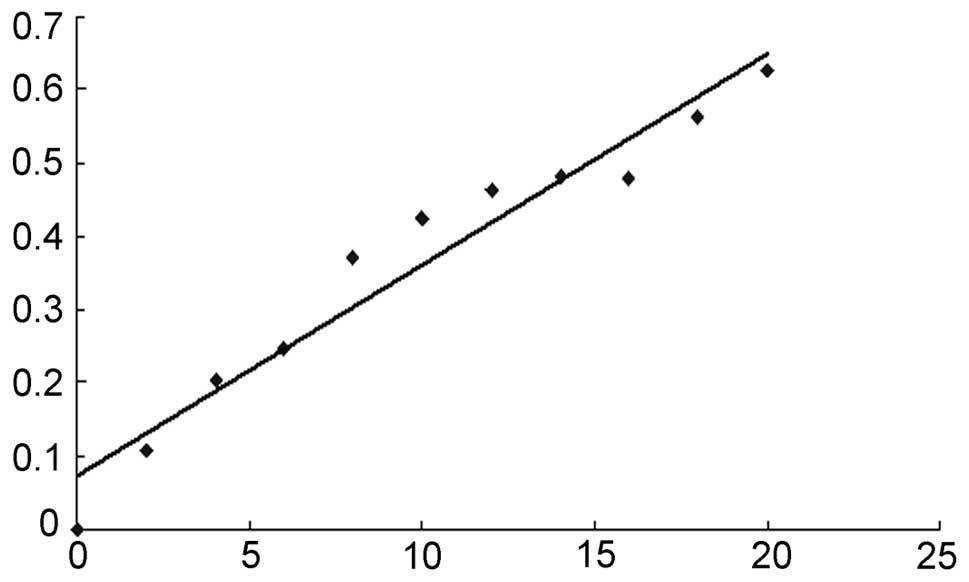
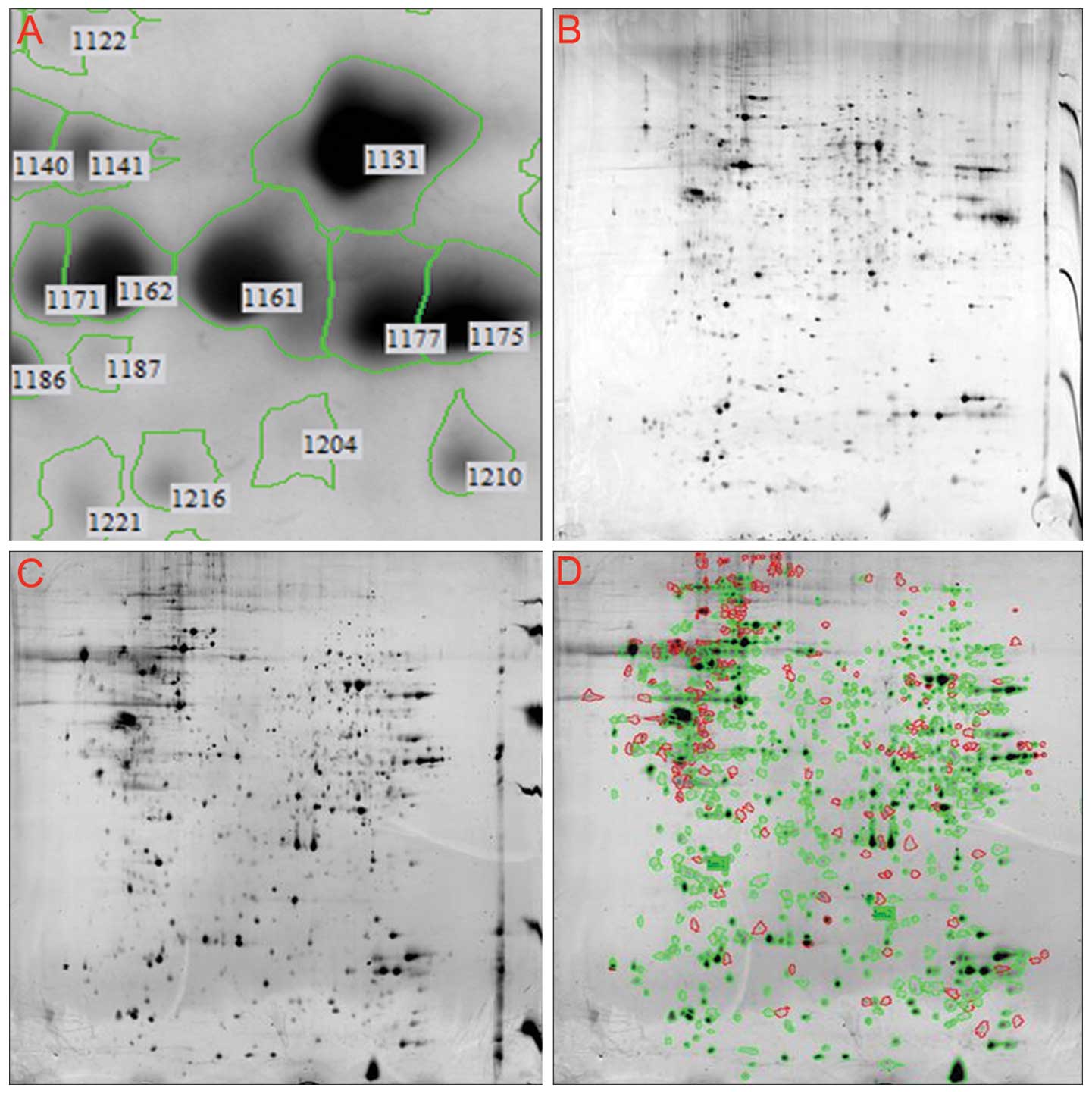
|
1
|
Gowri M, Jahan SK, Kavitha,
Prasannakumari, Madhumathi and Appaji L: Acute promyelocytic
leukemia with unusual karyotype. Indian J Hum Genet. 17:235–237.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo Y, Dolinko AV, Chinyengetere F, et al:
Blockade of the ubiquitin protease UBP43 destabilizes transcription
factor PML/RARα and inhibits the growth of acute promyelocytic
leukemia. Cancer Res. 70:9875–9885. 2010.PubMed/NCBI
|
|
3
|
Wang ZY and Chen Z: Acute promyelocytic
leukemia: from highly fatal to highly curable (Review). Blood.
111:2505–2515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kogan SC: Curing APL: differentiation or
destruction? Cancer Cell. 15:7–8. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lanotte M, Martin-Thouvenin V, Najman S,
Balerini P, Valensi F and Berger R: NB4, a maturation inducible
cell line with t(15;17) marker isolated from a human acute
promyelocytic leukemia (M3). Blood. 77:1080–1086. 1991.PubMed/NCBI
|
|
6
|
Wilkins MR, Appel RD, Van Eyk JE, et al:
Guidelines for the next 10 years of proteomics. Proteomics. 6:4–8.
2006.PubMed/NCBI
|
|
7
|
Müller K, Job C, Belghazi M, Job D and
Leubner-Metzger G: Proteomics reveal tissue-specific features of
the cress (Lepidium sativum L.) endosperm cap proteome and
its hormone-induced changes during seed germination. Proteomics.
10:406–416. 2010.PubMed/NCBI
|
|
8
|
Abu-Farha M, Elisma F, Zhou H, et al:
Proteomics: from technology developments to biological applications
(Review). Anal Chem. 81:4585–4599. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Washburn MP, Wolters D and Yates JR III:
Large-scale analysis of the yeast proteome by multidimensional
protein identification technology. Nat Biotechnol. 19:242–247.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anderson NL, Polanski M, Pieper R, et al:
The human plasma proteome: a nonredundant list developed by
combination of four separate sources. Mol Cell Proteomics.
3:311–326. 2004.PubMed/NCBI
|
|
11
|
Andersen JS, Lam YW, Leung AK, et al:
Nucleolar proteome dynamics. Nature. 433:77–83. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rabilloud T: Two-dimensional gel
electrophoresis in proteomics: old, old fashioned, but it still
climbs up the mountains. Proteomics. 2:3–10. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wilkins MR, Gasteiger E, Sanchez JC,
Bairoch A and Hochstrasser DF: Two-dimensional gel electrophoresis
for proteome projects: the effects of protein hydrophobicity and
copy number. Electrophoresis. 19:1501–1505. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yates JR, Ruse CI and Nakorchevsky A:
Proteomics by mass spectrometry: approaches, advances, and
applications. Annu Rev Biomed Eng. 11:49–79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han X, Aslanian A and Yates JR III: Mass
spectrometry for proteomics. Curr Opin Chem Biol. 12:483–490. 2008.
View Article : Google Scholar
|
|
16
|
Doran P, Donoghue P, O’Connell K, Gannon J
and Ohlendieck K: Proteomic profiling of pathological and aged
skeletal muscle fibres by peptide mass fingerprinting (Review). Int
J Mol Med. 19:547–564. 2007.PubMed/NCBI
|
|
17
|
Concu R, Dea-Ayuela MA, Perez-Montoto LG,
et al: Prediction of enzyme classes from 3D structure: a general
model and examples of experimental-theoretic scoring of peptide
mass fingerprints of Leishmania proteins. J Proteome Res.
8:4372–4382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nesvizhskii AI, Vitek O and Aebersold R:
Analysis and validation of proteomic data generated by tandem mass
spectrometry (Review). Nat Methods. 4:787–797. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swaney DL, McAlister GC and Coon JJ:
Decision tree-driven tandem mass spectrometry for shotgun
proteomics. Nat Methods. 5:959–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Görg A, Obermaier C, Boguth G, et al: The
current state of two-dimensional electrophoresis with immobilized
pH gradients (Review). Electrophoresis. 21:1037–1053. 2000.
|
|
21
|
Pasquali C, Fialka I and Huber LA:
Preparative two-dimensional gel electrophoresis of membrane
proteins. Electrophoresis. 18:2573–2581. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
González-Díaz H, González-Díaz Y, Santana
L, Ubeira FM and Uriarte E: Proteomics, networks and connectivity
indices (Review). Proteomics. 8:750–778. 2008.
|
|
23
|
Hanash SM, Bobek MP, Rickman DS, et al:
Integrating cancer genomics and proteomics in the post-genome era.
Proteomics. 2:69–75. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wittmann-Liebold B, Graack HR and Pohl T:
Two-dimensional gel electrophoresis as tool for proteomics studies
in combination with protein identification by mass spectrometry.
Proteomics. 6:4688–4703. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gygi SP, Corthals GL, Zhang Y, Rochon Y
and Aebersold R: Evaluation of two-dimensional gel
electrophoresis-based proteome analysis technology. Proc Natl Acad
Sci USA. 97:9390–9395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oh-Ishi M and Maeda T: Disease proteomics
of high-molecular-mass proteins by two-dimensional gel
electrophoresis with agarose gels in the first dimension (Agarose
2-DE) (Review). J Chromatogr B Analyt Technol Biomed Life Sci.
849:211–222. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wisniewski JR, Zougman A, Nagaraj N and
Mann M: Universal sample preparation method for proteome analysis.
Nat Methods. 6:359–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Isola D, Marzban G, Selbmann L, Onofri S,
Laimer M and Sterflinger K: Sample preparation and 2-DE procedure
for protein expression profiling of black microcolonial fungi.
Fungal Biol. 115:971–977. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pluskal T, Castillo S, Villar-Briones A
and Oresic M: MZmine 2: modular framework for processing,
visualizing, and analyzing mass spectrometry-based molecular
profile data. BMC Bioinformatics. 11:3952010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahmed FE: Sample preparation and
fractionation for proteome analysis and cancer biomarker discovery
by mass spectrometry (Review). J Sep Sci. 32:771–798.
2009.PubMed/NCBI
|