Introduction
Cervical cancer is a malignant tumor and the second
most malignant cancer in females. It is a major threat to female
health worldwide. Globally, 500,000 new cases and >250,000
mortalities occur each year. These figures account for ~5% of all
cancer cases worldwide with ~80% of new cases reported in
developing countries (1). In
China, the annual incidence of new cervical cancer cases exceeds
130,000, accounting for 28.8% of new cases worldwide (2). An estimated 20,000 individuals
succumb to cervical cancer every year in China, and incidence is
increasing in young adults (3).
The most important risk factor for cervical cancer
is infection with human papilloma virus (HPV), which accounts for
50–70% of all cervical cancer cases worldwide. Oncoproteins encoded
by two early HPV genes, E6 and E7, are important for cell cycle
control. E6 and E7 are required for malignant transformation and
maintenance of malignant phenotypes and are crucial for the
development and progression of cervical cancer (4–8).
HPV16 E7 binds to key tumor suppressors and inhibits their
activity. One of the most important targets of HPV16 E7 is the
retinoblastoma protein (pRb) family which contains pRb, p107 and
p130. In normal cells, pRb proteins are major regulators of the
cell cycle, binding directly to the E2F transcription factor and
negatively regulating its activity, thus inhibiting expression of
E2F target genes important for cell cycle progression (9–16).
In HPV16 E7-overexpressing cells, HPV16 E7 binds to pRb via its CR3
region. This binding induces pRb degradation through the
ubiquitin-proteasome system and releases E2F into the cytosol
(17–20). The free E2F translocates to the
nucleus, activates the transcription of its target genes and
promotes cell transformation. Therefore, suppression of HPV16 E7
expression is likely to inhibit cell growth and induce apoptosis
and senescence, which may limit the growth of cancer cells.
RNA interference (RNAi) has become widely used as an
experimental tool to analyze gene function and holds great promise
in the field of gene therapy in cancer. However, several
limitations restrict its use in basic research and clinical
application. First, siRNA is not stable and is easily degradated by
enzymes. Second, the delivery of siRNA into cells is a great
challenge. Although liposome and cationic polymers have been used
as carriers for siRNA delivery, these reagents are toxic to cells
and not suitable for in vivo transfection. Chitosan is
derived from chitin, the most abundant biopolymer in nature
following cellulose and is a biologically safe, non-toxic,
biodegradable and biocompatible polymer. It contains abundant -NH2
groups and is therefore positively charged at specific pH levels,
enabling it to complex with negatively charged nanoparticles
(21,22). In the present study, chitosan was
utilized as a carrier for delivery of HPV16 E7 siRNA into CaSki
cells constitutively expressing HPV16 E6 and E7. The effect of
chitosan/siRNA nanoparticles on the induction of apoptosis in these
cells was examined. Results indicate a potential use of
chitosan/siRNA complexes in the treatment of diseases, including
cervical cancer.
Materials and methods
Materials
Chitosan was purchased from Jinan Haidebei Marine
Bioengineering Co., Ltd. (Jinan, China). The degree of
deacetylation was 86%. The following siRNA oligos for HPV16 E7 were
used: sense, GCATGGAGATACACCTACA and antisense, TGTAGGTGTATCTCCATGC
(synthesized by Shanghai Generay Biotech Co., Ltd., Shanghai,
China). The study was approved by the ethics committee of the Third
Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan
Province, China.
Preparation and characterization of
chitosan/siRNA nanoparticles
Chitosan was dissolved in aqueous acetic acid (0.1 M
sodium acetate/0.1 M acetic acid, pH 4.5) to prepare various
concentrations of chitosan solution (25–300 μg/ml). Chitosan/siRNA
nanoparticles were prepared by adding a chitosan solution drop-wise
to an equal volume of siRNA solution (20 μg/ml) and incubating at
room temperature for 30 min. The chitosan was complexed with siRNA
at a weight ratio of 1.25:1–15:1. The size and ζ potential of
nanoparticles were measured using the submicron particle analysis
system 4700 (Beckman Coulter Inc., Miami, FL, USA) and the
Zetasizer Nano S (Malvern Instruments, Malvern, UK),
respectively.
Measurement of siRNA loading
efficiency
Chitosan and siRNA were mixed and the mixture was
centrifuged and the absorbance of supernatant was measured at 260
nm to determine the concentration of free siRNA. The loading
efficiency of siRNA was calculated by comparing the amount of siRNA
that was not present in the supernatant to the amount of total
siRNA.
Gel retardation assay
The binding of siRNA to chitosan was determined by
electrophoresis using a 4% agarose gel (low melting point).
Nanoparticles with various chitosan/siRNA weight ratios were loaded
onto the gel and subjected to electrophoresis. siRNA was visualized
by ultraviolet light.
Serum stability assay
Chitosan/siRNA nanoparticles (~5 μg siRNA, 200 μl)
were incubated with an equal volume of 20% fetal bovine serum (FBS)
in Dulbecco’s modified Eagle’s medium (DMEM) at 37°C. At various
time points (0, 0.5, 2, 4, 7, 24, 48 and 72 h), 30 μl mixture was
saved and stored at −20°C.
Characterization of the biological
activity of chitosan/siRNA nanoparticles
CaSki cells were seeded in 96-well plates at a
density of 3×104 cells/well and cultured in DMEM
containing 10% FBS (no antibiotics) for 24 h prior to transfection.
Chitosan/siRNA particles were added directly into the culture
medium and the cells were cultured for an additional 24–48 h prior
to examination by fluorescence microscopy.
Cell toxicity assay
Toxicity of chitosan was determined by the cell
viability of chitosan/siRNA nanoparticles, as described previously
(23).
TUNEL staining
CaSki cells were seeded in 96-well plates at a
density of 3×104 cells/well and cultured in DMEM
containing 10% FBS (no antibiotics) for 24 h prior to transfection.
Chitosan/siRNA particles were added directly to the culture medium
and the cells were cultured for an additional 24–48 h. Cell death
was detected using an in situ Cell Death Detection kit
(Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China).
Western blot analysis
CaSki cells were seeded in 6-well plates at a
density of 4×104 cells/well. Following plating (24 h),
cells were fed with fresh complete media and the chitosan/siRNA
nanoparticles were added to the media. Following an additional 48
h, cells were harvested with RIPA buffer. Samples were subjected to
SDS-PAGE and immunoblotted with antibodies against HPV16 E7 and
β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Statistical analysis
Data were analyzed using SPSS 11.0 (SPSS Inc.,
Chicago, IL, USA) and expressed as mean ± SE. P<0.05 was
considered to indicate a statistically significant difference.
Results
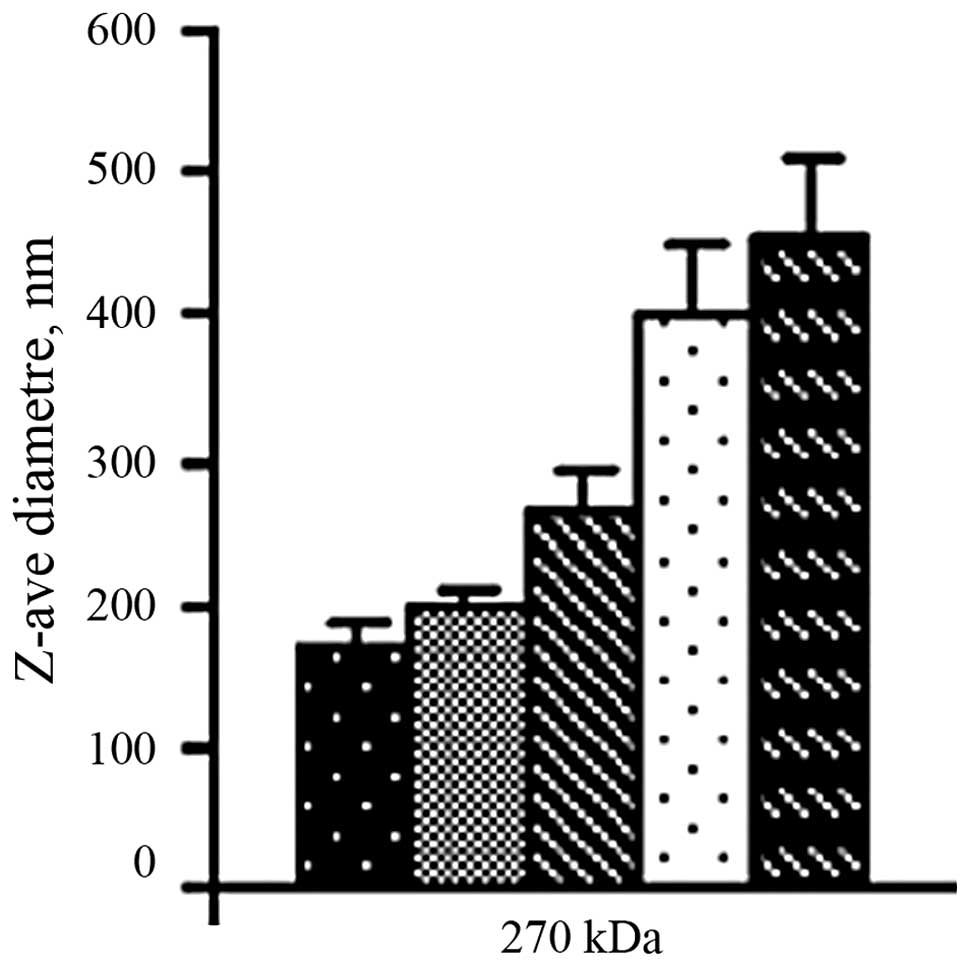
Size of chitosan/siRNA nanoparticles
The chitosan/siRNA particles formed by simple
complexation had a diameter between 185 and 465 nm and the size
increased with the increasing weight ratio of chitosan to siRNA
(Fig. 1).
Surface charge
As revealed in Table
I, the surface charge of chitosan/siRNA particles increased
with increasing chitosan concentration (the amount of siRNA
remained constant). Increased chitosan concentration increased the
positive charge of the particles, preventing aggregation of the
particles and enhancing their interaction with negatively charged
cell membranes.
 | Table IAlterations in ζ potential of
nanoparticles with varied weight ratio of chitosan to siRNA. |
Table I
Alterations in ζ potential of
nanoparticles with varied weight ratio of chitosan to siRNA.
| Chitosan
concentration (μg/ml) | ζ potential (mV) |
|---|
| 25 | −11 |
| 50 | −0.8 |
| 100 | 51 |
| 200 | 54 |
| 300 | 55 |
Interaction of siRNA with chitosan
Since chitosan and siRNA carry opposite charges,
they are attracted to one another in solutions with specific pH
values. Complete attachment of siRNA to chitosan was observed when
chitosan and siRNA were mixed at a weight ratio of 100:1 (Fig. 2). The loading efficiency of siRNA
was 72±1.5%.
Stability of siRNA in serum
Naked siRNA was not stable in serum and was
susceptible to enzyme digestion. When complexed with chitosan, the
rate of degradation was markedly reduced (Fig. 3), indicating that chitosan protects
siRNA from nuclease attack.
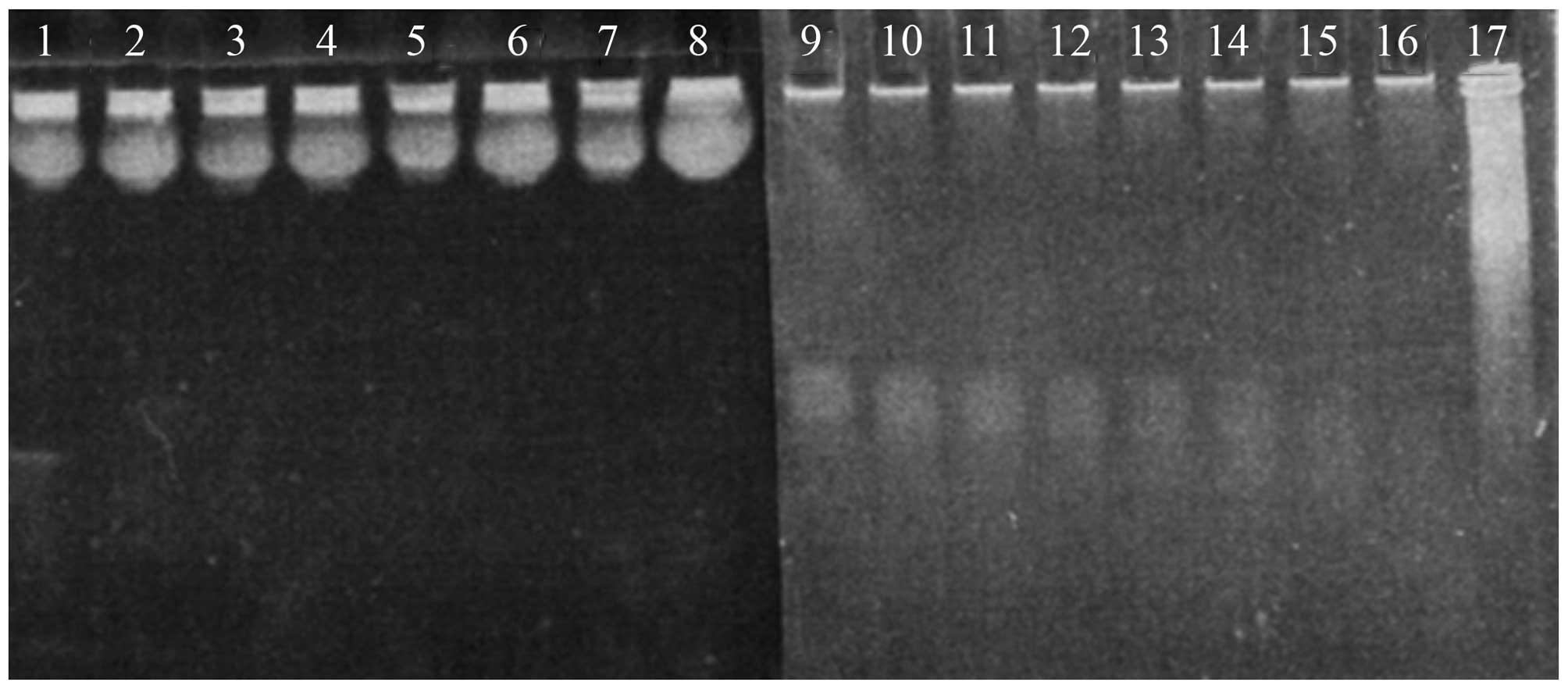 | Figure 3Degradation of naked siRNA and
chitosan-binding siRNA in 20% FBS-containing media at various time
points (lanes 1–8 and lanes 9–17 are 0, 0.5, 1, 2, 4, 7, 24, 48 and
72 h, respectively). Naked siRNA is completely degradated within 30
min, whereas siRNA in chitosan/siRNA particles remains after 72-h
incubation. FBS, fetal bovine serum. |
Biological activity of chitosan/siRNA
nanoparticles
To examine the transfection efficiency of
chitosan/siRNA nanoparticles, chitosan was complexed with
fluorescence-labeled HPV16 E7 siRNA and their accumulation in CaSki
cells was monitored. As demonstrated in Fig. 4, chitosan/siRNA particles were
efficiently tranfected into cells following 24-h incubation.
Protein levels of HPV16 E7 in CaSki cells were analyzed by western
blot analysis and identified to be significantly downregulated
(Fig. 5; P<0.05), indicating
that chitosan/HPV16 E7 nanoparticles suppress expression of HPV16
E7.
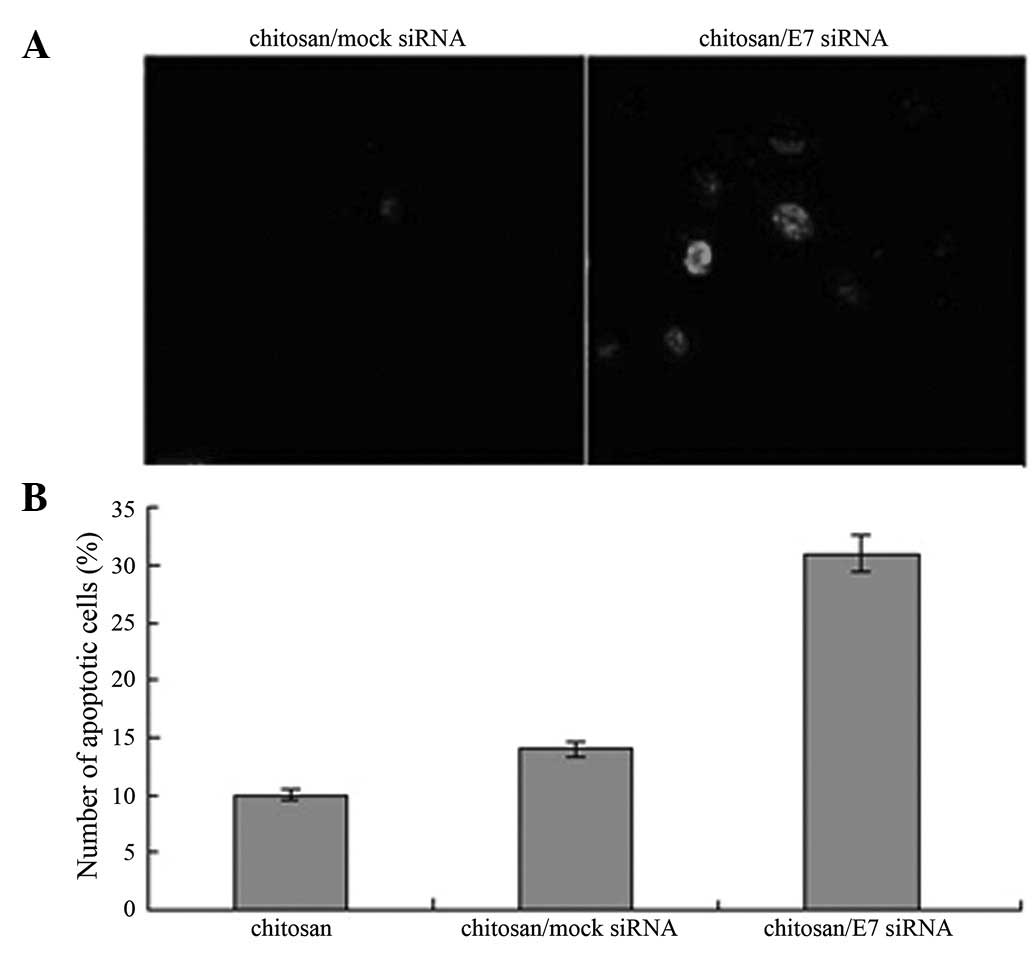
Induction of apoptosis in CaSki cells by
chitosan/HPV16 E7 nanoparticles
To examine the effect of chitosan/HPV16 E7
nanoparticles on cell apoptosis, apoptotic cells were detected
using the TUNEL assay in cells treated with chitosan/HPV16 E7
nanoparticles. A significantly higher number of apoptotic cells
were detected in cells treated with chitosan/HPV16 E7 nanoparticles
compared with cells treated with chitosan/mock siRNA particles
(Fig. 6; P<0.05).
Discussion
In the present study, chitosan/siRNA nanoparticles
were prepared by simple complexation (24). The size and shape of nanoparticles
is critical for efficient transfection of mammalian cells and
distribution of nanoparticles in living cells (25). Previous studies have reported that
nanoparticles exhibit higher levels of intracellular uptake
compared with microparticles (26–28).
This property is crucial for gene transfer, since the uptake of
chitosan/DNA nanoparticles and their release from lysosomes are
rate-limiting steps in this process (29,30).
Similar to DNA and oligodeoxyribonucleotides, siRNA is also taken
up by cells (31). However, RNAi
is not induced if siRNA fails to reach the cytoplasm (31). Using a carrier aids siRNA transfer
into the intracellular compartment and protects it from enzyme
degradation in lysosomes, thus efficiently inducing RNAi. In the
present study, chitosan/siRNA nanoparticles were prepared with a
size <500 nm in diameter which were easily taken up by cells.
The diameter of the nanoparticles increases with the increasing
weight ratio of chitosan to siRNA. Therefore, nanoparticles of
suitable sizes were prepared by adjusting the weight ratio of
chitosan to siRNA.
Binding of siRNA to chitosan was demonstrated by gel
retardation assay. The retarded migration of siRNA in agarose gel
revealed binding of siRNA to chitosan. However, this binding is not
as tight as that of DNA to chitosan, since DNA, but not siRNA, is
concentrated by low concentration chitosan (25 μg/ml), indicating
that siRNA binds to chitosan in a different manner to that of DNA
to chitosan. Previously, the size of chitosan/DNA nanoparticles
following concentration was reported to be 1,000 times smaller than
that without concentration (32,33)
and the minimal size of DNA for concentration was 800 bp (32,34–36).
In contrast to DNA, linearized siRNA is much shorter (21 bp). This
property may account for the weak interaction of siRNA with
chitosan. Since the size of nanoparticles remains unchanged
following complexation, multiple, but not single siRNA may complex
with chitosan.
The major cause of cervical cancer is infection with
high-risk HPV. The integration of viral DNA into human genomes
leads to the constitutive expression of oncoproteins E6 and E7,
altering the cell cycle, immortalizing cells and causing cancer.
Therefore, suppression of E7 expression may reverse the
transformation process and induce apoptosis or senescence.
Chitosan/HPV16 E7 siRNA nanoparticles were efficiently transfected
into the cells (Fig. 4) and were
found to suppress HPV16 E7 expression (Fig. 5). In addition, TUNEL staining
revealed that apoptosis was induced in the CaSki cells. These
results are consistent with previous studies. Chang et
al(37) demonstrated that
siRNA-mediated suppression of HPV E6 and E7 inhibited the growth of
tumor cells from cervical cancer. Sima et al(38) reported that HPV16 E7 shRNA inhibits
E6 and E7 expression and induces apoptosis in cancer cells via
activation of p53, p21 and Rb. More recently, Guo et
al(23) screened a phage
display peptide library, identifying a heptapeptide which promotes
degradation of E7 and prevent formation of E7/pRb complexes. This
peptide induced G1 phase arrest by restoration of pRb
activity, reinstating its ability to inhibit E2F activity. In
addition, downregulation of E7 was reported to increase levels of
p53 and induce apoptosis. Results of the current and previous
studies indicate that suppression of HPV16 E7 by chitosan/siRNA
nanoparticles inhibits growth of tumor cells and induces their
apoptosis, which may serve as a potential therapy for cervical
cancer.
References
|
1
|
Ma B, Roden R and Wu TC: Current status of
human papillomavirus vaccines. J Formos Med Assoc. 109:481–483.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shao J, Chen ZY, Wang J and Ma R:
Quantification of human papilloma virus DNA in the plasma of
patients with cervical cancer. Clin J Surg Oncol. 2:288–291.
2010.
|
|
3
|
Qiao YL, Zhang WH, Li L, et al: The
cross-sectional comparative research of cervical cancer gene
screening method. Acta Acad Med Sin. 24:50–53. 2002.
|
|
4
|
Liu X, Roberts J, Dakic A, Zhang Y and
Scheleqel R: HPV E7 contributes to the telomerase activity of
immortalized and tumorigenic cells and augments E6-induced hTERT
promoter function. Virology. 375:611–623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chu NR, Wu HB, Wu T, Boux LJ, Sieqel MI
and Mizzen LA: Immunotherapy of a human papillomavirus (HPV) type
16 E7-expressing tumour by administration of fusion protein
comprising mycobacterium bovis bacille calmette-guerin (BCG) hsp65
and HPV16 E7. Clin Exp Immunol. 121:216–225. 2000. View Article : Google Scholar
|
|
6
|
Kaufmann AM, Stern PL, Rankin EM, et al:
Safety and immunogenicity of TA-HPV, a recombinant vaccinia virus
expressing modified human papillomavirus (HPV)-16 and HPV-18 E6 and
E7 genes, in women with progressive cervical cancer. Clin Cancer
Res. 8:3676–3685. 2002.
|
|
7
|
Veldman T, Horikawa I, Barrett JC and
Schleqel R: Transcriptional activation of the telomerase hTERT gene
by human papillomavirus type 16 E6 oncoprotein. J Virol.
75:4467–4472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishimura A, Nakahara T, Ueno T, et al:
Requirement of E7 oncoprotein for viability of HeLa cells. Microbes
Infect. 8:984–993. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dyson N, Howley PM, Münger K and Harlow E:
The human papilloma virus-16 E7 oncoprotein is able to bind to the
retinoblastoma gene product. Science. 243:934–937. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boyer SN, Wazer DE and Band V: E7: protein
of human papilloma virus-16 induces degradation of retinoblastoma
protein through the ubiquitin-proteasome pathway. Cancer Res.
56:4620–4624. 1996.PubMed/NCBI
|
|
11
|
Gonzalez SL, Stremlau M, He X, Basile JR
and Münqer K: Degradation of the retinoblastoma tumor suppressor by
the human papillomavirus type 16 E7 oncoprotein is important for
functional inactivation and is separable from proteasomal
degradation of E7. J Virol. 75:7583–7591. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gammoh N, Grm HS, Massimi P and Banks L:
Regulation of human papillomavirus type 16 E7 activity through
direct protein interaction with the E2 transcriptional activator. J
Virol. 80:1787–1797. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Chen W and Roman A: The E7
proteins of low- and high-risk human papillomaviruses share the
ability to target the pRB family member p130 for degradation. Proc
Natl Acad Sci USA. 103:437–442. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Caldeira S, Dong W and Tommasino M:
Analysis of E7/Rb associations. Methods Mol Med. 119:363–379.
2005.PubMed/NCBI
|
|
15
|
Wu EW, Clemens KE, Heck DV and Münger K:
The human papillomavirus E7 oncoprotein and the cellular
transcription factor E2F bind to separate sites on the
retinoblastoma tumor suppressor protein. J Virol. 67:2402–2407.
1993.
|
|
16
|
Alani RM and Münger K: Human
papillomaviruses and associated malignancies. J Clin Oncol.
116:330–337. 1998.
|
|
17
|
Cobrinik D: Pocket proteins and cell cycle
control. Oncogene. 24:2796–2809. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dimova DK and Dyson NJ: The E2F
transcriptional network: old acquaintances with new faces.
Oncogene. 24:2810–2826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Helt AM and Galloway DA: Destabilization
of the retinoblastoma tumor suppressor by human papillomavirus type
16 E7 is not sufficient to overcome cell cycle arrest in human
keratinocytes. J Virol. 75:6737–6747. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Sampath A, Raychaudhuri P and
Baqchi S: Both Rb and E7 are regulated by the ubiquitin proteasome
pathway in HPV-containing cervical tumor cells. Oncogene.
20:4740–4749. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen M, Gao S, Dong M, et al:
Chitosan/siRNA nanoparticles encapsulated in PLGA nanofibers for
siRNA delivery. ACS Nano. 6:4835–4844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holzerny P, Ajdini B, Heusermann W, Bruno
K, Schuleit M, Meinel L and Keller M: Biophysical properties of
chitosan/siRNA polyplexes: profiling the polymer/siRNA interactions
and bioactivity. J Control Release. 157:297–304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo C, Liu K, Zheng Y, Luo H, Chen H and
Huang L: Apoptosis induced by an antagonist peptide against HPV16
E7 in vitro and in vivo via restoration of p53. Apoptosis.
16:606–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shu XZ and Zhu KJ: The influence of
multivalent phosphate structure on the properties of ionically
cross-linked chitosan films for controlled drug release. Eur J
Pharm Biopharm. 54:235–243. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gref R, Domb A, Quellec P, Blunkc T,
Müllerd RH, Verbavatze JM and Langerf R: The controlled intravenous
delivery of drugs using PEG-coated sterically stabilized
nanospheres. Adv Drug Deliv Rev. 16:215–233. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bivas-Benita M, Romeijn S, Junginger HE
and Borchard G: PLGA-PEI nanoparticles for gene delivery to
pulmonary epithelium. Eur J Pharm Biopharm. 58:1–6. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Panyam J and Labhasetwar V: Biodegradable
nanoparticles for drug and gene delivery to cells and tissue. Adv
Drug Deliv Rev. 55:329–347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zauner W, Farrow NA and Haines AM: In
vitro uptake of polystyrene microspheres: effect of particle size,
cell line and cell density. J Control Release. 71:39–51. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang M, Fong CW, Khor E and Lim LY:
Transfection efficiency of chitosan vectors: effect of polymer
molecular weight and degree of deacetylation. J Control Release.
106:391–406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chan V, Mao HQ and Leong KW: Effect of
chitosan molecular weight in gene delivery process. Abs Pap Am Chem
Soc. 221:U347. 2001.
|
|
31
|
Rozema DB and Lewis DL: siRNA delivery
technologies for mammalian systems. Targets. 2:253–260. 2003.
View Article : Google Scholar
|
|
32
|
Keller M: Lipidic carriers of RNA/DNA
oligonucleotides and polynucleotides: what a difference a
formulation makes. J Control Release. 103:537–540. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tam P, Monck M, Lee D, et al: Stabilized
plasmid-lipid particles for systemic gene therapy. Gene Ther.
7:1867–1874. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bloomfield VA, He S, Li AZ and Arscott PB:
Light scattering studies on DNA condensation. Biochem Soc Trans.
19:4961991.PubMed/NCBI
|
|
35
|
Bloomfield VA: Condensation of DNA by
multivalent cations: considerations on mechanism. Biopolymers.
31:1471–1481. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bloomfield VA: DNA condensation. Curr Opin
Struct Biol. 6:334–341. 1996. View Article : Google Scholar
|
|
37
|
Chang JT, Kuo TF, Chen YJ, et al: Highly
potent and specific siRNAs against E6 or E7 genes of HPV16- or
HPV18-infected cervical cancers. Cancer Gene Ther. 17:827–836.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sima N, Wang W, Kong D, et al: RNA
interference against HPV16 E7 oncogene leads to viral E6 and E7
suppression in cervical cancer cells and apoptosis via upregulation
of Rb and p53. Apoptosis. 13:273–281. 2008. View Article : Google Scholar : PubMed/NCBI
|