Introduction
Retroviruses, found in all mammals and in a wide
range of other vertebrates, provide unique opportunities to study
the biology and evolution of virus-host relationships.
Occasionally, infection of a germline cell by a retrovirus may lead
to an integrated provirus that is passed to the offspring and
inherited in a Mendelian fashion; known as an endogenous retrovirus
(ERV). Human endogenous retroviruses (HERVs) constitute ~8% of the
human genome and are distributed in ~700,000 different loci
(1). Expression of HERVs has been
associated with several positive physiological functions as well as
certain diseases, although their role as an etiological agent, a
possible contributing factor or a disease marker remains to be
established (2).
The proviral structure of HERV elements mainly
consists of
5′LTR-gag-pro-pol-env-3′LTR, in which
the 4 genes (gag, group-specific antigen; pro,
protease; pol, polymerase; and env, envelope) encode
structural/functional proteins essential to a replication-competent
retrovirus (3). The long terminal
repeats (LTRs) at both ends differentiate HERVs from other
retrotransposons, such as long interspersed nuclear elements
(LINEs). The majority of the remnants of HERVs are simply isolated
LTR copies, with the internal sequence having been lost during
integration or via homologous recombination. HERV families are
defined by different criteria, such as homogeneity to their
exogenous counterparts, sequence similarity of the pol genes
and the primer binding site (PBS) immediately downstream of the
5′LTR. H family HERV (HERV-H) contain a PBS with a sequence similar
to human tRNAHis(4).
HERV-H is one of the most abundant endogenous
retroviral families in the human genome, consisting of full-length
elements (~100 copies, of which only 18 are relatively complete)
(3), elements deleted in
pol and env [800–900 copies, of which only 3 have
been identified to contain intact env open reading frames
(ORFs)] (5), and solitary LTRs
(~1,000 copies) (6). HERV-H was
inserted into primate genomes prior to the divergence of New and
Old World monkeys, and the elements lacking in pol and
env were integrated with the Old World monkey lineage
(5,7,8).
HERV-H env fragments are found on human chromosomes 1, 2, 3,
4, 5, 6, 7, 9, 10, 11, 12, 14, 15, 16, 17, 18, 19, 20, X and Y.
These new HERV-H env fragments showed 82–99% sequence
similarity to that of HERV-H. The HERV-H members evolved by
intra-chromosomal spread during hominid radiation (9).
Using the inter-retrotransposon amplified
polymorphism (IRAP) technique, which is one of the
retrotransposon-based markers, regions flanked by two
LTR-retrotransposons or solo LTRs are amplified (10). In this method, polymorphisms are
detected by the presence or absence of the PCR product, where the
lack of amplification indicates the absence of a retrotransposon at
that particular region (11). IRAP
was used to investigate genetic relationships between different and
related species (12), for gene
mapping (13) and for the
characterization of the somaclonal variants in plants (14–16).
ERVs differ between host species, most likely in
correlation with differences in expression conditions, and in the
evolutionary and environmental history of each host (17). There are very limited data on
environmentally and developmentally (ED) regulated genomic
variation. Further research is required to understand the
cytogenetic, DNA-structural and gene regulation basis of ED-genomic
variation, and to study the importance of ED-genomic variation for
population genetics, adaptation, domestication, evolution, medical
science, plant science and agriculture (18).
In recent years, there have been a number of studies
on the different expression profiles of HERV-H in various clinical
situations (3,19,20).
However, there are no data on HERV-H provirus integration
variations between healthy individuals. In this study, we report
the HERV-H integration polymorphisms between healthy
individuals.
Materials and methods
Subjects
This study included 20 individuals (10 males and 10
females; age, 18–30 years) with different ethnic origins (Turkey,
Azerbaijan, Indonesia, China and Somalia; Table I). The study design was approved as
a Master’s Thesis (Project no. T-17640) by the Istanbul University,
Istanbul, Turkey. The blood samples were collected by the Istanbul
University Medico-Social Center, Istanbul, Turkey. Blood samples
were obtained by the Istanbul University Medico-Social Center,
Turkey, with the written consent of the healthy individuals.
 | Table IEvaluation forms of subjects. |
Table I
Evaluation forms of subjects.
| | | | Exposure to |
|---|
| | | |
|
|---|
| Subject | Gender | Age (years) | Ethnic group | Radiation | Infectious
disease |
|---|
| 1 | M | 28 | Asia
(Azerbaijan) | x | |
| 2 | M | 21 | Asia
(Azerbaijan) | x | |
| 3 | M | 19 | Asia
(Azerbaijan) | x | x |
| 4 | F | 24 | Asia
(Azerbaijan) | | x |
| 5 | M | 26 | Asia
(Azerbaijan) | | |
| 6 | F | 23 | Asia
(Azerbaijan) | | |
| 7 | M | 28 | Asia
(Azerbaijan) | x | |
| 8 | M | 28 | Asia (Iran -
Azerbaijan) | x | |
| 9 | F | 23 | Asia (Turkey) | | |
| 10 | F | 25 | Asia (China-Eastern
Turkmenistan) | x | x |
| 11 | F | 23 | Asia (Turkey) | | |
| 12 | M | 22 | Asia (Turkey) | | x |
| 13 | F | 22 | Asia (Turkey) | x | |
| 14 | M | 22 | Asia (Turkey) | | |
| 15 | F | 22 | Asia (Turkey) | x | |
| 16 | M | 23 | Asia (Turkey) | | x |
| 17 | F | 20 | Asia (Turkey) | | |
| 18 | F | 20 | Asia
(Indonesia) | x | |
| 19 | F | 22 | Africa
(Somali) | | |
| 20 | M | 24 | Asia (Turkey) | x | |
Genomic DNA isolation
Genomic DNA was extracted from blood samples using a
High Pure PCR Template Preparation kit (Roche Diagnostics,
Mannheim, Germany). The DNA concentrations were measured using a
spectrophotometer (Nanodrop2000) and genomic DNA samples were
separated on an EtBr-stained, 1% agarose gel to estimate the
quality of the DNA.
IRAP analysis
HERV-H integration polymorphisms were investigated
using the IRAP technique. LTR primers were used for IRAP analysis.
The primers shown in Table II
were designed using the online primer design site of Integrated DNA
Technologies (http://eu.idtdna.com/PrimerQuest/Home/Index). A pair
of primers was designed for each LTR7A, LTR7B and LTR7C region.
HERV-H LTR sequences were obtained from a relevant database
(www.girinst.org, AC D11078). PCR was performed using a
thermal cycler (Techne, TC3000) in a total volume of 20 μl,
containing 75 ng of template DNA, 10 μM of forward and reverse
primers and SapphireAmp Master Mix (2X). PCR conditions were as
follows: initial denaturation at 95°C (3 min) followed by 30 cycles
of denaturation at 94°C (30 sec), annealing at 65°C for LTR7A,
68.2°C for LTR7B and 67°C for LTR7C (30 sec) and extension at 72°C
(3 min). The reaction was completed by an additional extension at
72°C for 5 min. A total of 20 μl of IRAP-PCR products were mixed
with 4 μl 6X loading buffer (10 mM Tris-HCl, 60 mM EDTA, pH 8.0,
0.3% bromophenol blue, 60% glycerol) and resolved by 2% agarose gel
electrophoresis at 180 V for 2 h in 1X TAE buffer (90 mM Tris, 90
mM boric acid and 2 mM EDTA, pH 8.0). A molecular weight marker
(GeneRuler™ DNA Ladder Mix, SM0331, Fermentas, Waltham, MA, USA)
was also loaded to determine the size of the amplicons. After
running, the gels were photographed on a UV transilluminator and
analyzed visually using Kodak Molecular Imaging Software (1994–2007
Carestream Health, Inc., Rochester, NY, USA).
 | Table IIPrimers used for IRAP analyses for
HERV-H integration polymorphism study. |
Table II
Primers used for IRAP analyses for
HERV-H integration polymorphism study.
| Primers | G + C ratio
(%) | Tm (°C) | Sequence
(5′-3′) |
|---|
| LTR7A |
| Forward | 50 | 72 |
TGTTTGGTGGTCTCTTCACACGGA |
| Reverse | 50 | 72 |
ATGGTATGGCTTAGCTTGGGCTCA |
| LTR7B |
| Forward | 50 | 72 |
TGTTTGGTGGTCTCTTCACACGGA |
| Reverse | 50 | 72 |
ATGATGGCTTAGCTTGGGCTCAGA |
| LTR7C |
| Forward | 50 | 72 |
TTGCACCCAAGTGAATAAACGGCC |
| Reverse | 50 | 72 |
TTACAATGGCTGAGCTTCGGCTCA |
Statistical analysis
Polymorphism rates were calculated using Jaccard
similarity coefficient and the scores were evaluated statistically
by ANOVA test. P<0.05 was considered to indicate a statistically
significant difference.
Results
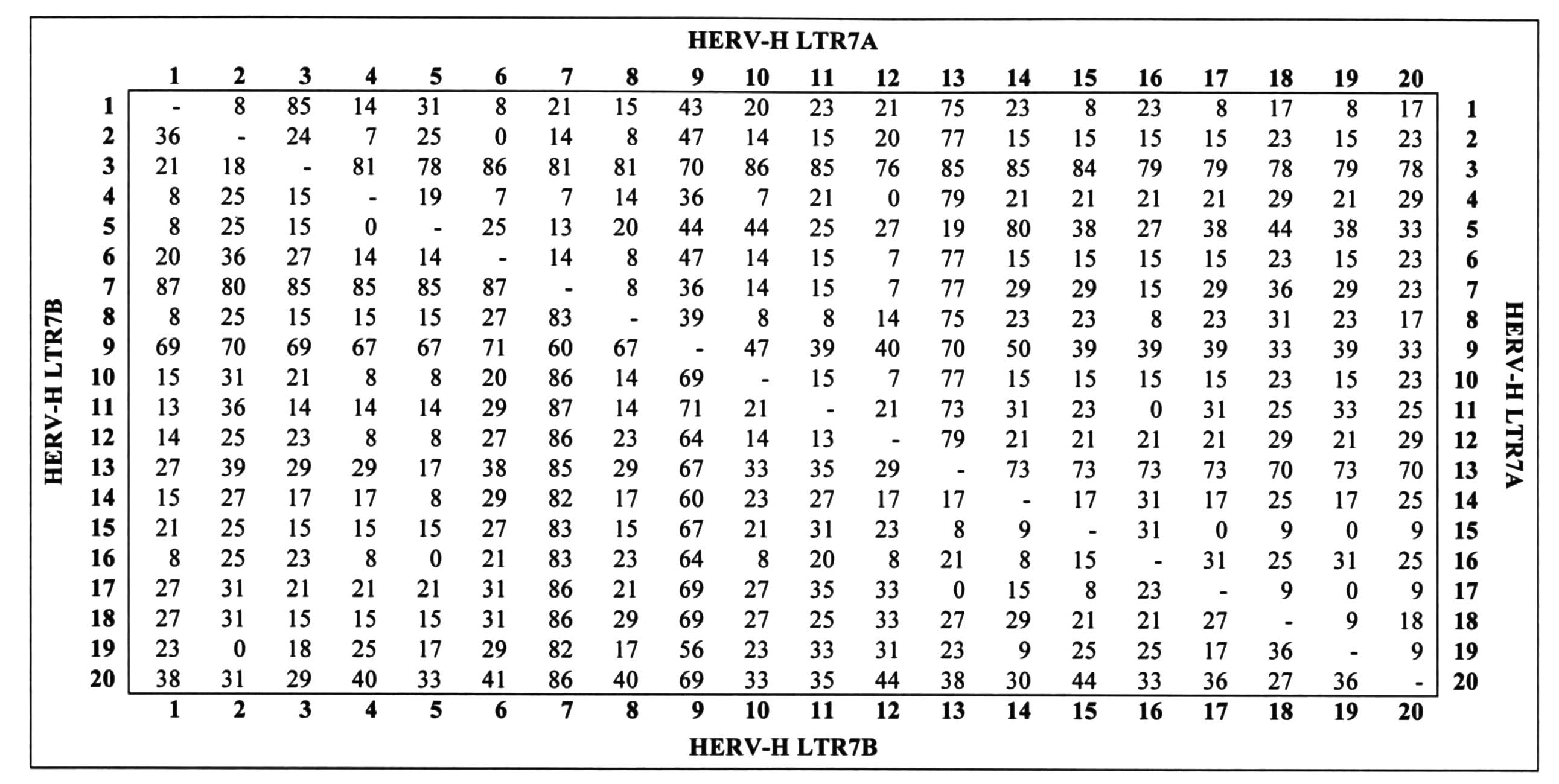
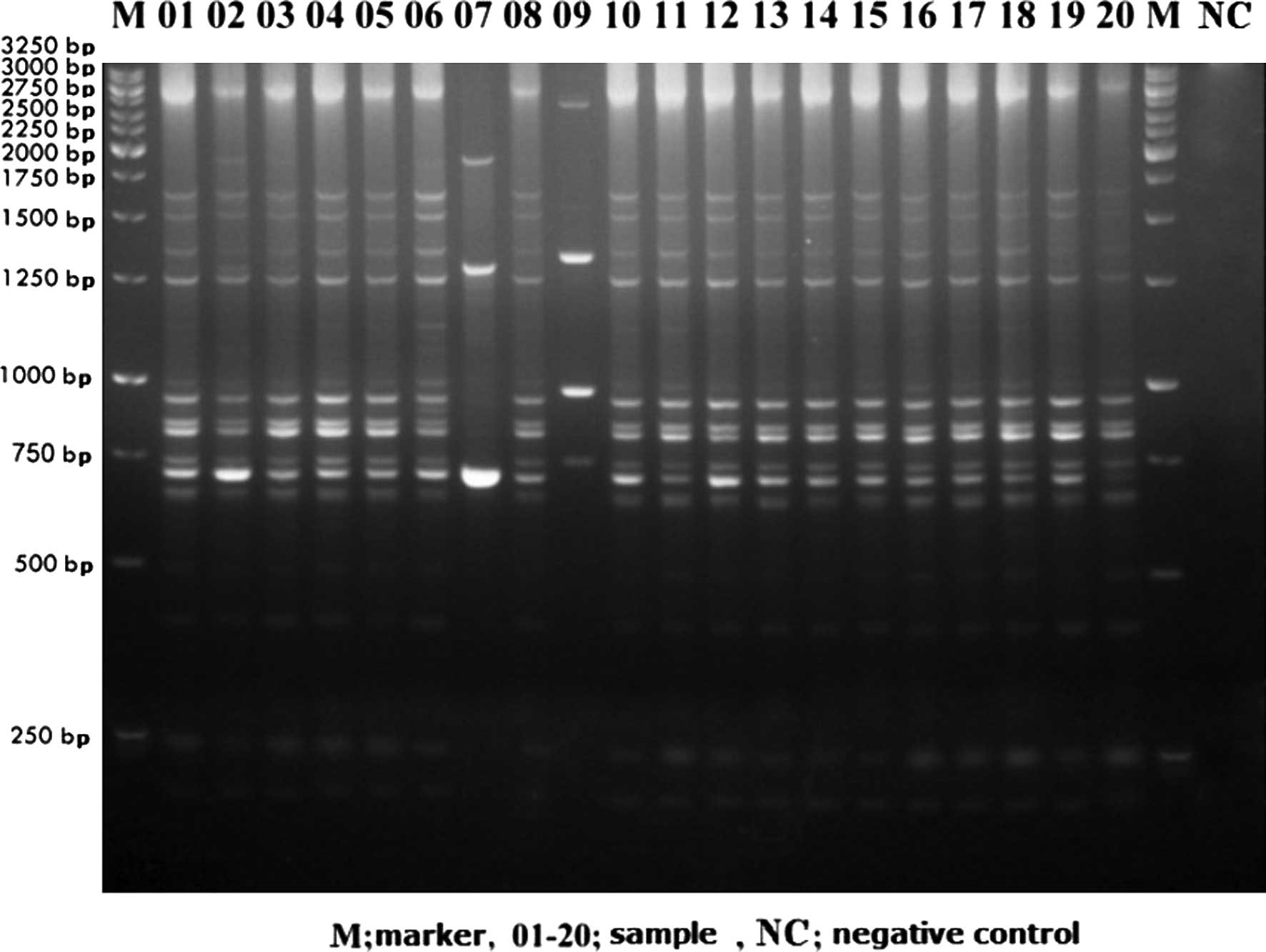
IRAP-PCR analyses using HERV-H LTR7A primers with 20
human DNA samples are shown in Fig.
1. Kodak Molecular Imaging Software analyses are presented in
Fig. 2. As can be observed in
Fig. 2, polymorphisms between
individuals were between 0–86%. Under the same experimental
conditions with HERV-H LTR7B primers, we also observed
polymorphisms (Fig. 3). Using
Kodak Molecular Imaging Software, the polymorphism rate between the
individuals was calculated to be between 0–87% with HERV-H LTR7B
(Fig. 2). These polymorphism rates
were used to calculate the p value by ANOVA test. The polymorphism
rates between individuals were found to be significant (P≤0.05).
However, we were not able to detect any amplification products
using HERV-H LTR7C primers. No significant data were obtained by
comparison of the ethnic groups. In the Azerbaijani group,
variations were calculated to be between 2.65–9.73% in terms of
HERV-H LTR7A and 1.40–5.63% in terms of HERV-H LTR7B.
Discussion
In the present study, LTR7A (450 bp), LTR7B (445 bp)
and LTR7C (471 bp) regions of HERV-H were analyzed by IRAP-PCR and
the integration patterns between the individuals were compared.
Blood samples from 20 individuals of various ethnic origins were
used for determination of the integration variations at the genomic
level. Isolated genomic DNA was screened using 3 pairs of primers
covering LTR regions of the HERV-H gene. The IRAP technique was
performed for polymorphism analyses. We observed integration
polymorphism patterns in all the tested individuals. To date, there
has been no report on HERV-H integration polymorphisms between
individuals; however, there are limited reports on integration
polymorphisms of HERV-K families (21–23).
Recently, database documentation relating to Alu retrotransposon
insertion polymorphisms (RIPs) was presented (24).
Insertion polymorphism of retroelements (REs) has
also attracted considerable attention due to certain advantageous
features that make REs useful tools for human population genetic
studies (25–27). These include known ancestral state
(absence of the RE), stability of insertion (there are no
mechanisms for removing inserted repeats) and relatively easy
detection. Moreover, an event of two independent integrations into
one and the same locus is very improbable, making this type of
polymorphism essentially homoplasy-free (28,29).
Previously, a number of cases of RIPs have been studied for
evolutionarily young groups of short interspersed elements (SINEs)
and long interspersed elements (LINEs) (30,31).
Although insertion polymorphisms of HERVs appear to be a rare
event, in our study we observed a high number of insertion
polymorphisms between individuals in terms of HERV-H insertion. We
were not able to observe significant differences between ethnic
groups, since we observed different degrees of polymorphisms
between all the test subjects.
The HERV-H family is the largest group among the
HERVs (19,32) with ~100 copies of full-length
HERV-H elements, 800–900 copies of HERV-H lacking pol and
env genes and solitary LTRs (up to ~1000 copies) in the
human genome (6,19). HERV-H elements integrated into the
primate genome 40 million years ago and their numbers have been
increasing over the past 30–35 million years (8,19,32).
Our results showed that HERV-H is still active in the human genome.
These data are also important from an evolutionary point of
view.
HERV-H contains regions of the gene caused by
mutations known to be due to integration into the human genome. An
important gene mutation of retroviruses in the env ORF was
detected (33). The total size of
HERV-H is 9 kb in the human genome and HERV-H has ORFs for
gag, pro, pol and env genes. LTR
sequences have variants of 1.1% due to mutations in the env
gene. Research on the env gene has demonstrated new
alternative reading frames (8,22)
and env expression was observed in normal tissues and colon
tumors.
To date, HERV-H research has been focused in
particular on investigating the relationship between neurological
diseases, cancer and gene expression (19,20).
In our study we were able to observe, for the first time, that
major genomic variations between individuals derive from the
integration of various elements. These polymorphic HERVs may be
associated with diseases. A gene that has an RIP may have potential
alternative transcripts, a promoter activity of alternative splice
sites, or create a poly-A signal (35).
IRAP is a valuable technique for searching the
retrotransposon movements in the plant kingdom (11,15).
We also demonstrated that this valuable technique can be
successfully applied to human research. Although the experiment was
conducted with a limited number of subjects, we were able to
observe polymorphisms and variants of inter-retrotransposon
regions. Our data may contribute to the understanding of the
variation of HERV-H in human populations. This study provides new
research areas on possible integration sites, and the existence of
infectious HERV-H variants may also be considered.
Acknowledgements
This study was supported by the Istanbul University
Research Foundation Project (no. T-17640).
References
|
1
|
Lander ES, Linton LM, Birren B, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jern P and Coffin JM: Effects of
retroviruses on host genome function. Annu Rev Genet. 42:709–732.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jern P, Sperber GO, Ahlsén G and Blomberg
J: Sequence variability, gene structure, and expression of
full-length human endogenous retrovirus H. J Virol. 79:6325–6337.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tristem M: Identification and
characterization of novel human endogenous retrovirus families by
phylogenetic screening of the human genome mapping project
database. J Virol. 74:3715–3730. 2000. View Article : Google Scholar
|
|
5
|
de Parseval N, Casella J, Gressin L and
Heidmann T: Characterization of the three HERV-H proviruses with an
open envelope reading frame encompassing the immunosuppressive
domain and evolutionary history in primates. Virology. 279:558–569.
2001.PubMed/NCBI
|
|
6
|
Hirose Y, Takamatsu M and Harada F:
Presence of env genes in members of the RTVL-H family of
human endogenous retrovirus-like elements. Virology. 192:52–61.
1993.
|
|
7
|
Goodchild N, Wilkinson DA and Mager D:
Recent evolutionary expansion of a subfamily of RTVL-H human
endogenous retrovirus-like elements. Virology. 196:778–788. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderssen S, Sjøttem E, Svineng G and
Johansen T: Comparative analyses of LTRs of the ERV-H family of
primate-specific retrovirus-like elements isolated from marmoset,
African green monkey, and man. Virology. 234:14–30. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi JM and Kim HS: Evolutionary
implications of human endogenous retrovirus HERV-H family. J Hum
Genet. 49:215–219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schulman AH and Kalendar R: A movable
feast: diverse retrotransposons and their contribution to barley
genome dynamics. Cytogenet Genome Res. 110:598–605. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalendar R, Grob T, Regina M, Suoniemi A
and Schulman A: IRAP and REMAP: two new retrotransposon-based DNA
fingerprinting techniques. Theor Appl Genet. 98:704–711. 1999.
View Article : Google Scholar
|
|
12
|
Guo D, Zhang H and Luo Z: Genetic
relationships of Diospyros kaki Thunb. and related species
revealed by IRAP and REMAP analysis. Plant Sci. 170:528–533.
2006.
|
|
13
|
Manninen O, Kalendar R, Robinson J and
Schulman AH: Application of BARE-1 retrotransposon markers
to the mapping of a major resistance gene for net blotch in barley.
Mol Gen Genet. 264:325–334. 2000.
|
|
14
|
Muhammad AJ and Othman RY:
Characterization of Fusarium wilt-resistant and
Fusarium wilt-susceptible somaclones of banana cultivar
rastali (Musa AAB) by random amplified polymorphic DNA and
retrotransposon markers. Plant Mol Biol Rep. 23:241–249. 2005.
|
|
15
|
Evrensel C, Yılmaz S, Temel A and
Gozukirmizi N: Variations in BARE-1 insertion patterns in
barley callus cultures. Genet Mol Res. 10:980–987. 2011.
|
|
16
|
Bayram E, Yilmaz S, Hamat-Mecbur H,
Kartal-Alacam G and Gozukirmizi N: Nikita retrotransposon
movements in callus cultures of barley (Hordeum vulgare L.).
POJ. 5:211–215. 2012.
|
|
17
|
Grandbastien MA, Audeon C, Bonnivard E, et
al: Stress activation and genomic impact of Tnt1 retrotransposons
in Solanaceae. Cytogenet Genome Res. 110:229–241. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li XQ: Developmental and environmental
variation in genomes. Heredity (Edinb). 102:323–329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi JM, Kim HM and Kim HS: Human endogenous
retrovirus HERV-H family in human tissues and cancer cells:
expression, identification and phylogeny. Cancer Lett. 231:228–239.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laska MJ, Brudek T, Nissen KK, Christensen
T, Møller-Larsen A, Petersen T and Nexø BA: Expression of HERV-Fc1,
a human endogenous retrovirus, is increased in patients with active
multiple sclerosis. J Virol. 86:3713–3722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mamedov I, Lebedev Y, Hunsmann G,
Khusnutdinova E and Sverdlov E: A rare event of insertion
polymorphism of a HERV-K LTR in the human genome. Genomics.
84:596–599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Belshaw R, Dawson AL, Woolven-Allen J,
Redding J, Burt A and Tristem M: Genomewide screening reveals high
levels of insertional polymorphism in the human endogenous
retrovirus family HERV-K (HML2): implications for present-day
activity. J Virol. 79:12507–12514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moyes D, Griffiths DJ and Venables PJ:
Insertional polymorphisms: a new lease of life for endogenous
retroviruses in human disease. Trends Genet. 23:326–333. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang P and Tang W: Database documentation
of retrotransposon insertion polymorphisms. Front Biosci (Elite
Ed). 4:1542–1555. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Batzer MA and Deininger PL: Alu repeats
and human genomic diversity. Nat Rev Genet. 3:370–379. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Watkins WS, Rogers AR, Ostler C, et al:
Genetic variation among world populations: inference from 100 Alu
insertion polymorphisms. Genome Res. 13:1607–1618. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bamshad MJ, Wooding S, Watkins WS, Ostler
CT, Batzer MA and Jorde LB: Human population genetic structure and
inference of group membership. Am J Hum Genet. 72:578–589. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roy-Engel AM, Carroll ML, El-Sawy M, Salem
AH, Garber RK, Nguyen SV, Deininger PL and Batzer MA:
Non-traditional Alu evolution and primate genomic diversity. J Mol
Biol. 316:1033–1040. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vincent BJ, Myers JS, Ho HJ, Kilroy GE,
Walker JA, Watkins WS, Jorde LB and Batzer MA: Following the LINEs:
an analysis of primate genomic variation at human-specific LINE-1
insertion sites. Mol Biol Evol. 20:1338–1348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carroll ML, Roy-Engel AM, Nguyen SV, et
al: Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and
their contribution to human genomic diversity. J Mol Biol.
311:17–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Myers JS, Vincent BJ, Udall H, et al: A
comprehensive analysis of recently integrated human Ta L1 elements.
Am J Hum Genet. 71:312–326. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mager DL and Freeman JD: HERV-H endogenous
retroviruses: prescence in the New World branch but amplification
in the Old World primate lineage. Virology. 213:395–404. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lindeskog M, Mager DL and Blomberg J:
Isolation of a human endogenous retroviral HERV-H element with an
open env reading frame. Virology. 258:441–450. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang AY, Gulden PH, Woods AS, et al: The
immunodominant major histocompatibility complex class I-restricted
antigen of a murine colon tumor derives from an endogenous
retroviral gene product. Proc Natl Acad Sci USA. 93:9730–9735.
1996. View Article : Google Scholar
|
|
35
|
Sin HS, Huh JH, Kim WY, et al: Long
terminal repeats of human endogenous retrovirus H family provide
alternative poly-adenylation signals to NADSYN1 gene. Korean J
Genet. 29:395–401. 2007.
|