Introduction
Mitochondria are unique organelles as they contain
their own DNA. The mitochondrial genome is a double-stranded
circular DNA which is composed of coding and non-coding regions
(1,2). Mitochondria are important in energy
metabolism, oxidative stress and cell apoptosis. Mitochondrial
replication and synthesis are regulated by multiple factors.
Replication of mitochondrial DNA (mtDNA) may be important in the
maintenance of mtDNA copy number. The displacement loop (D-loop)
region is the major control site for mtDNA replication and
transcription (3). The D-loop
region is increasingly susceptible to oxidative damage and
electrophilic attack (4,5), thus it is more prone to mutation. It
has been acknowledged that exercise causes an increase in the
skeletal muscle mitochondrial enzyme content and activity (6). Perturbations in mitochondrial content
and function have been linked to a wide variety of diseases in
multiple tissues, and exercise may serve as a potent approach by
which to prevent and treat these pathologies (5). However, no study has been conducted
to evaluate the association effects of exercise on mtDNA content
and mutations. In this study, we investigated the effects of
exercise intensity on mtDNA alterations of copy numbers and
mutations in the gastrocnemii of mice.
Materials and methods
Animals and exercise protocol
Fifty male C57BL/6 mice, with an average weight of
15±2 g, were used in this experiment. All animal use and protocols
were approved by the Sichuan University, Sichuan, China. Animals
were 4 weeks old at the beginning of the study. All animals were
maintained at room temperature on a 12 h light-dark cycle, with
free access to standard chow and water. The animals were randomly
divided into five groups (n=10); group A, 10 min of swimming per
day; group B, 30 min; group C, 60 min; group D, 90 min and control
group (group K). All mice in the exercise groups swam five times
per week for 20 weeks. All animals were sacrificed 48 h after the
last swimming session in order to examine the effect of total
exercise, but not the effects resulting from the last swim. The
gastrocnemii were carefully dissected, frozen in liquid nitrogen
and stored at −80°C until further analysis.
Detection of mtDNA content
Total DNA was isolated using the TIANamp Genomic DNA
kit (DP110413; TianGen Biotechnology, Beijing, China). The mtDNA
content was measured using real-time PCR. The primers used are
listed in Table I. A standard
curve was constructed using known concentration genomic DNA as a
standard. QPCR was performed using an ABI 7900HT Fast Real-Time PCR
System (Applied Biosystems, Carlsbad, CA, USA). The PCR conditions
were: 2 min at 94°C, followed by 40 cycles of 15 sec at 94°C, 15
sec at 58°C and 20 sec at 68°C; and a melting curve was
constructed. The mtDNA content was measured using the standard
curve.
 | Table IPrimers used for real-time PCR. |
Table I
Primers used for real-time PCR.
| Fragment | Primer sequences
(5′-3′) | Product size
(bp) |
|---|
| D-loop region | F:
CCAACTAGCCTCCATCTCATACTTC | 1185 |
| R:
GGACCAAACCTTTCTCTTTATGGGA | |
| mtDNA | F:
CACTCCTCGTCCCCATTCTA | 112 |
| R:
ATGCCGTATGGACCAACAAT | |
Detection of mtDNA mutations
PCR was performed for the whole length of the mtDNA
D-loop region, which was amplified using a 2X Taq PCR master mix
(KT201-02; TianGen) and the primers listed in Table I. The PCR conditions were as
follows: pre-denaturation for 4 min at 95°C, 30 cycles of 45 sec at
95°C, 55 sec at 58°C and 60 min at 72°C, with a final extension of
10 min at 72°C. PCR products for TA cloning, the positive bacilli,
were sequenced using delivery sequencing.
Statistical analysis
All data are expressed as the mean ± standard error
of the mean (SEM). Statistical analysis was performed using the
Wilcoxon signed-rank test and Fisher's exact probability test with
SPSS 13.0. P<0.01 was considered to indicate a statistically
significant difference.
Results
The mtDNA D-loop regions were successfully amplified
(Fig. 1). Homology analysis of the
sequencing results was conducted at http://blast.ncbi.nlm.nih.gov. The results detected
mutations, monobase insertions and deletions in the control group
and groups C and D which underwent 60 and 90 min of exercise,
respectively. There was no mutation detected in group A which
underwent 10 min of exercise. In group B which underwent 30 min of
exercise only one point mutation was identified (Table II).
 | Table IIMutations in the mtDNA D-loop regions
in the gastrocnemus of mice. |
Table II
Mutations in the mtDNA D-loop regions
in the gastrocnemus of mice.
| Nucleotide site | AY172335 sequence in
GeneBank | Control group | Exercise group |
|---|
|
|
|---|
| K | A | B | C | D |
|---|
| 15560 | C | Ins C | | | | |
| 16032 | G | - | | | | |
| 16232 | A | - | | | - | - |
| 16236 | T | T | | | - | |
| 16293 | T | T | | C | | |
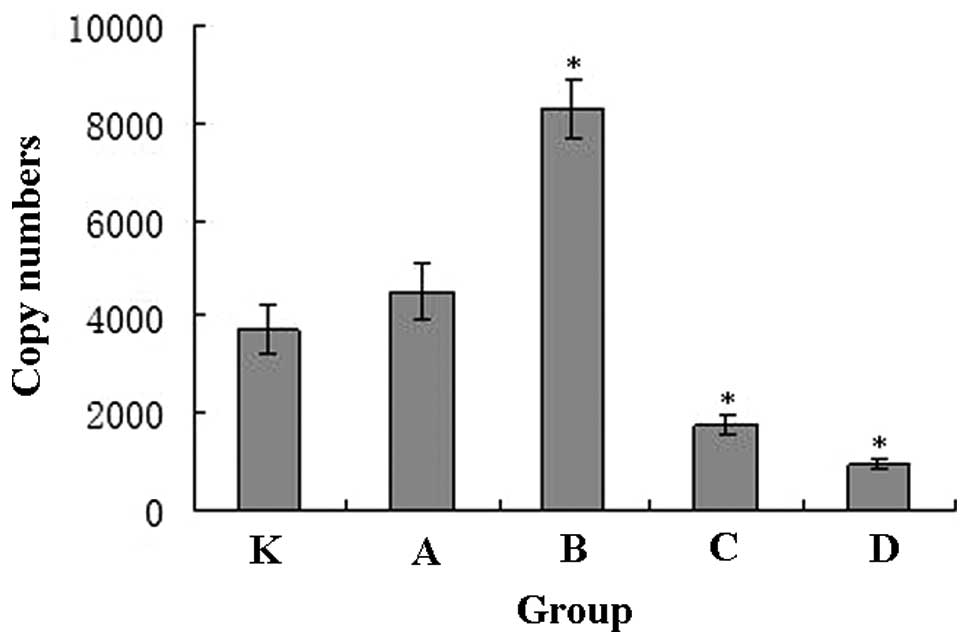
The melting curve of the real-time PCR products
demonstrated a single peak, indicating the high specificity of the
amplification. The standard curve correlation coefficient was 0.994
and the slope was −3.325. Using the slope, the Ct values and
concentrations of genomic DNA were used to calculate the mtDNA
contents of each group (Fig. 2).
The mtDNA content in groups A and B, which underwent 10 and 30 min
of exercise, was significantly increased, while the mtDNA content
in groups C and D, which underwent 60 and 90 min of exercise, was
significantly decreased.
Discussion
Mitochondria are important in energy metabolism,
oxidative stress and cell apoptosis. mtDNA is susceptible to damage
by environmental carcinogens and endogenous reactive oxygen species
(ROS) (3,7), resulting in mtDNA mutations or base
deletions. It has been confirmed that the mtDNA copy number and
mtDNA mutations are related to disease and aging. Recent research
has demonstrated that the mtDNA copy number is negatively
correlated with tumor cell differentiation. The alteration in the
mtDNA copy number may be an early event in oncogenesis. The mtDNA
copy number decreases during aging. Mutated mtDNA may be
preferentially amplified, and this increase might become clinically
relevant after deconditioning (8).
Considerable progress has been achieved towards the understanding
of basic mitochondrial genetics, including the correlation between
inherited mutations and disease phenotypes, and the identification
of acquired mtDNA mutations during aging and cancer. The mtDNA
D-loop is a non-coding sequence of the mitochondrial genome that is
implicated in mtDNA replication and transcription. The D-loop
region is more susceptible to oxidative damage and electrophilic
attack.
In this study, we sequenced the mtDNA D-loop region
of mouse gastrocnemii and detected a point mutation, monobase
insertion and deletion in the control group, the 60 min exercise
group and the 90 min exercise group, respectively. Extensive
exercise training can alter mutations in the mtDNA D-loop region of
mouse gastrocnemus cells, suggesting that 10 and 30 min of exercise
intensity is more appropriate than other exercise intensities for
reducing mouse gastrocnemus mtDNA D-loop mutations, while
high-intensity sports have a negative impact. Research has
demonstrated that the accumulation of mtDNA mutations and apoptosis
markers are closely related, therefore it has been speculated that
the accumulation of mtDNA mutations caused by apoptosis, may be a
core mechanism involved in aging and disease. In this study, 10 and
30 min per day of exercise training increased the relative mtDNA
copy number, however, 60 and 90 min per day of exercise
significantly reduced the mtDNA copy number; these results were in
accordance with the results of the mtDNA D-loop region mutations.
mtDNA mutations have the ability to cause injury to respiratory
chain subunit protein synthesis, formation of a defective
respiratory chain and reduction in the number of normal
mitochondria (9). Accumulation of
mtDNA mutations may cause apoptosis associated with reduced
mitochondrial copy number. The direct impact of reduced
mitochondrial copy number may reduce mitochondrial oxidative
phosphorylation, resulting in decreased productivity, and promotion
of ROS generation, resulting in decreased cell function and
apoptosis and promotion in the occurrence of aging and disease.
Moderate exercise can reduce damage caused by mtDNA mutations and
increase the number of normal mitochondria. It may enhance the
activity of mitochondria to prevent apoptosis, aging and
diseases.
In conclusion, our study demonstrated that regular
exercise could alter mtDNA mutations and mtDNA copy number. This
may provide a new approach to enhance the function of mitochondrial
oxidative phosphorylation for the prevention of disease and aging.
Further investigations are required.
Acknowledgements
This study was supported by the Sichuan University
Scientific Research Foundation for Young Teachers
(2009SCU11145).
References
|
1
|
Shen L, Fang H, Chen T, et al: Evaluating
mitochondrial DNA in cancer occurrence and development. Ann NY Acad
Sci. 1201:26–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bayona-Bafaluy MP, Acín-Pérez R, Mullikin
JC, et al: Revisiting the mouse mitochondrial DNA sequence. Nucleic
Acids Res. 31:5349–5355. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suzuki M, Toyooka S, Miyajima K, et al:
Alterations in the mitochondrial displacement loop in lung cancers.
Clin Cancer Res. 9:5636–5641. 2003.PubMed/NCBI
|
|
4
|
Mambo E, Gao X, Cohen Y, et al:
Electrophile and oxidant damage of mitochondrial DNA leading to
rapid evolution of homoplasmic mutations. Proc Natl Acad Sci USA.
100:1838–1843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park HW, Ahn Y, Jeong MH, et al: Chronic
atrial fibrillation associated with somatic mitochondrial DNA
mutations in human atrial tissue. J Clin Pathol. 60:948–950. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Little JP, Safdar A, Benton CR and Wright
DC: Skeletal muscle and beyond: the role of exercise as a mediator
of systemic mitochondrial biogenesis. Appl Physiol Nutr Metab.
36:598–607. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lievre A, Chapusot C, Bouvier AM, et al:
Clinical value of mitochondrial mutations in colorectal cancer. J
Clin Oncol. 23:3517–3525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taylor RW and Turnbull DM: Mitochondrial
DNA mutations in human disease. Nat Rev Genet. 6:389–402. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Richter CJ, Park BN and Ames BN: Normal
oxidative damage to mitochondrial and nuclear DNA is extensive.
Proc Natl Acad Sci USA. 85:6465–6467. 1988. View Article : Google Scholar : PubMed/NCBI
|