Introduction
Cholangiocarcinoma (CCA), originating from the
epithelial cells lining the biliary duct, is a rare but devastating
malignancy (1,2). CCAs are clinically silent in the
majority of cases (3). Radical
resection of the tumor is applicable only in a minority of patients
due to late clinical presentation and diagnosis (4). Moreover, the recurrence rate
following resection is extremely high (5). Although advanced CCAs may respond to
chemotherapy, there are no standard treatments for the palliation
of advanced disease (3). The
overall survival rate of the disease is poor (6).
Although a number of risk factors have been
identified, including primary sclerosing cholangitis (PSC),
infestation with liver flukes, toxic compounds, congenital
disorders, hepatolithiasis, viral hepatitis, alcohol abuse,
smoking, obesity and diabetes (4,7),
only a minority of CCA patients have known risk factors. Up to 90%
of patients presenting with CCA have no identifiable risk factors
(4,6).
CCA, in common with certain other cancers, is caused
by gene-environment interactions. Carcinogens induce tumorigenesis
in genetically susceptible individuals (8,9).
Genomic DNA is continuously attacked by a large number of agents
that damage DNA. However, cancers only occur in a small proportion
of people since DNA damage is normally repaired by genome
surveillance mechanisms and DNA repair pathways, including
homologous recombination (HR), base excision repair (BER),
nucleotide excision repair (NER) and mismatch repair (MMR)
(10). Polymorphisms of DNA repair
genes may affect the quantity and activity of the resulting protein
and the DNA repair capacity. The possession of specific alleles of
polymorphic genes may increase susceptibility to cancers and
facilitate cancer development in normal or exposed individuals
(11).
The BER pathway has a principal role in the repair
of mutations caused by oxidized or reduced bases. Human oxoguanine
glycosylase 1 (hOGG1), MutY homolog (MUTYH, MYH),
apurinic/apyrimidinic endonuclease-1 (APEX1, APE1) and X-ray
cross-complementing group 1 (XRCC1) are key proteins in the BER
pathway (12–14). We have previously analyzed hOGG1
Ser326Cys and XRCC1 codon 194 and 399 polymorphisms in CCAs and did
not find an association between polymorphisms of these genes and
susceptibility to CCA (15,16).
MYH is involved in the repair of mutations caused by
oxidized bases (13,14). Harmful environmental agents may
cause the production of reactive oxygen species (ROS), including
hydroxyl radicals, in the body (14). Oxidative DNA damage occurs when the
production of ROS exceeds the antioxidant capacity of the cell
(17). 8-Hydroxy-2′-deoxyguanosine
(8-oxoG) is one of the most abundant and stable products of
oxidative DNA damage and may cause G:C to T:A transversions
(14,17). The BER pathway is responsible for
repairing 8-oxoG lesions. MYH is a DNA glycosylase that removes
adenine paired with 8-oxoG (13).
Biallelic loss of MYH is associated with somatic G:C to T:A
transversions in the adenomatous polyposis coli (APC) gene and is
responsible for polyp formation and colorectal cancer (18). However, the role of MYH in the
tumorigenesis of CCA is largely unknown. It has been reported that
one SNP in the MYH gene (rs3219476) was associated with an
increased risk of treatment-related mortality (TRM) in recipients
who took alkylating agents with or without ionizing radiation for
pretransplant conditioning regimens and underwent allogeneic
hematopoietic cell transplant (HCT) for the treatment of
hematologic malignancies (19).
However, the relationship between MYH rs3219476 polymorphism and
the risk of CCA has not been reported. In this study, we evaluated
the influence of polymorphisms of two adjacent SNPs (rs3219476 and
rs3219472) in the MYH gene on the risk of CCA in a Chinese
population.
Materials and methods
Subjects
A hospital-based case-control study of CCA was
conducted in the Second Affiliated Hospital of Nanjing Medical
University between May 2007 and June 2010. Written informed consent
was obtained from all participants and the study was approved by
the Ethics Committee of the Hospital. The cases were 59 CCA
patients aged from 27 to 86 years with an average of 64.7±15.5
years. Among the patients, 37 cases were male and 22 were female.
The patients were diagnosed with CCA according to clinical
presentations and imaging, including computerized tomography (CT),
magnetic resonance imaging (MRI), magnetic resonance
cholangiopancreatography (MRCP) and endoscopic retrograde
cholangiopancreatography (ERCP) (20). Ten cases of CCA diagnosis were
confirmed by histology. None of the cases had a history of any
cancer other than CCA. A total of 100 controls with no current or
previous diagnosis of cancer or other major diseases were
recruited. Among the controls, 59 were male, 41 were female, and
the average age was 61.7±10.2 years. All subjects were unrelated
Han Chinese living in Jiangsu province.
Blood samples and DNA isolation
Venous blood (3 ml) was collected from each of the
CCA patients or controls. After collection, the whole blood was
immediately stored at −80°C until use. Genomic DNA was extracted
from the blood samples by a routine phenol-chloroform method.
Genotyping of MYH rs3219476 and rs3219472
polymorphisms
MYH genotypes were detected using a polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP)
technique. MYH rs3219476 and rs3219472 were amplified to form
undigested fragments of 363 and 176 bp, respectively, using primers
designed by ourselves with a web-based PCR-RFLP designing tool, SNP
PCR-RFLP assay design at http://bioapp.psych.uic.edu/SNP_cutter.htm. The
primers used were as follows: forward 5′-GGA AGG AAG TGA CGT GAT
CTG G-3′ and reverse 5′-TGG TGG TCT CTT CAC ATG GAC A-3′ for
rs3219476; forward 5′-GTA AGA GCT ACA AGG CAG-3′ and reverse 5′-GCT
TAA ACA CCT CAA GTG-3′ for rs3219472. Briefly, 50 ng genomic DNA
was amplified in a volume of 50 μl containing 1X PCR buffer, 1.5 mM
MgCl2, 0.2 mM of each dNTP, 0.5 μM of each primer and
1.25 units Taq DNA Polymerase (Takara Biotechnology, Dalian,
China). The PCR conditions were a 5 min denaturation step at 94°C;
followed by 40 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C
for 30 sec; and a final extension at 72°C for 10 min. After
amplification, 8 μl PCR products were digested with BtsCI at
50°C or Tsp509I at 65°C for the genotyping of rs3219476 or
rs3219472, respectively, for 4 h in a final volume of 20 μl
containing 1X NE buffer and 0.5 μl (2.5 units) restriction enzyme
(New England Biolabs, Beverly, MA, USA). The digested fragments
were separated by electrophoresis on 2% (w/v) agarose gel and
visualized by ethidium bromide staining using a DL2000 DNA Marker
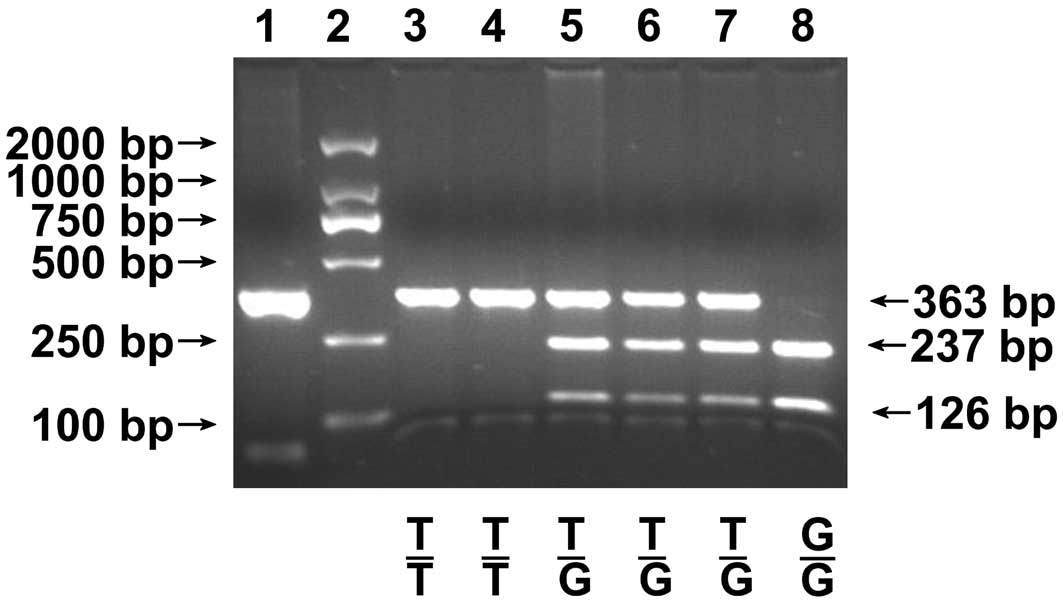
(Takara BioTechnology). All experiments included positive and
negative controls. For rs3219476, the wild-type genotype T/T had no
BtsCI site and was characterized by a 363 bp fragment on the
gel; the variant genotype G/G was characterized by 237 and 126 bp
fragments; and the heterozygous genotype T/G was characterized by
363, 237 and 126 bp fragments (Fig.
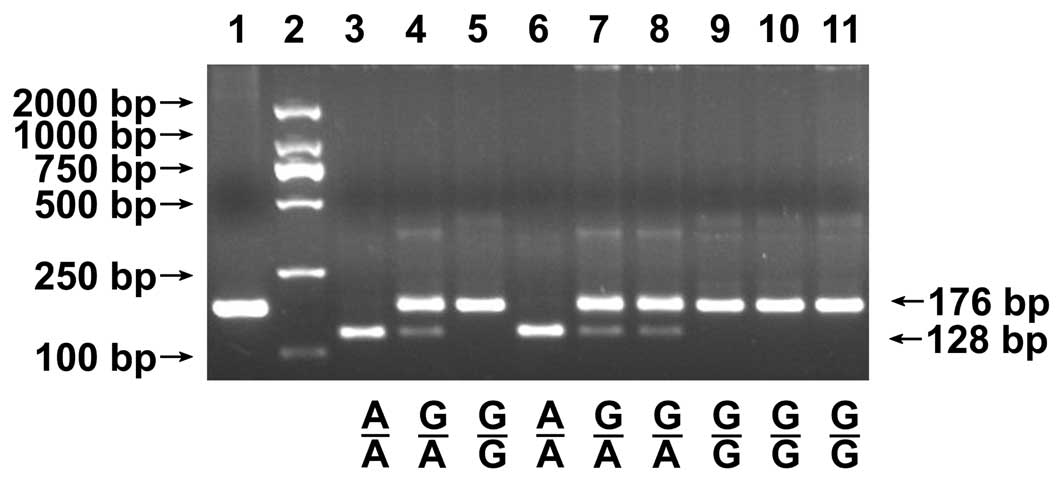
1). In the genotyping of rs3219472, the complete digestion of
PCR products produced 176 bp fragments for the G allele and 128 bp
and 48 bp fragments for the A allele (the 48 bp fragment was too
small to resolve accurately; Fig.
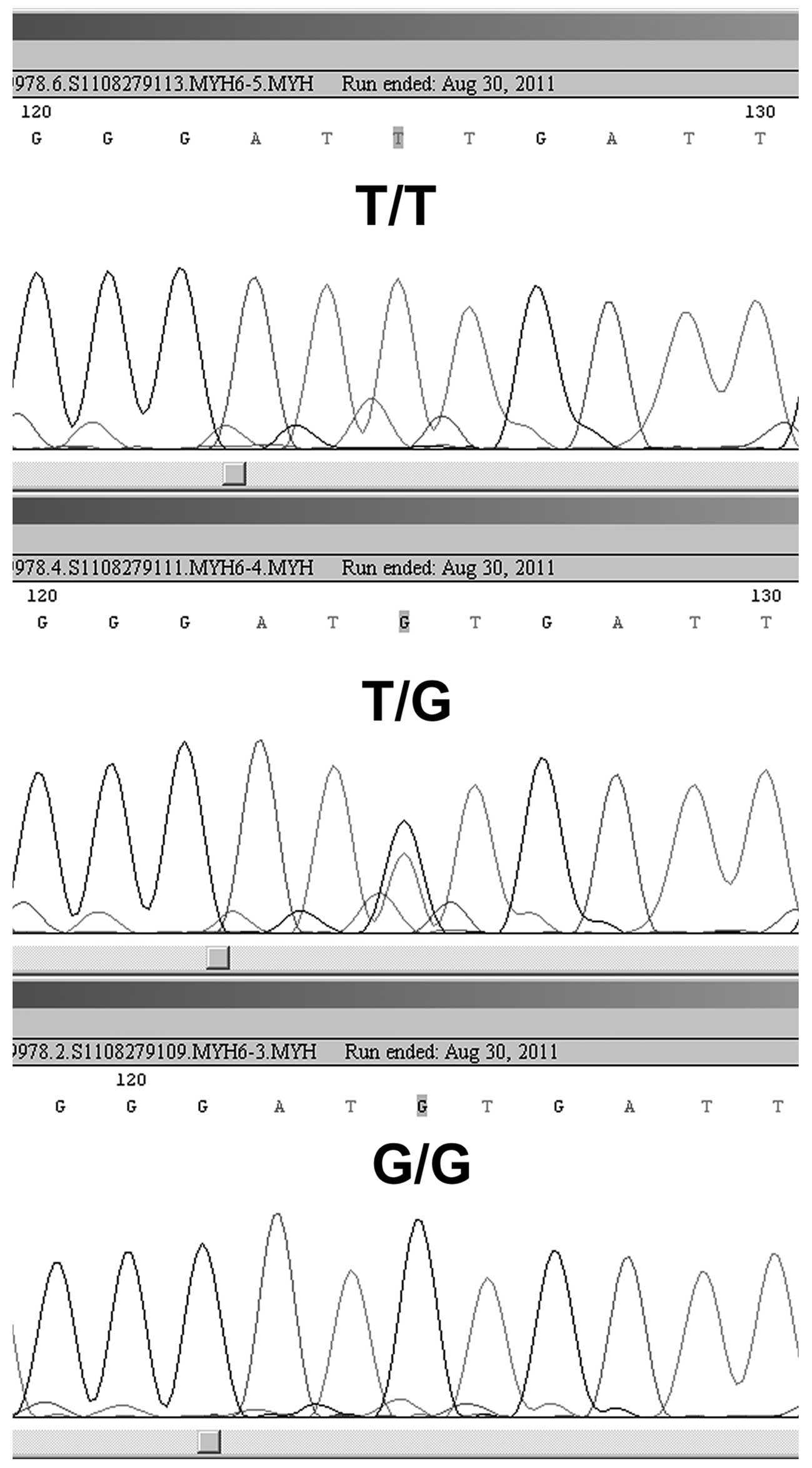
2). DNA sequencing was performed to confirm the genotypes in
certain cases (Figs. 3 and
4).
Statistical analysis
Mean and standard deviations are presented in cases
of continuous variables. Differences between the means of the two
continuous variables were evaluated by the Student’s t test. The
Hardy-Weinberg equilibrium was tested using a web-based program,
OEGE (Online Encyclopedia for Genetic Epidemiology) at http://www.oege.org/software/hwe-mr-calc.shtml, to
compare the observed genotype frequencies with the expected
frequencies among the control subjects. The Chi-square test was
used to compare the gender distribution and test the association
between the genotypes and alleles in relation to the cases and
controls. P<0.05 was considered to indicate a statistically
significant result. The odds ratio (OR) and 95% confidence interval
(95% CI) were calculated to estimate the associations between the
genotypes of MYH and the risk of CCA. Statistical analyses were
carried out using SPSS version 11.0 software (SPSS, Chicago, IL,
USA) and Microsoft Excel 2003 (Microsoft Corporation, Seattle,
USA).
Results
The case and control groups were not statistically
different with respect to age (t=1.335, P=0.185) and gender
(χ2=0.214, P=0.644). The frequencies of the MYH
rs3219472 genotypes associated with CCA are shown in Table I. For rs3219472, the genotype
frequencies of G/G, G/A and A/A were 47.5, 32.2 and 20.3%,
respectively, in the CCA cases compared with 46.0, 47.0 and 7.0%,
respectively, in the controls. The genotype distribution in the
controls was within the Hardy-Weinberg equilibrium
(χ2=1.18, P>0.05). The frequency of the MYH rs3219472
A allele was 36.4% in the CCA group and 30.5% in the control group.
Statistically significant differences were observed in the genotype
frequencies of the MYH rs3219472 polymorphism between the control
group and the patients with CCA (χ2=7.499, P=0.024), but
no statistically significant differences were identified in the
allele frequencies of the MYH rs3219472 polymorphism between the
two groups (χ2=1.190, P=0.275). Compared with subjects
carrying the MYH G/G genotype, those with the A/A genotype had a
2.816-fold higher risk of CCA (OR=2.816, 95% CI=0.992–7.999,
P=0.047).
 | Table IDistribution of allele and genotype
frequencies of MutY homolog (MYH) rs3219472 polymorphism in
cholangiocarcinoma (CCA) patients and healthy controls. |
Table I
Distribution of allele and genotype
frequencies of MutY homolog (MYH) rs3219472 polymorphism in
cholangiocarcinoma (CCA) patients and healthy controls.
| CCA, n (%) | Controls, n (%) | OR (95% CI) | P-value |
|---|
| Genotypes |
| G/G | 28 (47.5) | 46 (46.0) | 1 (−) | - |
| G/A | 19 (32.2) | 47 (47.0) | 0.664
(0.326–1.351) | 0.258 |
| A/A | 12 (20.3) | 7 (7.0) | 2.816
(0.992–7.999) | 0.047 |
| G/A+A/A | 31 (52.5) | 54 (54.0) | 0.943
(0.495–1.797) | 0.859 |
| Alleles |
| G | 75 (63.6) | 139 (69.5) | 1 (−) | - |
| A | 43 (36.4) | 61 (30.5) | 1.306
(0.808–2.113) | 0.275 |
As shown in Table
II, for rs3219476, the genotype frequencies of T/T, T/G and G/G
were 42.4, 33.9 and 23.7%, respectively, in the CCA cases compared
with 26.0, 58.0 and 16.0%, respectively, in the controls. The
genotype distribution in the controls was within the Hardy-Weinberg
equilibrium (χ2=2.95, P>0.05). The frequency of the
MYH rs3219476 G allele was 40.7% in the CCA group and 45.0% in the
control group. Statistically significant differences were observed
in the genotype frequencies of the MYH rs3219476 polymorphism
between the control group and the patients with CCA
(χ2=8.670, P=0.013), but no statistically significant
differences were identified in the allele frequencies of the MYH
rs3219476 polymorphism between the two groups (χ2=0.564,
P=0.453). Compared with subjects carrying the MYH T/T genotype,
those with the T/G genotype had a reduced risk of CCA (OR=0.359,
95% CI=0.17–0.758, P=0.006).
 | Table IIDistribution of allele and genotype
frequencies of MutY homolog (MYH) rs3219476 polymorphism in
cholangiocarcinoma (CCA) patients and healthy controls. |
Table II
Distribution of allele and genotype
frequencies of MutY homolog (MYH) rs3219476 polymorphism in
cholangiocarcinoma (CCA) patients and healthy controls.
| CCA, n (%) | Controls, n (%) | OR (95% CI) | P-value |
|---|
| Genotypes |
| T/T | 25 (42.4) | 26 (26.0) | 1 (−) | - |
| T/G | 20 (33.9) | 58 (58.0) | 0.359
(0.17–0.758) | 0.006 |
| G/G | 14 (23.7) | 16 (16.0) | 0.91
(0.369–2.246) | 0.838 |
| T/G+G/G | 34 (57.6) | 74 (74.0) | 0.478
(0.241–0.946) | 0.033 |
| Alleles |
| T | 70 (59.3) | 110 (55.0) | 1 (−) | - |
| G | 48 (40.7) | 90 (45.0) | 0.838
(0.529–1.329) | 0.453 |
Discussion
The MYH gene excises adenine bases from the DNA
backbone at sites where adenine is inappropriately paired with
guanine, cytosine or 8-oxoG (19).
Although multiple mutations (Y165C, G382D, R260Q and H434D) or
single nucleotide polymorphisms (rs3219484 and rs3219489) have been
identified in the MYH gene (21,22),
no associations have been identified between these variants and the
susceptibility to CCA (14,22).
Furthermore, the Y165C and G382D mutations are rarely detected in
Chinese Han populations (23,24).
The two SNPs analyzed in our study, rs3219472 and rs3219476, are
located in the intron region near the 5′-UTR of the MYH gene. In
this study, significantly increased CCA risk was found in
individuals with the homozygous variant genotype for rs3219472, but
no increased risk was identified in those with the homozygous
variant genotype for rs3219476. Furthermore, a reduced risk of CCA
was found in those with heterozygous genotype for rs3219476.
The minor allele frequency (MAF) for rs3219476
(0.450) in the controls in the current study were comparable to
those in the HapMap-HCB and CHB_GENO_PANEL databases (MAF=0.444 and
0.443, respectively), which are based on the Chinese Han
population. However, the MAF for rs3219472 (0.305) in the controls
in the current study was slightly different from that in the
HAPMAP-CHB database (MAF=0.363), which is also based on the Chinese
Han population, reflecting sampling errors and regional differences
between the different data. It has been reported that MYH rs3219476
polymorphism was associated with 1-year TRM in HCT patients who
accepted ionizing radiation and alkylating agents that damaged DNA
(19). However, the effect of MYH
rs3219476 and rs3219472 polymorphism on the risk of CCA is unknown.
To our knowledge, this is the first study to evaluate the ability
of MYH rs3219476 and rs3219472 polymorphisms to predict the
susceptibility to CCA. Our findings suggest that male Han Chinese
with the MYH A/A genotype in rs3219472 have a higher susceptibility
to CCA. It may be a biomarker for screening individuals at high
risk of developing the disease.
However, our data may be biased by the relatively
small number of subjects and requires validation in larger samples.
As CCA is a rare disease (3),
multiple center cooperation would be necessary to recruit enough
CCA patients for investigation. Further studies of the precise
mechanisms by which MYH polymorphism affects the risk of CCA are
merited.
Acknowledgements
This study was supported by the Natural Science
Foundation of Jiangsu Province, No. BK2007260.
References
|
1
|
Isomoto H: Epigenetic alterations
associated with cholangiocarcinoma (Review). Oncol Rep. 22:227–232.
2009.PubMed/NCBI
|
|
2
|
Petrowsky H and Hong JC: Current surgical
management of hilar and intrahepatic cholangiocarcinoma: the role
of resection and orthotopic liver transplantation. Transplant Proc.
41:4023–4035. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valle J, Wasan H, Palmer DH, et al:
Cisplatin plus gemcitabine versus gemcitabine for biliary tract
cancer. N Engl J Med. 362:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gatto M, Bragazzi MC, Semeraro R, et al:
Cholangiocarcinoma: update and future perspectives. Dig Liver Dis.
42:253–260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen FZ, Zhang BY, Feng YJ, et al: Current
research in perineural invasion of cholangiocarcinoma. J Exp Clin
Cancer Res. 29:242010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meza-Junco J, Montano-Loza AJ, Ma M, Wong
W, Sawyer MB and Bain VG: Cholangiocarcinoma: has there been any
progress? Can J Gastroenterol. 24:52–57. 2010.PubMed/NCBI
|
|
7
|
Ustundag Y and Bayraktar Y:
Cholangiocarcinoma: a compact review of the literature. World J
Gastroenterol. 14:6458–6466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shields PG and Harris CC: Cancer risk and
low-penetrance susceptibility genes in gene-environment
interactions. J Clin Oncol. 18:2309–2315. 2000.PubMed/NCBI
|
|
9
|
Galvan A, Ioannidis JP and Dragani TA:
Beyond genome-wide association studies: genetic heterogeneity and
individual predisposition to cancer. Trends Genet. 26:132–141.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martin SA, Hewish M, Lord CJ and Ashworth
A: Genomic instability and the selection of treatments for cancer.
J Pathol. 220:281–289. 2010.PubMed/NCBI
|
|
11
|
Jiang J, Zhang X, Yang H and Wang W:
Polymorphisms of DNA repair genes: ADPRT, XRCC1, and XPD and cancer
risk in genetic epidemiology. Methods Mol Biol. 471:305–333. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
David SS, O’Shea VL and Kundu S:
Base-excision repair of oxidative DNA damage. Nature. 447:941–950.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kasahara M, Osawa K, Yoshida K, et al:
Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal
cancer and smoking in a Japanese population. J Exp Clin Cancer Res.
27:492008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baudhuin LM, Roberts LR, Enders FT, et al:
MYH Y165C and G382D mutations in hepatocellular carcinoma and
cholangiocarcinoma patients. J Cancer Res Clin Oncol. 132:159–162.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, You SH, He W, et al: hOGG1
Ser326Cys polymorphisms and genetic susceptibility of
cholangiocarcinomas. Journal of Medical Postgraduates.
23:1156–1159. 2010.(In Chinese).
|
|
16
|
You SH, Wang X, Huang S, Gao Y, Xia JR and
Fan ZN: XRCC1 polymorphisms and genetic susceptibility of
cholangiocarcinomas. Chinese Journal of Clinical Research.
23:1068–1072. 2010.(In Chinese).
|
|
17
|
Tsukino H, Hanaoka T, Otani T, et al:
hOGG1 Ser326Cys polymorphism, interaction with environmental
exposures, and gastric cancer risk in Japanese populations. Cancer
Sci. 95:977–983. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jones S, Emmerson P, Maynard J, et al:
Biallelic germline mutations in MYH predispose to multiple
colorectal adenoma and somatic G:C->T:A mutations. Hum Mol
Genet. 11:2961–2967. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thyagarajan B, Lindgren B, Basu S, et al:
Association between genetic variants in the base excision repair
pathway and outcomes after hematopoietic cell transplantations.
Biol Blood Marrow Transplant. 16:1084–1089. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van Beers BE: Diagnosis of
cholangiocarcinoma. HPB (Oxford). 10:87–93. 2008.
|
|
21
|
Sieber OM, Lipton L, Crabtree M, et al:
Multiple colorectal adenomas, classic adenomatous polyposis, and
germ-line mutations in MYH. N Engl J Med. 348:791–799. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Forsbring M, Vik ES, Dalhus B, et al:
Catalytically impaired hMYH and NEIL1 mutant proteins identified in
patients with primary sclerosing cholangitis and
cholangiocarcinoma. Carcinogenesis. 30:1147–1154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang L, Zhou M, Zhu M, et al: Variation
analysis of the MYH gene associated with sporadic colorectal
cancer. Carcinogenesis, Teratogenesis and Mutagenesis. 21:435–438.
2009.(In Chinese).
|
|
24
|
Yang L, Zhu M, Chen SQ, et al: Research on
MUTYH gene polymorphism associated with colorectal cancer. Shandong
Medical Journal. 49:3–5. 2009.(In Chinese).
|