Introduction
Gastric cancer is the second most common cause of
cancer-related mortality in Asia and worldwide (1,2).
Surgery remains the mainstay of curative treatment. However,
following radical surgery, the majority of gastric cancer patients
develop local or distant recurrence (3). Efforts to improve these poor outcomes
have focused on developing effective postoperative systemic and
regional adjuvant therapies. Several meta-analyses of postoperative
adjuvant trials have demonstrated a significant benefit for
chemotherapy-treated patients (4).
However, these therapies are often limited by varying degrees of
survival benefits and debilitating toxicities. As a result,
pharmacogenetics, the study of specific genetic or molecular
signatures that may be predictive of treatment outcomes, has gained
considerable interest.
Oxaliplatin (OXA) is a third-generation
diaminocyclohexane platinum compound that inhibits DNA replication
by forming adducts between two adjacent guanines or an adjacent
guanine and adenine. The adducts formed by OXA appear to be more
effective at inhibiting DNA synthesis compared with cisplatin
adducts (5). Numerous studies have
confirmed the activity and tolerability of the combination of OXA
and 5-fluorouracil (5-FU) modulated with leucovorin (LV)
administered to patients with gastric cancer (6–9).
However, resistance to OXA remains a major obstacle to further
improvement of the response rate. DNA repair capacity is considered
to be a crucial molecular pathway implicated in the resistance to
platinum-based chemotherapy (10).
Nucleotide excision repair (NER) is the primary DNA repair
mechanism for the removal of bulky, helix-distorting DNA adducts,
including those generated by platinum-based chemotherapy (11,12),
while the base excision repair (BER) system mainly repairs the
small lesions around the damaged bases or single-strand breaks
(SSBs) (13,14). Single nucleotide polymorphisms in
DNA repair genes may be correlated with DNA-repair capacity and may
affect the response to platinum-based chemotherapy (10).
The multifunctional detoxifying glutathione
S-transferase (GST) enzymes have been specifically implicated in
the metabolism of platinum drugs (15) and the polymorphisms of the GST
family may affect platinum efficacy by lowering the intracellular
concentrations of drugs.
Numerous genomic polymorphisms in genes encoding
DNA-repair enzymes and detoxification enzymes have been shown to be
correlated with the response to platinum-based chemotherapy. Among
which, the four most commonly studied gene polymorphisms correlated
with the sensitivity of cancer cells to platinum-based chemotherapy
are excision repair cross-complimentary group 1 (ERCC1) Asp118Asp,
X-ray repair cross-complementing protein 1 (XRCC1) Arg399Gln,
xeroderma pigmentosum group D (XPD) Lys751Gln and glutathione
S-transferase π 1 (GSTP1) Ile105Val. However, the association
between these gene polymorphisms and the clinical end points are
controversial. In addition, little is known with regard to the
correlation between these putative markers and survival or toxicity
in gastric cancer patients receiving OXA-based adjuvant
chemotherapy. The aim of the present study was to assess the
correlation between the four SNPs and survival/toxicity in a series
of consecutive resected gastric cancer patients treated with
OXA/5-FU adjuvant chemotherapy.
Materials and methods
Patients and treatment protocols
Blood samples were obtained from 126 patients with
stage Ib-III disease, who were recruited during the period between
October 2004 and March 2007 and underwent surgery at the Department
of Gastroenterological Surgery, Changzhou Tumor Hospital (Jiangsu,
China). The patients comprised of 90 males and 36 females (range,
30–78 years of age; median age, 57 years). None of the patients had
previously received chemotherapy. This study was approved by the
local ethics committee of The Changzhou Tumor Hospital (Changzhou,
China) and written informed consent was obtained from all patients.
Following surgery, all patients received ≥8 cycles of 85
mg/m2 OXA plus 20 mg/m2 LV on the first day
of treatment, followed by 5-FU via a 400 mg/m2 bolus,
and a 22-h continuous infusion of 600 mg/m2 5-FU on days
1–2 at 2-week intervals. Side effects were graded according to the
United States National Cancer Institute (NCI) Common Toxicity
Criteria, version 2.0 (16). OXA
dose reductions of 25% were performed in cases of grade 4
hematological toxicity, febrile neutropenia and persistent
paresthesias (P14 d) or painful, temporary paresthesias (7–14 d).
If hematological and non-hematological toxicities had not recovered
prior to the next treatment cycle, the OXA dose was delayed for a
maximum of 2 weeks. If these toxicities had not recovered by that
time, patients were removed from the study. All patients received a
cumulative OXA dose of ≥500 mg/m2. Prophylactic use of
hematological growth factors was not permitted.
Genotyping
DNA extractions from peripheral blood samples were
performed using the QiaAmp kit (Qiagen, Valencia, CA, USA). SNPs in
ERCC1 Asp118Asp, XPD Lys751Gln, XRCC1 Arg399Gln and GSTP1 Ile105Val
(Table I) were assessed by a 5′
nuclease allelic discrimination assay (Applied Biosystems, Foster
City, CA, USA) using a fluorescent temperature cycler (icycler iQ
Multicolor Real-time PCR Detection system; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Each reaction contained the template DNA
and a final concentration of 1X TaqMan PCR Master Mix (Applied
Biosystems, Foster City, CA, USA), 300 nM of each primer, 100 nM of
wild-type probe (Applied Biosystems) and 100 nM of variant probe
(Applied Biosystems). The PCR conditions were 50°C for 2 min and
95°C for 15 min, followed by 45 cycles at 95°C for 15 sec and 60°C
for 1 min. For each SNP, sequencing was performed using an ABI 3730
Genetic Analyzer (Applied Biosystems). Those with concordant
results from the two analyses were included in the final data
analysis.
 | Table IGenetic markers evaluated in this
study. |
Table I
Genetic markers evaluated in this
study.
| Polymorphism
substitution | dbSNP | NCBI Ref Seq | Exon | Genotype | Amino acid |
|---|
| ERCC Asn118Asn | rs11615 | NM_001983 | 4 | C/T | Asn/118 |
| XRCC Arg399Gln | rs25487 | NM_006297 | 10 | G/A | Gln/399 |
| XPD Lys751Gln | rs13181 | NM_00400 | 23 | A/G | Val/751 |
| GSTP1
Ile105Val | rs1695 | NM_000852 | 5 | A/G | Val/105 |
Follow-up
Interim history, physical examination, hematological
studies, carcinoembryonic antigen levels and whole-body computed
tomography were performed every 2 months in the first year and
every 6 months thereafter. Patients underwent upper endoscopy 3
months following surgery and every 6 months thereafter. The
recurrences or metastases of gastric carcinoma were confirmed by
cytology and biopsy, surgery or whole-body computed tomography. The
Union for International Cancer Control (UICC) staging system (7th
version) was used for the classification of each case. The study
was conducted in a blind fashion, so that the patient outcome was
unknown to the investigators performing the molecular analysis.
Relapse-free survival (RFS) was the time from study entry until
disease recurrence, mortality or the day of the last follow-up
visit (whichever occurred first). Overall survival (OS) was the
time from study entry until the date of death, regardless of the
cause, or the most recent documented follow-up.
Statistical analysis
Statistical significance was based on a two-sided
significance level of 0.05. All analyses were performed with SPSS
(version 13.0; SPSS Inc., Chicago, IL, USA). The correlation
between different genotypes and the clinical variables or toxicity
to chemotherapy were tested by the χ2 test or Fisher’s
exact test (two-sided), as appropriate. The Kaplan-Meier survival
method was used to estimate the survival curves, and the log-rank
test was used to analyze univariate distributions for RFS and OS.
The prognostic significance of the different gene SNPs following
adjustment for other prognostic factors was assessed using the Cox
proportional hazards regression model.
Results
Patients
A total of 126 gastric cancer patients comprised of
90 males and 36 females (range, 30–78 years of age; median age, 57
years). Of the total number of patients, 15.08% had stage Ib and
stage II disease, and 84.92% had stage III disease at the time of
diagnosis. The Eastern Cooperative Oncology Group performance
status (ECOG PS) was 0–1 in 90 patients and 2 in 36 patients at the
time of accepting chemotherapy. Detailed demographic and disease
characteristics are listed in Table
II. The median RFS time (M-RFS) was 12 months (range, 2–56
months), and the median survival time (MST) was 21 months (range,
5–56 months). The patient characteristics and their outcomes were
unknown to the investigators performing the genetic analysis. The
genotyping results were disclosed to the clinical investigators
following data analysis.
 | Table IIFactors correlated with survival in
patients receiving surgery followed by oxaliplatin-based adjuvant
chemotherapy. |
Table II
Factors correlated with survival in
patients receiving surgery followed by oxaliplatin-based adjuvant
chemotherapy.
| Characteristic | n | M-RFS (months) | P-value | MST (months) | P-value |
|---|
| Gender | | | 0.177 | | 0.513 |
| Male | 90 | 16 | | 27 | |
| Female | 36 | 12 | | 24 | |
| Age (years) | | | 0.553 | | 0.912 |
| ≤57 | 54 | 13 | | 24 | |
| >57 | 72 | 15 | | 26 | |
| ECGO PS | | | <0.001 | | <0.001 |
| 0 or 1 | 90 | 39 | | 45 | |
| >2 | 36 | 5 | | 15 | |
| Tumor
differentiation | | | 0.147 | | 0.112 |
| Well
differentiated | 28 | 21 | | | |
| Moderately
differentiated | 80 | 16 | | 27 | |
|
Undifferentiated | 18 | 5 | | 14 | |
| Tumor location | | | 0.018 | | 0.020 |
| Proximal
stomach | 40 | 12 | | 18 | |
| Stomach body | 31 | | | | |
| Distal
stomach | 55 | 13 | | 24 | |
| Staging | | | <0.001 | | <0.001 |
| Ib + II | 19 | | | | |
| III | 107 | 12 | | 21 | |
| ERCC1-118 | | | <0.001 | | <0.001 |
| C/C | 81 | 45 | | | |
| C/T+T/T | 45 | 5 | | 15 | |
| XRCC1-399 | | | 0.001 | | <0.001 |
| G/G | 71 | 8 | | 18 | |
| A/G+A/A | 55 | 47 | | | |
| XPD-751 | | | 0.639 | | 0.769 |
| A/A | 107 | 15 | | 26 | |
| A/G | 19 | 12 | | 24 | |
| GSTP1-105 | | | 0.033 | | 0.019 |
| A/A | 86 | 12 | | 21 | |
| A/G+G/G | 40 | 47 | | | |
| CEA (ng/ml) | | | <0.001 | | <0.001 |
| ≤5 | 79 | 47 | | | |
| >5 | 47 | 6 | | 16 | |
Genotype frequencies of polymorphisms of
ERCC1, XRCC1, XPD and GSTP1
The results of the genotyping of ERCC1-118,
XRCC1-399, XPD-751 and GSTP1-105 were available for all 126
patients. The wild genotype (C/C) of ERCC1 Asp118Asp was observed
in 81 patients (64.29%), while the heterozygous variant (C/T) was
present in 36 patients (28.57%) and the homozygous variant (T/T)
was present in 9 patients (7.14%). The wild genotype (G/G) of XRCC1
Arg399Gln was observed in 71 patients (56.35%) and the heterozygous
variant (G/A) in 33 patients (26.19%), whereas the homozygous
variant (A/A) was present in 6 patients (4.76%). The wild type
(A/A) of GSTP1 Ile105Val was present in 86 patients (68.25%), the
heterozygous variant (A/G) was observed in 35 patients (27.78%),
and the homozygous variant (G/G) was present in 5 patients (3.97%).
The wild genotype (A/A) of XPD Lys751Gln was observed in 107
patients (84.92%), while the heterozygous variant (A/C) was present
in 19 patients (15.08%), and the homozygous variant of codon 751 in
the XPD gene was not observed in any patient. Genotype frequencies
for ERCC1, XRCC1, XPD and GSTP1 polymorphisms were demonstrated to
be in Hardy-Weinberg equilibrium. No significant correlations were
observed between any of these polymorphisms and age, gender, ECOG
status, initial tumor stage and grade.
Correlation between polymorphisms and
survival
With regard to RFS, the three variable ERCC1
Asp118Asp, XRCC1 Arg399Gln and GSTP1 Ile105Val SNPs demonstrated a
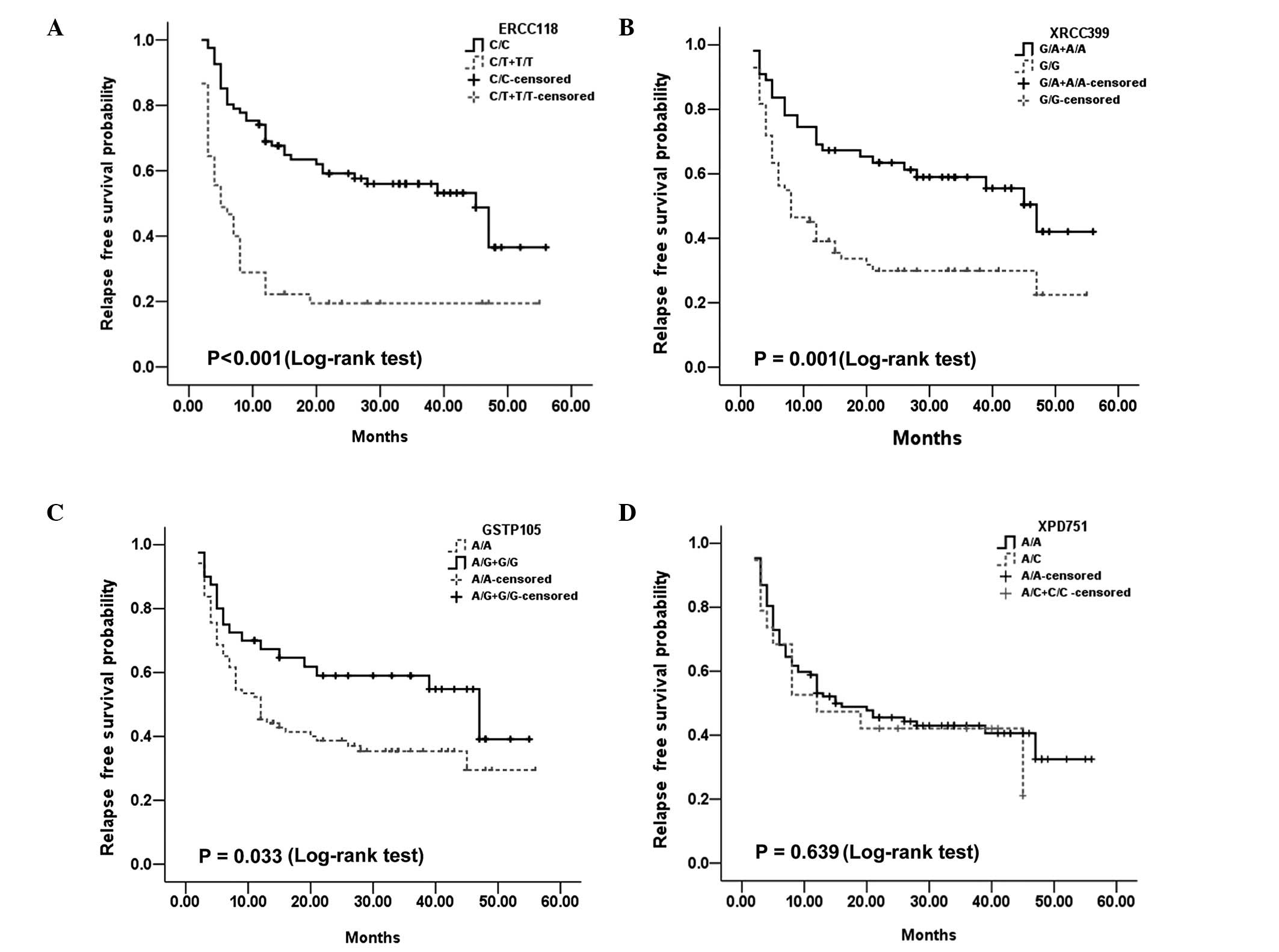
predictive value. For ERCC1 Asp118Asp, the M-RFS was 5 months for
TT and C/T cases, and 45 months for C/C patients (P<0.001;
Fig. 1A). For XRCC1 Arg399Gln, the
M-RFS was 47 months for AA and A/G cases, and 8 months for G/G
patients (P=0.001). Additionally, for GSTP1 Ile105Val, the M-RFS
was 47 months for G/G and A/G cases, and 12 months for AA patients
(P=0.033). Both ERCC1 Asp118Asp and XRCC1 Arg399Gln remained
significant in the multivariate analysis. For ERCC1 Asp118Asp, TT
and C/T patients had a 2.22-fold increased risk of recurrence
compared with C/C patients (RR, 1.392–3.540; P=0.001). In addition,
for XRCC1 Arg399Gln, compared with the G/G cases, the patients with
heterozygous and homozygous polymorphic variants (A/G and A/A) had
a decreased risk of recurrence by 0.499-fold (RR, 0.298–0.836;
P=0.008).
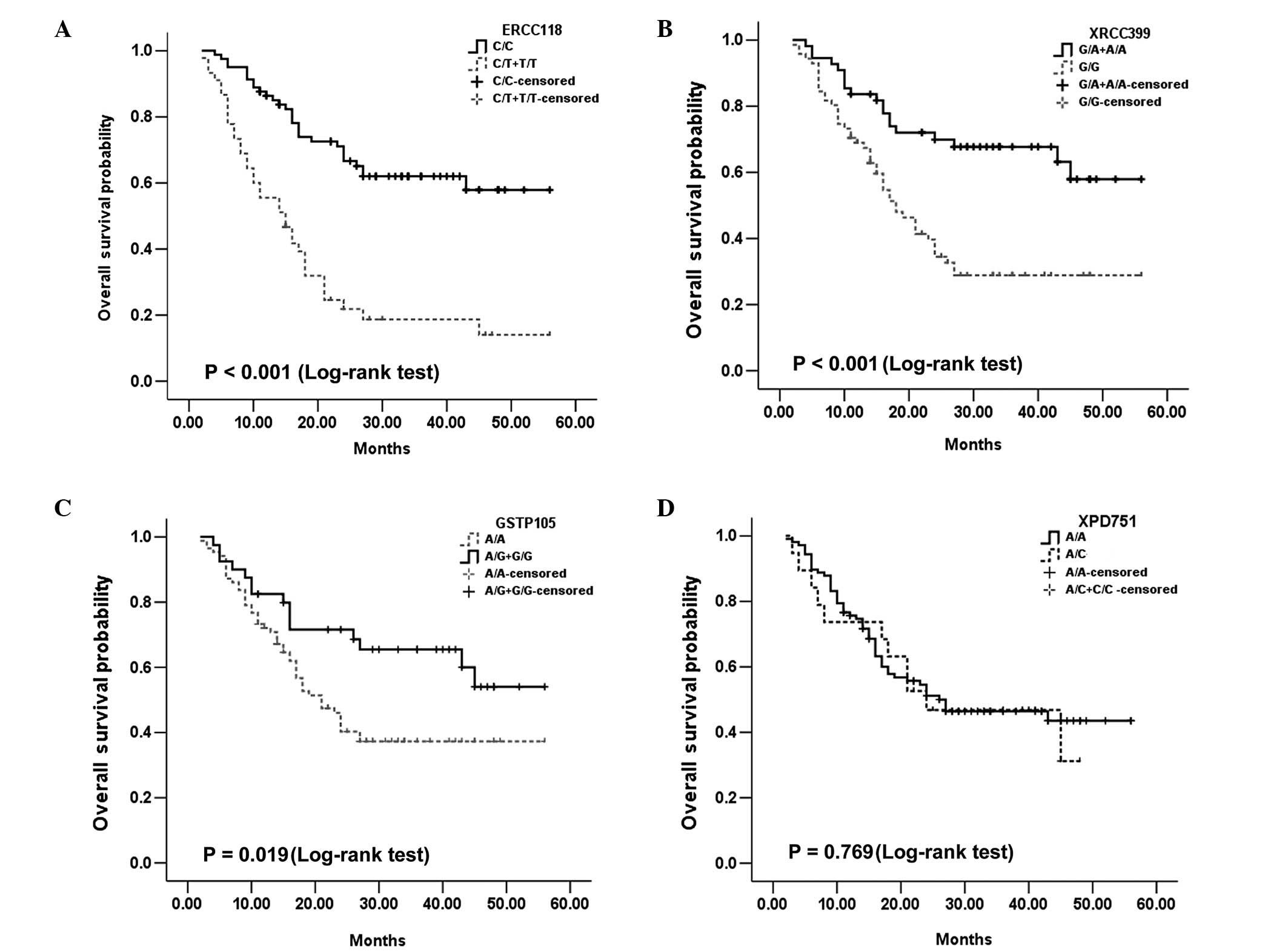
As for OS, the ERCC1 Asp118Asp, XRCC1 Arg399Gln and
GSTP1 Ile105Val SNPs also retained their significant predictive
value. For ERCC1-118, the median OS was 15 months for T/T and C/T,
and undefined for C/C cases (P<0.001; Fig. 2A). For the XRCC1 Arg399Gln, the
median OS was 18 months for G/G patients and undefined for AA and
A/G patients (P<0.001; Fig.
2B). Additionally, for GSTP1 Ile105Val, the median OS was not
defined for G/G and A/G cases, and 21 months for AA patients
(P=0.019; Fig. 2D). Both ERCC1
Asp118Asp and XRCC1 Arg399Gln remained significant in the
multivariate Cox survival analysis [P=0.001; HR=2.262; 95%
confidence interval (CI), 1.369–3.738; and P=0.02; HR=0.508; 95%
CI, 0.288–03.898, respectively; Table III].
 | Table IIIHazard ratios for relapse-free
survival and overall survival. |
Table III
Hazard ratios for relapse-free
survival and overall survival.
| RFS | OS |
|---|
|
|
|
|---|
| Prognostic
factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| ERCC1-118 | | | 0.001 | | | 0.001 |
| C/C | 1 | | | 1 | | |
| C/T+T/T | 2.220 | 1.392–3.540 | | 2.262 | 1.369–3.738 | |
| XRCC1-399 | | | 0.008 | | | 0.020 |
| G/G | 1 | | | 1 | | |
| A/G+A/A | 0.499 | 0.298–0.836 | | 0.508 | 0.288–0.898 | |
| Staging | | | 0.012 | | | 0.027 |
| Ib + II | 1 | | | 1 | | |
| III | 13.142 | 1.782–96.919 | | 9.648 | 1.297–71.781 | |
| CEA (ng/ml) | | | | | | 0.005 |
| ≤5 | 1 | | 0.017 | 1 | | |
| >5 | 1.844 | 1.114–3.053 | | 2.213 | 1.279–3.829 | |
| ECOG PS | | | 0.006 | | | 0.018 |
| 0 or 1 | | | | | | |
| 2 | 1.994 | 1.221–3.254 | | 1.855 | 1.111–3.097 | |
Other factors that were significantly correlated
with RFS and OS in the univariate analysis using the Kaplan-Meier
survival curves and the log-rank test were ECOG PS, tumor location,
tumor stage and the levels of serum carcinoembryonic antigen
(Table II). Gender, age and tumor
differentiation were not significant prognostic factors for RFS and
OS. ECOG PS, stage and serum carcinoembryonic antigen remained
significant prognostic factors correlated with RFS and OS in the
Cox proportional hazards regression model multivariate analysis
(Table III).
Correlation between polymorphisms and
toxicity
We analyzed whether the previously mentioned four
gene SNPs were correlated with the severe OX/5-FU regimen-related
toxicities in all 126 patients. There were no significant
correlations between the ERCC1-118 and XPD751 polymorphisms with
grade 3 or 4 toxicity. However, carrying at least one variant XRCC1
Arg399Gln and GSTP1 Ile105Val allele was significantly correlated
with grade 3 or 4 hematological toxicity (P=0.029 and P<0.001,
respectively). In particular, carrying at least one variant GSTP1
Ile105Val allele also remained significantly associated with grade
3 or 4 gastrointestinal toxicity and neurotoxicity (P=0.002 and
P=0.018, respectively).
Discussion
The ability of cancer cells to recognize and repair
DNA damage and to enhance detoxification by the GST pathway may
contribute to tumor resistance to platinum-based chemotherapy
(17,18). In the present study, we selected
four putative molecular parameters to determine whether these
markers were partly responsible for sensitivity and toxicity to
OXA-based chemotherapy used as adjuvant treatment in gastric
cancer. Our findings supported the hypothesis that pharmacogenetic
profiling may be useful for predicting the prognosis of survival
and the toxicity associated with OXA-based adjuvant chemotherapy in
gastric cancer patients. The univariate analysis revealed that
ERCC1-118, XRCC1-399 and GSTP1-105 polymorphisms were significantly
correlated with RFS (P<0.001, P=0.001 and P=0.033, respectively)
and OS (P<0.001, P<0.001 and P=0.019, respectively). The Cox
proportional hazards regression model multivariate analysis
suggested that ERCC1-118 and XRCC1-399 polymorphisms also retained
significant predictive value for RFS (P=0.001 and P=0.008,
respectively) and OS (P=0.001 and P=0.02, respectively). However,
we also demonstrated that carrying at least one variant XRCC1
Arg399Gln or GSTP1 Ile105Val allele was significantly correlated
with grade 3 or 4 hematological toxicity (P=0.029 and P<0.001,
respectively). In particular, carrying at least one variant GSTP1
Ile105Val allele remained significantly correlated with grade 3 or
4 gastrointestinal toxicity and neurotoxicity (P=0.002 and P=0.018,
respectively).
NER is the main mechanism in mammalian cells for the
removal of bulky, helix-distorting DNA adducts produced by platinum
agents (11). ERCC1 is an
important DNA repair gene with critical roles in the NER pathway,
which is the most important system for repairing a wide variety of
structural DNA lesions, including bulky DNA adducts (19). Several studies have demonstrated
that patients, including those with gastric cancer, with low ERCC1
expression were more sensitive to platinum-based chemotherapy
(20–23). In vitro experiments have
also demonstrated that cells with low ERCC1 expression were more
sensitive to platinum derivatives or alkylating agents (24). Functional variants of the gene may
alter the levels of ERCC1 gene expression. Chang et
al(25) revealed that higher
ERCC1 protein expression levels were correlated with the variant
ERCC1-118 T allele, which may lead to resistance to platinum
derivatives. This may explain the lower RFS and OS times observed
in the individuals in our study of FOLFOX4 adjuvant chemotherapy
(Tables II and III; Figs.
1 and 2) and other studies
where patients have been treated with platinum-based chemotherapy
(26–28). However, Yu et al obtained
contrary results in studies of ovarian cell lines, where the ERCC1
codon 118 C-T substitution was associated with reduced levels of
ERCC1 mRNA and protein expression (29). The contrasting results from
different clinical studies of the ERCC-1 polymorphism (30) may be due to insufficient sample
sizes; with more definitive results likely to be achieved through a
large sample, multicenter prospective studies should be conducted
in the future. We did not observe a correlation between the ERCC-1
polymorphism and overall toxicity, which suggests that ERCC1 may be
not involved in adverse reactions to FOLFOX4 treatment, and is
consistent with the results of a previous study (28).
XPD is another important component of NER. The
majority of studies have demonstrated that variance in the DNA
sequence of the ERCC2/XPD gene 751 was correlated with impaired DNA
repair activity (31,32), while one study demonstrated the
opposite results (33). A
significant correlation has also been observed between homozygosity
for the wild-type XPD 751 allele (Lys/Lys) and an improved response
to FU-OXA in metastatic colorectal cancer (34). By contrast, another study in
patients with stage III and IV gastric cancer treated with surgery
following radiation therapy plus FU/LV-based chemotherapy obtained
the opposite result; patients with the wild-type XPD-751 allele
(Lys/Lys) were more likely to have relapse compared with those with
Lys/Gln and Gln/Gln genotypes (35). In the present study, we did not
observe a significant correlation between XPD-751 polymorphism and
clinical outcome in gastric cancer patients treated with FOLFOX4
adjuvant chemotherapy, which is consistent with a previous study in
non-small cell lung cancer (36).
At the same time, no C/C genotype was detected in the present
study, which is consistent with the findings of studies examining
the Chinese population (37,38).
These results suggest that the polymorphisms may differ according
to ethnicity.
Base excision repair, another critical DNA repair
mechanism, is also important in the response to platinum-based
therapy. XRCC1 is a key player in the BER pathway. In vitro
assays have demonstrated that reduced DNA repair capacity is
associated with the XRCC1-399 polymorphism, and the rate of
irradiation-specific DNA repair decreased with an increasing number
of variant XRCC1 Arg399Gln alleles (39,40).
Several studies have demonstrated a correlation between XRCC1-399
G-A substitution with improved outcomes in patients with solid
tumors treated with platinum-based chemotherapy (41–43).
Consistent with the above results, in the present study, patients
carrying at least one variant XRCC1-399A allele had a better
prognosis, which may have been correlated with enhanced sensitivity
to OXA-based chemotherapy. We also identified that variance in the
XRCC1 Arg399Gln allele was significantly correlated with grade 3 or
4 hematological toxicity, which may have been due to the less
proficient DNA repair activity. However, opposite results
demonstrated an improved survival for patients with the XRCC1-399 G
allele receiving platinum-based chemotherapy for colorectal, lung,
esophageal, gastric and cervical carcinoma (26,30,38,44–50).
Several studies have also demonstrated that no statistically
significant correlation was identified between the XRCC1 codon 399
polymorphism and survival or toxicities correlated with
platinum-based chemotherapy. The aforementioned conflicting results
may be due to different study populations, chemotherapy regimens
and genotyping methods.
GSTs participate in the detoxification of a variety
of chemotherapeutics, including platinum. The GSTP1-105A allele may
be correlated with lower GSTP1 enzyme activity in the tumor tissue
(51). In the present study, we
verified that patients with GSTP1-105A allele variants not only
exhibited longer relapse-free (P<0.01) and overall (P<0.01)
survival times; however, also had a higher incidence of grade 3/4
cumulative neuropathy, gastrointestinal toxicity and hematological
toxicity following different cycles of treatment, which is partly
in accordance with the results of Stoehlmacher et
al(52). The aforementioned
results may be correlated with the reduced metabolism and slower
removal of chemotherapeutic agents, which yields a longer RFS and
OS; however, leads to toxicity of platinum-based chemotherapy.
Although our findings supported the theory that
ERCC1 Asp118Asp, XRCC1 Arg399Gln and GSTP1 Ile105Val polymorphisms
may be useful for predicting the prognosis of survival and the
toxicity associated with OXA-based adjuvant chemotherapy in gastric
cancer patients, the limitations of our study must be acknowledged.
These include insufficient sample sizes and a single unit
population. Therefore, larger sample sizes, multicenter prospective
studies and even basic functional studies are required to confirm
the results and identify the biological basis of these
findings.
In conclusion, the results of the present study
indicate that pharmacogenetic profiling may be useful for
predicting the prognosis of survival and the toxicity associated
with the OXA-based adjuvant chemotherapy in gastric cancer
patients.
Acknowledgements
The authors would like to thank the personnel at the
Changzhou Tumor Hospital (Changzhou, China) in our study. The study
was partly supported by the Science and Technology Planning Project
of Changzhou, Jiangsu (CS20092025); the Research of Health
Department in Jiangsu (Z201221); the Science and Technology
Planning Project of Changzhou Health Bureau, Jiangsu (QN201106 and
ZD201203); the 333 Talents Training Project of Jiangsu Province;
the Key Medical Innovation Talents Training Project of Changzhou,
Jiangsu and the Project of Jiangsu Province Sanitation Innovation
Team.
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar
|
|
2
|
Leung WK, Wu MS, Kakugawa Y, et al:
Screening for gastric cancer in Asia: current evidence and
practice. Lancet Oncol. 9:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Macdonald JS: Treatment of localized
gastric cancer. Semin Oncol. 31:566–573. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carrato A, Gallego-Plazas J and
Guillen-Ponce C: Adjuvant therapy of resected gastric cancer is
necessary. Semin Oncol. 32:S105–S108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mamenta EL, Poma EE, Kaufmann WK,
Delmastro DA, Grady HL and Chaney SG: Enhanced replicative bypass
of platinum-DNA adducts in cisplatin-resistant human ovarian
carcinoma cell lines. Cancer Res. 54:3500–3505. 1994.PubMed/NCBI
|
|
6
|
Louvet C, André T, Tigaud JM, et al: Phase
II study of oxaliplatin, fluorouracil, and folinic acid in locally
advanced or metastatic gastric cancer patients. J Clin Oncol.
20:4543–4548. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Vita F, Orditura M, Matano E, et al: A
phase II study of biweekly oxaliplatin plus infusional
5-fluorouracil and folinic acid (FOLFOX-4) as first-line treatment
of advanced gastric cancer patients. Br J Cancer. 92:1644–1649.
2005.PubMed/NCBI
|
|
8
|
Al-Batran SE, Atmaca A, Hegewisch-Becker
S, et al: Phase II trial of biweekly infusional fluorouracil,
folinic acid, and oxaliplatin in patients with advanced gastric
cancer. J Clin Oncol. 22:658–663. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cavanna L, Artioli F, Codignola C, et al:
Oxaliplatin in combination with 5-fluorouracil (5-FU) and
leucovorin (LV) in patients with metastatic gastric cancer (MGC).
Am J Clin Oncol. 29:371–375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei Q, Frazier ML and Levin B: DNA repair:
a double-edged sword. J Natl Cancer Inst. 92:440–441. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reardon JT, Vaisman A, Chaney SG and
Sancar A: Efficient nucleotide excision repair of cisplatin,
oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-
platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer
Res. 59:3968–3971. 1999.
|
|
12
|
Reed E: Platinum-DNA adduct, nucleotide
excision repair and platinum based anti-cancer chemotherapy. Cancer
Treat Rev. 24:331–344. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goode EL, Ulrich CM and Potter JD:
Polymorphisms in DNA repair genes and associations with cancer
risk. Cancer Epidemiol Biomarkers Prev. 11:1513–1530.
2002.PubMed/NCBI
|
|
14
|
Wood RD, Mitchell M, Sgouros J and Lindahl
T: Human DNA repair genes. Science. 291:1284–1289. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Frankmoelle WP, Medina JC, Shan B, Narbut
MR and Beckmann H: Glutathione S-transferase metabolism of the
antineoplastic pentafluorophenylsulfonamide in tissue culture and
mice. Drug Metab Dispos. 28:951–958. 2000.PubMed/NCBI
|
|
16
|
National Cancer Institute. NCI Common
Toxicity Criteria. http://ctep.cancer.gov.
Accessed October 1, 2004
|
|
17
|
Salinas AE and Wong MG: Glutathione
S-transferases - a review. Curr Med Chem. 6:279–309. 1999.
|
|
18
|
Gossage L and Madhusudan S: Cancer
pharmacogenomics: role of DNA repair genetic polymorphisms in
individualizing cancer therapy. Mol Diagn Ther. 11:361–380. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Silva IU, McHugh PJ, Clingen PH and
Hartley JA: Defining the roles of nucleotide excision repair and
recombination in the repair of DNA interstrand cross-links in
mammalian cells. Mol Cell Biol. 20:7980–7990. 2000.PubMed/NCBI
|
|
20
|
Weberpals J, Garbuio K, O’Brien A, et al:
The DNA repair proteins BRCA1 and ERCC1 as predictive markers in
sporadic ovarian cancer. Int J Cancer. 124:806–815. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Denlinger CS and Cohen SJ: Progress in the
development of prognostic and predictive markers for
gastrointestinal malignancies. Curr Treat Options Oncol. 8:339–351.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iqbal S, Stoehlmacher J and Lenz HJ:
Tailored chemotherapy for colorectal cancer: a new approach to
therapy. Cancer Invest. 22:762–773. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hofler H, Langer R, Ott K and Keller G:
Prediction of response to neoadjuvant chemotherapy in carcinomas of
the upper gastrointestinal tract. Adv Exp Med Biol. 587:115–120.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guichard S, Arnould S, Hennebelle I, Bugat
R and Canal P: Combination of oxaliplatin and irinotecan on human
colon cancer cell lines: activity in vitro and in
vivo. Anticancer Drugs. 12:741–751. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang PM, Tzeng CH, Chen PM, et al: ERCC1
codon 118 C-->T polymorphism associated with ERCC1 expression
and outcome of FOLFOX-4 treatment in Asian patients with metastatic
colorectal carcinoma. Cancer Sci. 100:278–283. 2009.
|
|
26
|
Kalikaki A, Kanaki M, Vassalou H, et al:
DNA repair gene polymorphisms predict favorable clinical outcome in
advanced non-small-cell lung cancer. Clin Lung Cancer. 10:118–123.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stoehlmacher J, Park DJ, Zhang W, et al: A
multivariate analysis of genomic polymorphisms: prediction of
clinical outcome to 5-FU/oxaliplatin combination chemotherapy in
refractory colorectal cancer. Br J Cancer. 91:344–354. 2004.
|
|
28
|
Ruzzo A, Graziano F, Loupakis F, et al:
Pharmacogenetic profiling in patients with advanced colorectal
cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol.
25:1247–1254. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu JJ, Lee KB, Mu C, et al: Comparison of
two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that
are equally resistant to platinum, but differ at codon 118 of the
ERCC1 gene. Int J Oncol. 16:555–560. 2000.PubMed/NCBI
|
|
30
|
Huang MY, Huang ML, Chen MJ, et al:
Multiple genetic polymorphisms in the prediction of clinical
outcome of metastatic colorectal cancer patients treated with
first-line FOLFOX-4 chemotherapy. Pharmacogenet Genomics. 21:18–25.
2011. View Article : Google Scholar
|
|
31
|
Spitz MR, Wu X, Wang Y, et al: Modulation
of nucleotide excision repair capacity by XPD polymorphisms in lung
cancer patients. Cancer Res. 61:1354–1357. 2001.PubMed/NCBI
|
|
32
|
Wolfe KJ, Wickliffe JK, Hill CE, Paolini
M, Ammenheuser MM and Abdel-Rahman SZ: Single nucleotide
polymorphisms of the DNA repair gene XPD/ERCC2 alter mRNA
expression. Pharmacogenet Genomics. 17:897–905. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lunn RM, Helzlsouer KJ, Parshad R, et al:
XPD polymorphisms: effects on DNA repair proficiency.
Carcinogenesis. 21:551–555. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei
DD, Groshen S and Lenz HJ: A Xeroderma pigmentosum group D gene
polymorphism predicts clinical outcome to platinum-based
chemotherapy in patients with advanced colorectal cancer. Cancer
Res. 61:8654–8658. 2001.
|
|
35
|
Zárate RN, Arias F, Bandres E, Cubedo E,
Malumbres R and García-Foncillas J: Xeroderma pigmentosum group D
751 polymorphism as a predictive factor in resected gastric cancer
treated with chemo-radiotherapy. World J Gastroenterol.
12:6032–6036. 2006.PubMed/NCBI
|
|
36
|
Tibaldi C, Giovannetti E, Vasile E, et al:
Correlation of CDA, ERCC1, and XPD polymorphisms with response and
survival in gemcitabine/cisplatin-treated advanced non-small cell
lung cancer patients. Clin Cancer Res. 14:1797–1803. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xing D, Tan W, Wei Q and Lin D:
Polymorphisms of the DNA repair gene XPD and risk of lung cancer in
a Chinese population. Lung Cancer. 38:123–129. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu B, Wei J, Zou Z, et al: Polymorphism
of XRCC1 predicts overall survival of gastric cancer patients
receiving oxaliplatin-based chemotherapy in Chinese population. Eur
J Hum Genet. 15:1049–1053. 2007. View Article : Google Scholar
|
|
39
|
Vodicka P, Kumar R, Stetina R, et al:
Genetic polymorphisms in DNA repair genes and possible links with
DNA repair rates, chromosomal aberrations and single-strand breaks
in DNA. Carcinogenesis. 25:757–763. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vodicka P, Stetina R, Polakova V, et al:
Association of DNA repair polymorphisms with DNA repair functional
outcomes in healthy human subjects. Carcinogenesis. 28:657–664.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pacetti P, Giovannetti E, Mambrini A, et
al: Single nucleotide polymorphisms and clinical outcome in
patients with biliary tract carcinoma treated with epirubicin,
cisplatin and capecitabine. Anticancer Res. 29:1835–1840. 2009.
|
|
42
|
Giachino DF, Ghio P, Regazzoni S, et al:
Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single
nucleotide polymorphisms in lung cancer. Clin Cancer Res.
13:2876–2881. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moreno V, Gemignani F, Landi S, et al:
Polymorphisms in genes of nucleotide and base excision repair: risk
and prognosis of colorectal cancer. Clin Cancer Res. 12:2101–2108.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Suh KW, Kim JH, Kim do Y, Kim YB, Lee C
and Choi S: Which gene is a dominant predictor of response during
FOLFOX chemotherapy for the treatment of metastatic colorectal
cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol. 13:1379–1385.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
de las Peñas R, Sanchez-Ronco M, Alberola
V, et al: Polymorphisms in DNA repair genes modulate survival in
cisplatin/gemcitabine-treated non-small-cell lung cancer patients.
Ann Oncol. 17:668–675. 2006.PubMed/NCBI
|
|
46
|
Bewick MA, Conlon MS and Lafrenie RM:
Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after
treatment for metastatic breast cancer. J Clin Oncol. 24:5645–5651.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yoon HH, Catalano PJ, Murphy KM, et al:
Genetic variation in DNA-repair pathways and response to
radiochemotherapy in esophageal adenocarcinoma: a retrospective
cohort study of the Eastern Cooperative Oncology Group. BMC Cancer.
11:1762011. View Article : Google Scholar
|
|
48
|
Cheng XD, Lu WG, Ye F, Wan XY and Xie X:
The association of XRCC1 gene single nucleotide polymorphisms with
response to neoadjuvant chemotherapy in locally advanced cervical
carcinoma. J Exp Clin Cancer Res. 28:912009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang ZH, Hua D and Du X: Polymorphisms in
p53, GSTP1 and XRCC1 predict relapse and survival of gastric cancer
patients treated with oxaliplatin-based adjuvant chemotherapy.
Cancer Chemother Pharmacol. 64:1001–1007. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Goekkurt E, Al-Batran SE, Hartmann JT, et
al: Pharmacogenetic analyses of a phase III trial in metastatic
gastroesophageal adenocarcinoma with fluorouracil and leucovorin
plus either oxaliplatin or cisplatin: a study of the
arbeitsgemeinschaft internistische onkologie. J Clin Oncol.
27:2863–2873. 2009. View Article : Google Scholar
|
|
51
|
Kweekel DM, Koopman M, Antonini NF, et al:
GSTP1 Ile105Val polymorphism correlates with progression-free
survival in MCRC patients treated with or without irinotecan: a
study of the Dutch Colorectal Cancer Group. Br J Cancer.
99:1316–1321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Stoehlmacher J, Park DJ, Zhang W, et al:
Association between glutathione S-transferase P1, T1, and M1
genetic polymorphism and survival of patients with metastatic
colorectal cancer. J Natl Cancer Inst. 94:936–942. 2002. View Article : Google Scholar : PubMed/NCBI
|