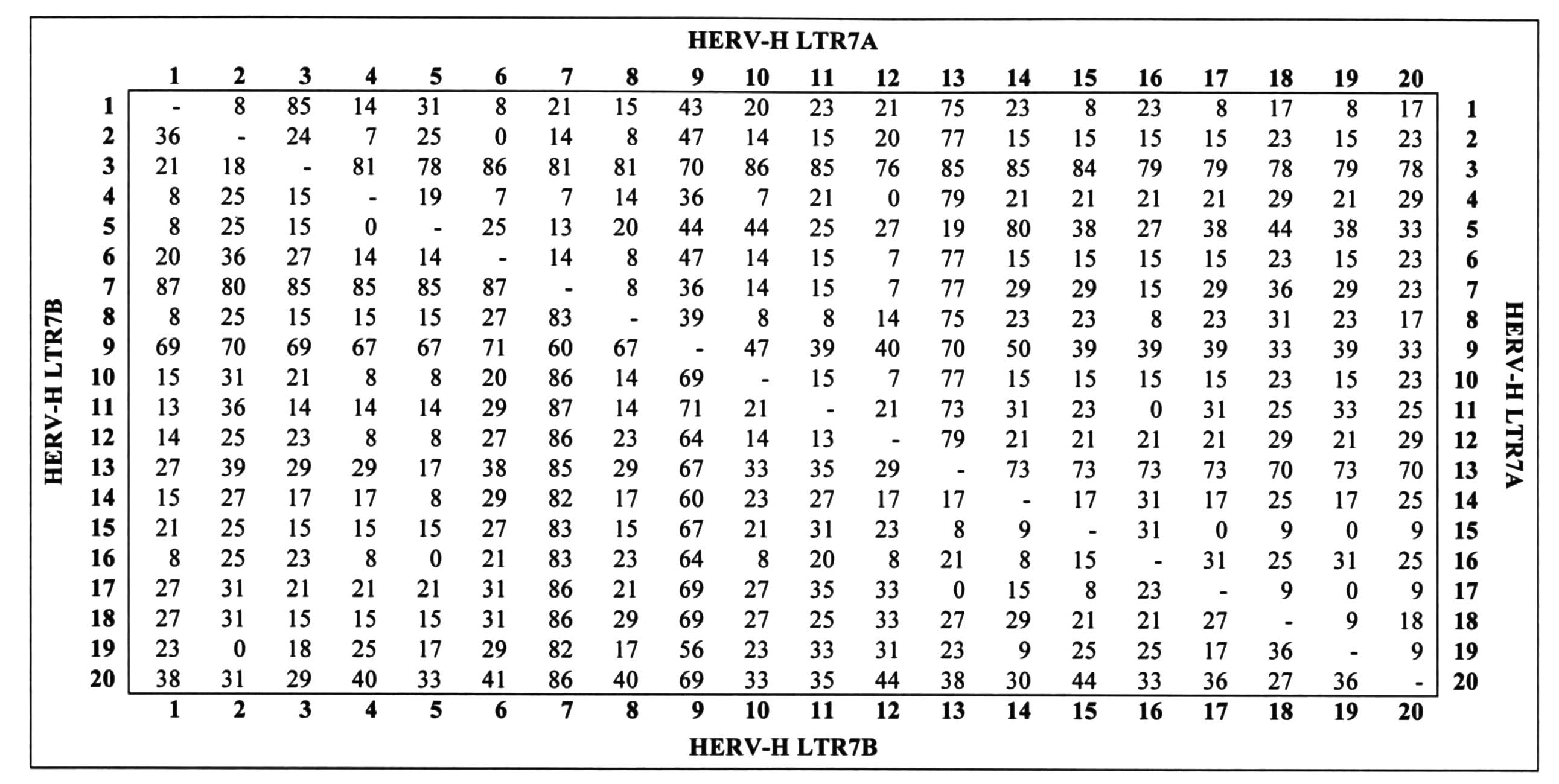
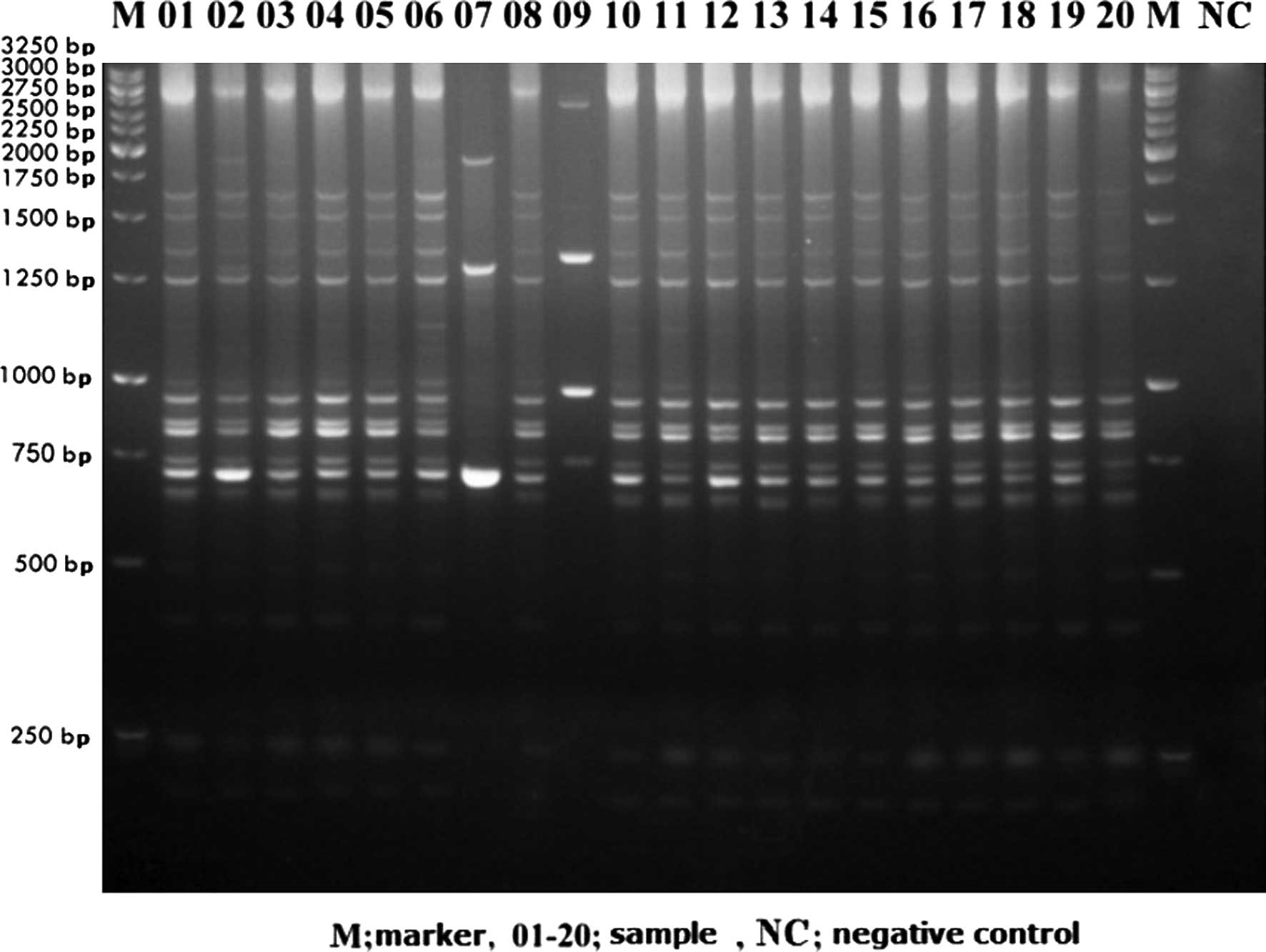
|
1
|
Lander ES, Linton LM, Birren B, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jern P and Coffin JM: Effects of
retroviruses on host genome function. Annu Rev Genet. 42:709–732.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jern P, Sperber GO, Ahlsén G and Blomberg
J: Sequence variability, gene structure, and expression of
full-length human endogenous retrovirus H. J Virol. 79:6325–6337.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tristem M: Identification and
characterization of novel human endogenous retrovirus families by
phylogenetic screening of the human genome mapping project
database. J Virol. 74:3715–3730. 2000. View Article : Google Scholar
|
|
5
|
de Parseval N, Casella J, Gressin L and
Heidmann T: Characterization of the three HERV-H proviruses with an
open envelope reading frame encompassing the immunosuppressive
domain and evolutionary history in primates. Virology. 279:558–569.
2001.PubMed/NCBI
|
|
6
|
Hirose Y, Takamatsu M and Harada F:
Presence of env genes in members of the RTVL-H family of
human endogenous retrovirus-like elements. Virology. 192:52–61.
1993.
|
|
7
|
Goodchild N, Wilkinson DA and Mager D:
Recent evolutionary expansion of a subfamily of RTVL-H human
endogenous retrovirus-like elements. Virology. 196:778–788. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderssen S, Sjøttem E, Svineng G and
Johansen T: Comparative analyses of LTRs of the ERV-H family of
primate-specific retrovirus-like elements isolated from marmoset,
African green monkey, and man. Virology. 234:14–30. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi JM and Kim HS: Evolutionary
implications of human endogenous retrovirus HERV-H family. J Hum
Genet. 49:215–219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schulman AH and Kalendar R: A movable
feast: diverse retrotransposons and their contribution to barley
genome dynamics. Cytogenet Genome Res. 110:598–605. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalendar R, Grob T, Regina M, Suoniemi A
and Schulman A: IRAP and REMAP: two new retrotransposon-based DNA
fingerprinting techniques. Theor Appl Genet. 98:704–711. 1999.
View Article : Google Scholar
|
|
12
|
Guo D, Zhang H and Luo Z: Genetic
relationships of Diospyros kaki Thunb. and related species
revealed by IRAP and REMAP analysis. Plant Sci. 170:528–533.
2006.
|
|
13
|
Manninen O, Kalendar R, Robinson J and
Schulman AH: Application of BARE-1 retrotransposon markers
to the mapping of a major resistance gene for net blotch in barley.
Mol Gen Genet. 264:325–334. 2000.
|
|
14
|
Muhammad AJ and Othman RY:
Characterization of Fusarium wilt-resistant and
Fusarium wilt-susceptible somaclones of banana cultivar
rastali (Musa AAB) by random amplified polymorphic DNA and
retrotransposon markers. Plant Mol Biol Rep. 23:241–249. 2005.
|
|
15
|
Evrensel C, Yılmaz S, Temel A and
Gozukirmizi N: Variations in BARE-1 insertion patterns in
barley callus cultures. Genet Mol Res. 10:980–987. 2011.
|
|
16
|
Bayram E, Yilmaz S, Hamat-Mecbur H,
Kartal-Alacam G and Gozukirmizi N: Nikita retrotransposon
movements in callus cultures of barley (Hordeum vulgare L.).
POJ. 5:211–215. 2012.
|
|
17
|
Grandbastien MA, Audeon C, Bonnivard E, et
al: Stress activation and genomic impact of Tnt1 retrotransposons
in Solanaceae. Cytogenet Genome Res. 110:229–241. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li XQ: Developmental and environmental
variation in genomes. Heredity (Edinb). 102:323–329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi JM, Kim HM and Kim HS: Human endogenous
retrovirus HERV-H family in human tissues and cancer cells:
expression, identification and phylogeny. Cancer Lett. 231:228–239.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laska MJ, Brudek T, Nissen KK, Christensen
T, Møller-Larsen A, Petersen T and Nexø BA: Expression of HERV-Fc1,
a human endogenous retrovirus, is increased in patients with active
multiple sclerosis. J Virol. 86:3713–3722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mamedov I, Lebedev Y, Hunsmann G,
Khusnutdinova E and Sverdlov E: A rare event of insertion
polymorphism of a HERV-K LTR in the human genome. Genomics.
84:596–599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Belshaw R, Dawson AL, Woolven-Allen J,
Redding J, Burt A and Tristem M: Genomewide screening reveals high
levels of insertional polymorphism in the human endogenous
retrovirus family HERV-K (HML2): implications for present-day
activity. J Virol. 79:12507–12514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moyes D, Griffiths DJ and Venables PJ:
Insertional polymorphisms: a new lease of life for endogenous
retroviruses in human disease. Trends Genet. 23:326–333. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang P and Tang W: Database documentation
of retrotransposon insertion polymorphisms. Front Biosci (Elite
Ed). 4:1542–1555. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Batzer MA and Deininger PL: Alu repeats
and human genomic diversity. Nat Rev Genet. 3:370–379. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Watkins WS, Rogers AR, Ostler C, et al:
Genetic variation among world populations: inference from 100 Alu
insertion polymorphisms. Genome Res. 13:1607–1618. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bamshad MJ, Wooding S, Watkins WS, Ostler
CT, Batzer MA and Jorde LB: Human population genetic structure and
inference of group membership. Am J Hum Genet. 72:578–589. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roy-Engel AM, Carroll ML, El-Sawy M, Salem
AH, Garber RK, Nguyen SV, Deininger PL and Batzer MA:
Non-traditional Alu evolution and primate genomic diversity. J Mol
Biol. 316:1033–1040. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vincent BJ, Myers JS, Ho HJ, Kilroy GE,
Walker JA, Watkins WS, Jorde LB and Batzer MA: Following the LINEs:
an analysis of primate genomic variation at human-specific LINE-1
insertion sites. Mol Biol Evol. 20:1338–1348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carroll ML, Roy-Engel AM, Nguyen SV, et
al: Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and
their contribution to human genomic diversity. J Mol Biol.
311:17–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Myers JS, Vincent BJ, Udall H, et al: A
comprehensive analysis of recently integrated human Ta L1 elements.
Am J Hum Genet. 71:312–326. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mager DL and Freeman JD: HERV-H endogenous
retroviruses: prescence in the New World branch but amplification
in the Old World primate lineage. Virology. 213:395–404. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lindeskog M, Mager DL and Blomberg J:
Isolation of a human endogenous retroviral HERV-H element with an
open env reading frame. Virology. 258:441–450. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang AY, Gulden PH, Woods AS, et al: The
immunodominant major histocompatibility complex class I-restricted
antigen of a murine colon tumor derives from an endogenous
retroviral gene product. Proc Natl Acad Sci USA. 93:9730–9735.
1996. View Article : Google Scholar
|
|
35
|
Sin HS, Huh JH, Kim WY, et al: Long
terminal repeats of human endogenous retrovirus H family provide
alternative poly-adenylation signals to NADSYN1 gene. Korean J
Genet. 29:395–401. 2007.
|