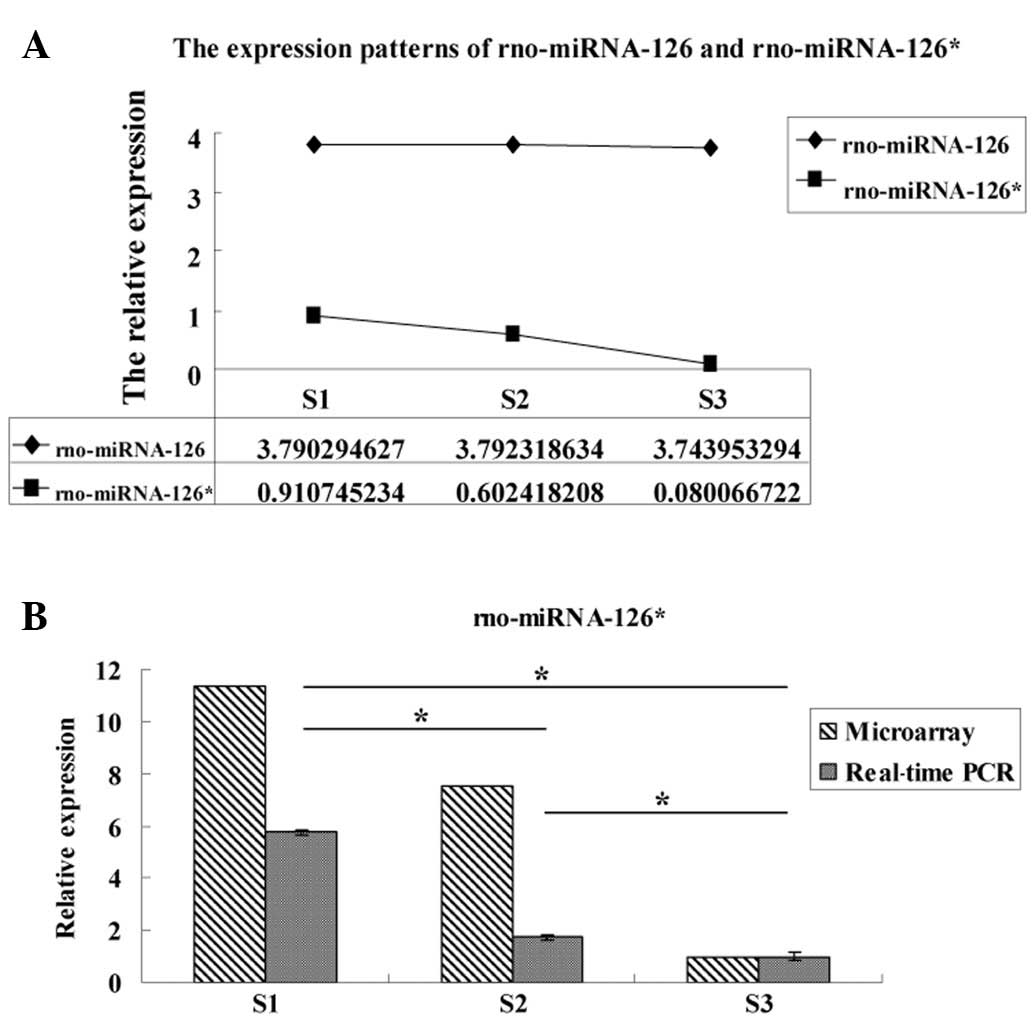
of the embryo (14,15). Therefore, we also prepared line
charts to show the expression patterns of rno-miRNA-126* and
rno-miRNA-126 in order to further study their possible effects in
the lung development of embryos (Fig.
4).
Real-time PCR
Due to the exhibition of marked differential
expression between rno-miRNA-126* and rno-miRNA-126 in the
microarray, we selected the former for analysis by real-time PCR
(Fig. 4). The results revealed
that the relative expression of rno-miRNA-126* determined by
real-time PCR was consistent with the microarray result, revealing
significant differences among the 3 groups (P<0.05; Fig. 4).
Discussion
In the present study, we used the latest microarray
technology to prepare a miRNA expression profile among three time
points of fetal lung development, with 202 differentially expressed
miRNAs being identified, including four different expression
patterns. Among these miRNAs, a number are reported for the first
time in the field of lung development, including rno-miRNA-126*,
rno-miRNA-186*, rno-miRNA-25, rno-miRNA-195 and rno-miRNA-107
(Table IV).
In addition to these newly identified miRNAs, we
also found certain classical ones associated with lung development,
for example, the Let-7 family and miRNA-17-92 cluster, and their
expression patterns are shown as line charts in Fig. 3. For several years, these two types
of miRNAs have been reported and studied repeatedly due to their
important roles in lung development and lung cancer. Johnson et
al(10) reported that Let-7 is
highly expressed in the developing lungs during mouse
embryogenesis. Furthermore, they demonstrated that the Let-7 family
directly regulates a number of key cell cycle proto-oncogenes,
including RAS, CDC25a, CDK6 and cyclin D, and thus may further
regulate cell proliferation by promoting the G1 phase into the S
phase. Compared with the high expression in lung development,
certain members of this family have been found to be deregulated in
specific processes involved in pathological mechanisms, including
pulmonary allergy, asthma and lung cancer (11). These findings indicate that the
Let-7 family is essential to lung homeostasis and development.
With regard to the miRNA-17-92 cluster, Lu et
al(12) firstly reported that
its expression is high at the embryo stage and steadily declines
during the development into adulthood. The authors further reported
that overexpression of the miRNA-17-92 cluster in murine models
resulted in abnormalities, in particular, terminal air sacs were
lacking and were replaced by undifferentiated and highly
proliferative pulmonary epithelium. By contrast, Ventura et
al(13) demonstrated that mice
deficient in the miRNA-17-92 cluster exhibited an altered phenotype
characterized by hypoplasia of the lung. Subsequently, these
findings were confirmed by Jevnaker et al(16) and Carraro et al(17), respectively. In this study, we also
screened these two important types of miRNA. Their expression
patterns are additionally presented as line charts. Moreover, the
expression trends are essentially in agreement with the former
studies (Fig. 3). The expression
of the Let-7 family increased gradually from group S3 (E16) to S1
(E21), while the expression of the miRNA-17-92 cluster manifested a
downward trend as the fetal lung grew.
Compared with these intensely investigated miRNAs,
certain other miRNAs, including miRNA-126* and miRNA-126, have
rarely been discussed in relation to lung development. miRNA-126*
and miRNA-126 originate from the same domain [both mapping to
intron-9 of the epidermal growth factor-like domain 7 gene (Egfl7)
(14)]; the common miRNA-126
(known as miRNA-126-3p) was firstly identified in mouse (18) and another less common miRNA-126 was
later revealed to be processed from the 5′-half of the same
pre-miRNA and was therefore renamed miRNA-126* (also known as
miRNA-126-5p). Although introns occupy a large portion of all
genomes of eukaryotes and have been generally considered to be
sequences that exist only to be removed and destroyed (19,20),
the finding of intronic miRNA has indicated that certain excised
introns may have particular functions. These include intron-9 in
the Egfl7 gene, which has been repeatedly demonstrated to be
expressed at high levels in the vasculature associated with tissue
proliferation and is abundant in specific organs, including the
lungs (14,21-23).
miRNA-126 and miRNA-126* are processed from the same
primary miRNA and located in the same intron of Egfl7, a well-known
epidermal growth factor domain gene (14). These two miRNAs have been found to
be highly enriched in endothelial cells derived from mouse
embryonic stem cells (15).
Consequently, they are likely to be critical in the regulation of
vascular integrity and angiogenesis (15,24).
During the past several years, related studies on miRNA-126 and
miRNA-126* have mostly been limited to the pathogenesis of cancers.
Due to miRNA-126 targeting the vascular endothelial growth factor A
(VEGFA), sprouty-related, EVH1 domain-containing protein 1
(Spred-1) and phosphatidylinositol 3-kinase (PI3K), its expression
has been found to be decreased in human breast cancer. By contrast,
in lung cancer, miRNA-126 has been shown to be upregulated to
decrease the growth rate of cell lines in vitro(25), partially through the VEGF/PI3K
signaling pathway (26). Moreover,
Liu et al(25) focused on
the miRNA-126 interaction with VEGF in lung cancer cells and three
other cancer cell lines infected with LV-miRNA-126, and revealed
that miRNA-126 efficiently reduced the expression of VEGF and
inhibited cell proliferation as a potential tumor suppressor.
Moreover, two more studies demonstrated that the role of miRNA-126
as a tumor suppressor in lung cancers may be due to its regulation
of chicken tumor virus no. 10 regulator of kinase (Crk), which has
been described in lung cancer and associated with increased tumor
invasiveness (27,28). These studies suggest that miRNA-126
and miRNA-126* have various roles in developmental angiogenesis as
well as in carcinogenesis (29).
Compared with that of miRNA-126, the role of
miRNA-126* is less understood, with the exception of its joint
function with miRNA-126 in VEGF and angiogenesis. Musiyenko et
al(30) reported that the
natural absence of Egfl7 leads to the absence of its intronic mRNA
(miRNA-126*), which indirectly promotes the invasiveness of LNCaP
prostate cancer cells by allowing the expression of protein. This
is one of the few studies concerning the function of miRNA-126*.
Until several years ago, it was assumed that one strand of the RNA
duplex preferentially binds to the silencing complex during the
processing of miRNA, whereas the other strand, i.e., miRNA* is
degraded (31), representing a
functionally irrelevant carrier strand. However, subsequently,
miRNA* species have been detected in increasing numbers with
large-scale small RNA sequencing efforts. In the case of
miRNA-126/miRNA-126*, they have been detected with high expression
levels in the same tissue (32).
In addition, the two strands of a miRNA pair are able to
functionally suppress the expression of their target genes
(33,34). Therefore, compared with common
miRNA-126, miRNA-126* may also have its own special functions
(29).
Since miRNA-126 and miRNA-126* originate from the
Egfl7 gene and share a common mRNA, in previous reports, their
expression appeared always to be generally in parallel (15,23).
Nevertheless, there are currently a few studies which report that
miRNA-126 expression is regulated independently of the EGFL7
protein (35), suggesting that
independent promoters may exist which regulate the expression of
miRNA-126 and miRNA-126*, further indicating that they may each
have respective roles in different signaling pathways and
physiological mechanisms. In the present study, following the
microarray screening, we found that miRNA-126 did not exhibit the
same differential expression as miRNA-126* in the three groups
(Fig. 4), verifying the above
hypothesis.
The results of a subsequent real-time PCR assay
demonstrated that miRNA-126* has significant differences in
expression among the three time points (group S3 → S2 → S1)
(Fig. 4). This is the first time
that the differential expression of miRNA-126* has been reported in
fetal lung development and, according to these results, we suggest
that miRNA-126* is important in the process of fetal lung
development, since its expression increases gradually as the lung
develops.
As previously stated, most of the studies on these
two miRNAs have been conducted in cancers. However, cells at an
early stage of development are known to share several
characteristics with carcinoma cells, including high and rapid
proliferation. The higher tissue levels of miRNA-126/126* in
carcinoma cells are closely associated with their role in
angiogenesis. Therefore, in the embryo, the two miRNAs may also
promote lung development through the same mechanism since these two
processes share the same characteristics. This hypothesis may also
help to explain the role of miRNA-126* in fetal lung development in
the rat embryo. Furthermore, we consider that these findings are
likely to lay a certain physiological foundation for studies
concerning neonatal lung developmental diseases, including RDS
(respiratory distress syndrome) and BPD (bronchopulmonary
dysplasia). Therefore, future studies are necessary.
Acknowledgements
The authors would like to thank Dr Shi for
assistance in the revision of this manuscript. This study was
funded by the Project Foundation of Jiangsu Province Health
Department (no. H200642).
References
|
1
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
4
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007. View Article : Google Scholar
|
|
6
|
Fabbri M, Garzon R, Cimmino A, et al:
MicroRNA-29 family reverts aberrant methylation in lung cancer by
targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA.
104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Williams AE, Moschos SA, Perry MM, Barnes
PJ and Lindsay MA: Maternally imprinted microRNAs are
differentially expressed during mouse and human lung development.
Dev Dyn. 236:572–580. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Williams AE, Perry MM, Moschos SA and
Lindsay MA: microRNA expression in the aging mouse lung. BMC
Genomics. 8:1722007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomankova T, Petrek M and Kriegova E:
Involvement of microRNAs in physiological and pathological
processes in the lung. Respir Res. 11:1592010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu Y, Thomson JM, Wong HY, Hammond SM and
Hogan BL: Transgenic overexpression of the microRNA miR-17-92
cluster promotes proliferation and inhibits differentiation of lung
epithelial progenitor cells. Dev Biol. 310:442–453. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ventura A, Young AG, Winslow MM, et al:
Targeted deletion reveals essential and overlapping functions of
the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Parker LH, Schmidt M, Jin SW, et al: The
endothelial-cell-derived secreted factor Egfl7 regulates vascular
tube formation. Nature. 428:754–758. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jevnaker AM, Khuu C, Kjøle E, Bryne M and
Osmundsen H: Expression of members of the miRNA17-92 cluster during
development and in carcinogenesis. J Cell Physiol. 226:2257–2266.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carraro G, El-Hashash A, Guidolin D, et
al: miR-17 family of microRNAs controls FGF10-mediated embryonic
lung epithelial branching morphogenesis through MAPK14 and STAT3
regulation of E-Cadherin distribution. Dev Biol. 333:238–250. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of tissue
specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naora H and Deacon NJ: Relationship
between the total size of exons and introns in protein-coding genes
of higher eukaryotes. Proc Natl Acad Sci USA. 79:6196–6200. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clement JQ, Qian L, Kaplinsky N and
Wilkinson MF: The stability and fate of a spliced intron from
vertebrate cells. RNA. 5:206–220. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soncin F, Mattot V, Lionneton F, Spruyt N,
Lepretre F, Begue A and Stehelin D: VE-statin, an endothelial
repressor of smooth muscle cell migration. EMBO J. 22:5700–5711.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fitch MJ, Campagnolo L, Kuhnert F and
Stuhlmann H: EGFL7, a novel epidermal growth factordomain gene
expressed in endothelial cells. Dev Dyn. 230:316–324. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Campagnolo L, Leahy A, Chitnis S,
Koschnick S, Fitchx MJ, Fallon JT, Loskutoff D, Taubman MB and
Stuhlmann H: EGFL7 is a chemoattractant for endothelial cells and
is upregulated in angiogenesis and arterial injury. Am J Pathol.
167:275–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang S, Aurora AB, Johnson BA, Qi X,
McAnally J, Hill JA, Richardson JA, Bassel-Duby R and Olson EN: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: MiR-126 restoration downregulate VEGF and inhibit the growth of
lung cancer cell lines in vitro and in vivo. Lung
Cancer. 66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu N, Zhang D, Xie H, et al:
Endothelial-specific intron-derived miR-126 is downregulated in
human breast cancer and targets both VEGFA and PIK3R2. Mol Cell
Biochem. 351:157–164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crawford M, Brawner E, Batte K, Yu L,
Hunter MG, Otterson GA, Nuovo G, Marsh CB and Nana-Sinkam SP:
MicroRNA-126 inhibits invasion in non-small cell lung carcinoma
cell lines. Biochem Biophys Res Commun. 373:607–612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Li X, Yang Q, Wang X, Zhou Y,
Jiang T, Ma Q and Wang YJ: The role of microRNA in human lung
squamous cell carcinoma. Cancer Genet Cytogenet. 200:127–133. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meister J and Schmidt MH: miR-126 and
miR-126*: new players in cancer. Sci World J. 10:2090–2100.
2010.
|
|
30
|
Musiyenko A, Bitko V and Barik S: Ectopic
expression of miR-126*, an intronic product of the vascular
endothelial EGF-like 7 gene, regulates prostein translation and
invasiveness of prostate cancer LNCaP cells. J Mol Med (Berlin).
86:313–322. 2008.
|
|
31
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saito Y, Friedman JM, Chihara Y, Egger G,
Chuang JC and Liang G: Epigenetic therapy upregulates the tumor
suppressor microRNA-126 and its host gene EGFL7 in human cancer
cells. Biochem Biophys Res Commun. 379:726–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Okamura K, Phillips MD, Tyler DM, Duan H,
Chou YT and Lai EC: The regulatory activity of microRNA* species
has substantial influence on microRNA and 3′ UTR evolution. Nat
Struct Mol Biol. 15:354–363. 2008.
|
|
35
|
Li X, Shen Y, Ichikawa H, Antes T and
Goldberg GS: Regulation of miRNA expression by Src and contact
normalization: effects on nonanchored cell growth and migration.
Oncogene. 28:4272–4283. 2009. View Article : Google Scholar : PubMed/NCBI
|