Introduction
Chirality is a basic characteristic of biochemical
systems. Numerous endogenous macromolecules are chiral, including
enzymes, carriers, receptors, plasma proteins and polysaccharides.
Chiral drugs and biochemical systems are stereoselective, which
causes variations in pharmacology and pharmacological
stereoselectivity, including pharmacodynamic and pharmacokinetic
stereoselectivity. These differences may lead to low levels of
effectiveness, ineffectiveness and even toxicity in clinical use.
Therefore, the US Food and Drug Administration (FDA) requires that
the pharmacological function of the monomers of chiral drugs be
cleared during the development and declaration of new drugs, making
the development of chiral drugs a hot topic. Little research has
been conducted with regard to chiral drugs due to the lack of
simple and effective research methods and strategies available. As
a result, investigators have only been able to deduce the mechanism
for chiral drugs indirectly. This has become a major bottleneck in
drug development. Proteomic techniques are providing new methods
and strategies for the study of drug mechanisms (1–3);
however, there have been few studies on applying this technique to
chiral drug development. The aim of the present study was to use
proteomic techniques to investigate chiral drug development.
Propranolol (PRO), a non-cardioselective
β-adrenergic antagonist, is a widely used drug in the treatment of
numerous disorders, including arrhythmia and angina. PRO has one
chiral center and two enantiomers [R(+)-PRO and S(−)-PRO]. We used
PRO as a pharmacokinetic model for elucidating chiral drug
mechanisms. PRO blocks the β-adrenergic receptor (β-AR)-mediated
signal pathway by competitively preventing its excitomotor
neurotransmitter from binding. The two PRO enantiomers were
previously found to have diverse biological effects; S(−)-PRO binds
to the β-receptor ~100 times more strongly than R(+)-PRO (4). However, the underlying mechanism of
the stereoselective effects of PRO remains unclear.
A differential protein expression profile is useful
for deducing the mechanism involved. Differentially expressed
proteins provide important information with regard to the
pharmacodynamic mechanism of the stereoselectivity of the chiral
drug. Two-dimensional gel electrophoresis (2-DE) and mass
spectrometry analysis have made this feasible. In the present
study, we selected PRO as a model drug and employed proteomic
approaches to characterize the proteomic differences between human
umbilical vein endothelial cells (HUVECs) treated with R(+)-PRO and
those treated with S(−)-PRO. Our aim was to provide new knowledge
and strategies for chiral drug development.
Materials and methods
Materials
Iodoacetamide, ammonium bicarbonate, trifluoroacetic
acid and α-cyano-4-hydroxycinnamic acid (CHCA) were purchased from
Sigma (St. Louis, MO, USA); acetonitrile (ACN) from Fisher (Fair
Lawn, NJ, USA); trypsin (modified, sequencing grade, lyophilized)
from Promega (Hercules, CA, USA); immobilized pH gradient strips,
acrylamide/bis (30%, 29:1) and all other reagents were purchased
from Bio-Rad (Hercules, CA, USA), unless otherwise stated.
Cell preparation
HUVECs were provided by the Institute of Burn
Research, Southwest Hospital of the Third Military Medical
University (Chongqing, China). They were cultured in RPMI-1640
medium (Gibco-BRL, Grand Island, NY, USA) supplemented with 10%
heat-inactivated (56°C, 30 min) fetal calf serum (Hyclone, Logan,
UT, USA). Two groups of cultured HUVECs were pretreated for 2 h
with 80 μmol/l of either R(+)-PRO or S(−)-PRO prior to proteomic
analysis. The study was approved by the ethics committee of the
First People's Hospital of Yunnan Province.
2-DE
Protein extraction and 2-DE were performed as
previously described (http://www.proteome.tmig.or.jp/2D/2DE_method.html)
with some modifications. In total, 107 cells were
harvested and lysed in 350 μl lysis buffer [7 M urea, 2 M thiourea,
4% w/v CHAPS, 1% Pharmalyte (pH 3.1; Amersham Biosciences,
Piscataway, NJ, USA), 65 mM dithiothreitol] and sonicated. After
centrifugation, the supernatant was collected and the protein
concentration was determined by Coomassie blue G-250 staining using
ovalbumin as a standard. The protein spots were subsequently
separated and quantified using the PDQuest 7.1 software program
(Bio-Rad). We performed 3 independent experiments and the unpaired
Student's t-test was used to determine whether the differences were
statistically significant. The spots that demonstrated >2-fold
differences in expression between HUVECs treated with R(+)-PRO and
those treated with S(−)-PRO were defined as differentially
expressed proteins.
Protein spot identification
The spots showing large differences in expression
were subjected to protein identification using the proteolytic
peptide fingerprinting method. The candidate protein spots in the
gel were digested with trypsin according to the manufacturer's
instructions. The digested protein was directly mixed with an equal
volume of 10 mg/ml CHCA and the mass spectra of the peptide
fragments were obtained on an ABI Voyager DE Pro MALDI-TOF MS
(Applied Biosystems, Inc., Carlsbad, CA, USA). Protein
identification was performed based on peptide fragment matching
using the MASCOT search program (http://www.matrixscience.com). P<0.05 was
considered to indicate successful protein identification. The
parameters used were as follows: Species, Homo sapiens;
enzyme, trypsin; mass values, monoisotopic; peptide mass tolerance,
±1.0 Da; peptide charge state, 1+; max missed cleavages, 1.
Confirmation of guanine
nucleotide-binding protein subunit β-2-like 1 (GBLP)
To confirm the difference in expression of GBLP
between HUVECs treated with R(+)-PRO and those treated with
S(−)-PRO, western blot analysis and confocal laser scanning
microscopy (LSCM) were employed. For western blot analysis, the
whole cell proteins were separated by 12% SDS-PAGE and then
transferred onto a PVDF membrane using a semi-dry transfer system.
After blocking with 5% skimmed milk, the membranes were initially
probed with rabbit anti-RACK1 (H-187) polyclonal antibody (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA; Cat. #sc-10775) and
then simultaneously with horseradish peroxidase-labeled anti-rabbit
IgG (H+L) polyclonal antibody (Pierce Biotechnology, Rockford, IL,
USA; Cat. #31460) and anti-β-tubulin (Epitomics, Inc., Cat.
#1879-1) monoclonal antibody. Finally, immunoreactive bands were
detected using enhanced chemiluminescence (ECL), developed and
scanned as TIF format images with a Kodak EDAS 290 scanner. For
LSCM, the cells were cultured on poly-L-lysine cover slips (BD
BioCoat, San Diego, CA, USA) and treated for 2 h with either
R(+)-PRO or S(−)-PRO. Once the cells were fixed with 4%
paraformaldehyde in PBS and blocked with normal serum, they were
probed sequentially with primary and fluorescently labeled
secondary antibodies (Sigma). The cover slips were mounted onto
glass slides and imaging was performed using a Leica laser scanning
confocal microscope (TCS SP2; Mannheim, Germany).
Statistical analysis
Statistical analysis was performed using SPSS 10.0
software. A two-sided probability test was used and P<0.05 was
considered to indicate a statistically significant difference.
Protein spots from the 2-D gels with at least a two-fold difference
(in either direction) were identified as being differentially
expressed. Differences in the protein expression of GBLP between
HUVECs treated with R(+)-PRO and those treated with S(−)-PRO were
determined with a t-test.
Results
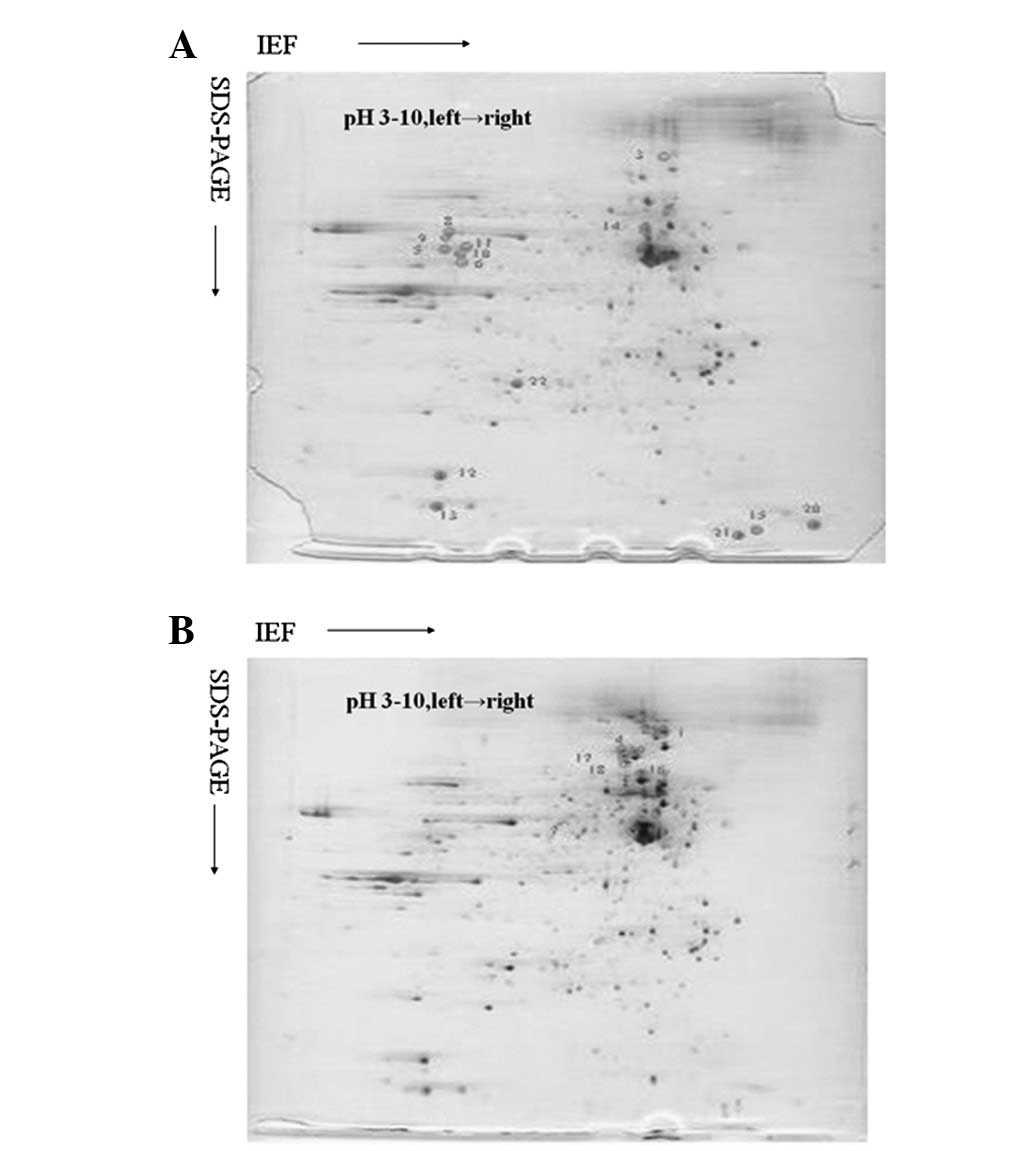
Analysis of the 2-D gel images
Representative silver nitrate-stained 2-D gel images
of protein extracts from HUVECs treated with R(+)-PRO and those
treated with S(−)-PRO revealed >750 protein spots in each gel.
The majority of the protein spots exhibited comparable protein
expression levels in the two groups. The density of each spot was
determined by the software and normalized against the total gel
density, which represented the total protein quantity in the cell
group. There were 22 spots with significant differences in the spot
optical densities between the HUVECs treated with R(+)-PRO and
those treated with S(−)-PRO (Fig.
1). Of these 22 differentially expressed proteins, 14 were
significantly upregulated by R(+)-PRO, while 8 were significantly
downregulated in the HUVECs treated with R(+)-PRO compared with
those treated with S(−)-PRO.
Identification of differentially
expressed proteins
Of the 22 proteins that were differentially
expressed between the two groups of cells, we were able to identify
10 using the MASCOT database (Table
I). These proteins were comprised of 4 intracellular signaling
molecules, 3 heat shock proteins, 3 metabolic enzymes and 1
cytoskeleton structural element, all of which are involved in
numerous essential cellular functions. Of these, 4 proteins were
upregulated and 6 were downregulated in the HUVECs treated with
R(+)-PRO compared with those treated with S(−)-PRO.
 | Table IDifferentially expressed proteins
identified by MALDI-TOF-MS. |
Table I
Differentially expressed proteins
identified by MALDI-TOF-MS.
| Category | Protein | Accession no. | Mra (Da) | pIa | Expression
status |
|---|
| Signaling
molecule | GBLP-HUMAN (Guanine
nucleotide-binding protein subunit β-2-like 1) (Receptor of
activated protein kinase C1) | P63244 | 35,055 | 7.60 | R>S |
| Signaling
molecule | RAB36-HUMAN
(Ras-related protein Rab-36) | Q2M390 | 36,299 | 8.05 | R>S |
| Signaling
molecule | KBTB2-HUMAN (Kelch
repeat and BTB domain-containing protein 2) (BTB and kelch
domain-containing protein 1) | Q8IY47 | 71,284 | 5.42 | S>R |
| Signaling
molecule | KLH34-HUMAN
(Kelch-like protein 34) | Q8N239 | 70,568 | 5.41 | S>R |
| Metabolic enzyme | TGM2-HUMAN
[Protein-glutamine γ-glutamyltransferase 2 (EC 2.3.2.13)] | P21980 | 77,280 | 5.11 | R>S |
| Metabolic enzyme | GCNT2-HUMAN
[N-acetyllactosaminide β-1,6-N-acetylglucosaminyl-transferase
(N-acetylglucosaminyltransferase)] | Q06430 | 45,825 | 8.46 | R>S |
| Heat shock | HSP90A-HUMAN [Heat
shock protein HSP 90-α (HSP 86)] | P07900 | 84,607 | 4.94 | S>R |
| Heat shock | HSP90B-HUMAN [Heat
shock protein HSP 90-β (HSP 84)] | P08238 | 83,212 | 4.97 | S>R |
| Heat shock | GRP75-HUMAN
[Stress-70 protein, mitochondrial precursor (75 kDa
glucose-regulated protein)] | P38646 | 73,635 | 5.87 | S>R |
| Cytoskeleton | KLC18-HUMAN (Keratin,
type I cytoskeletal 18) | P05783 | 48,029 | 5.34 | S>R |
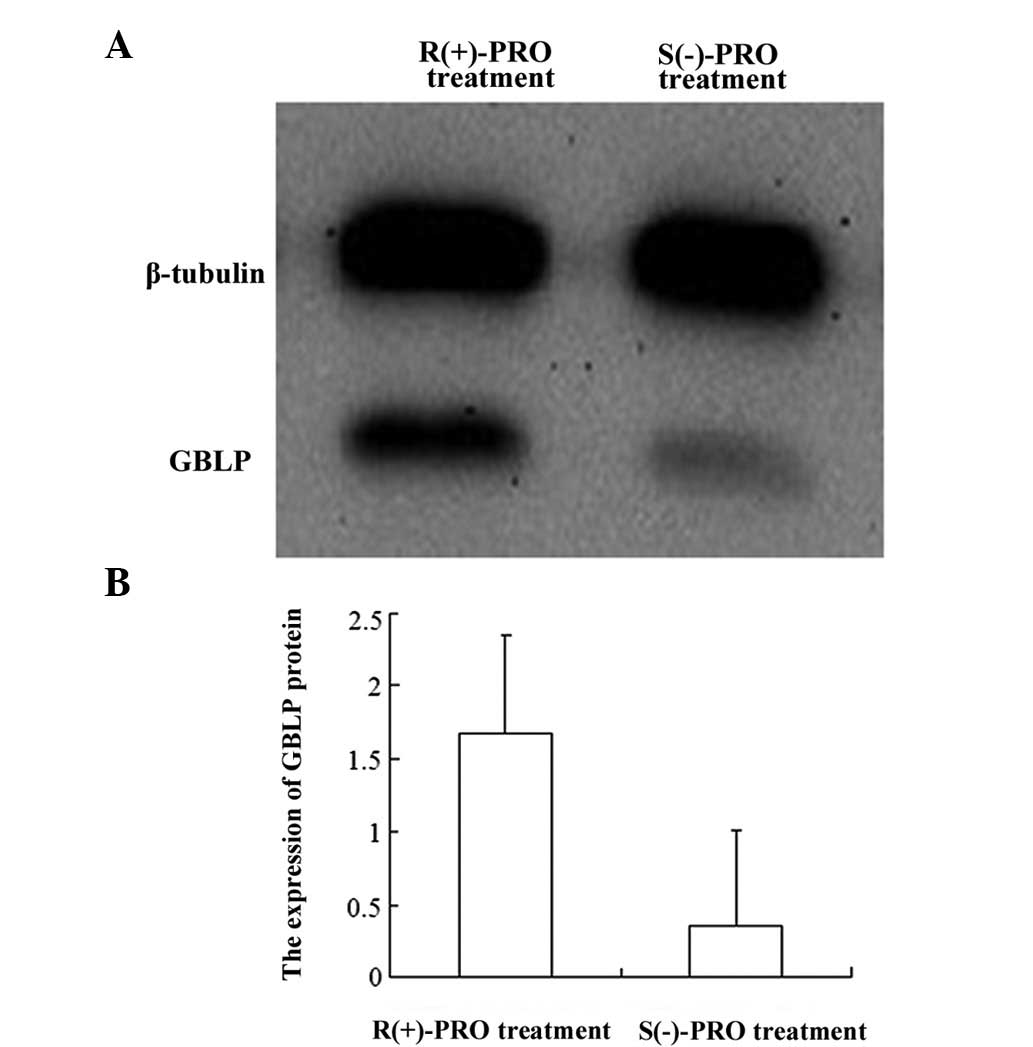
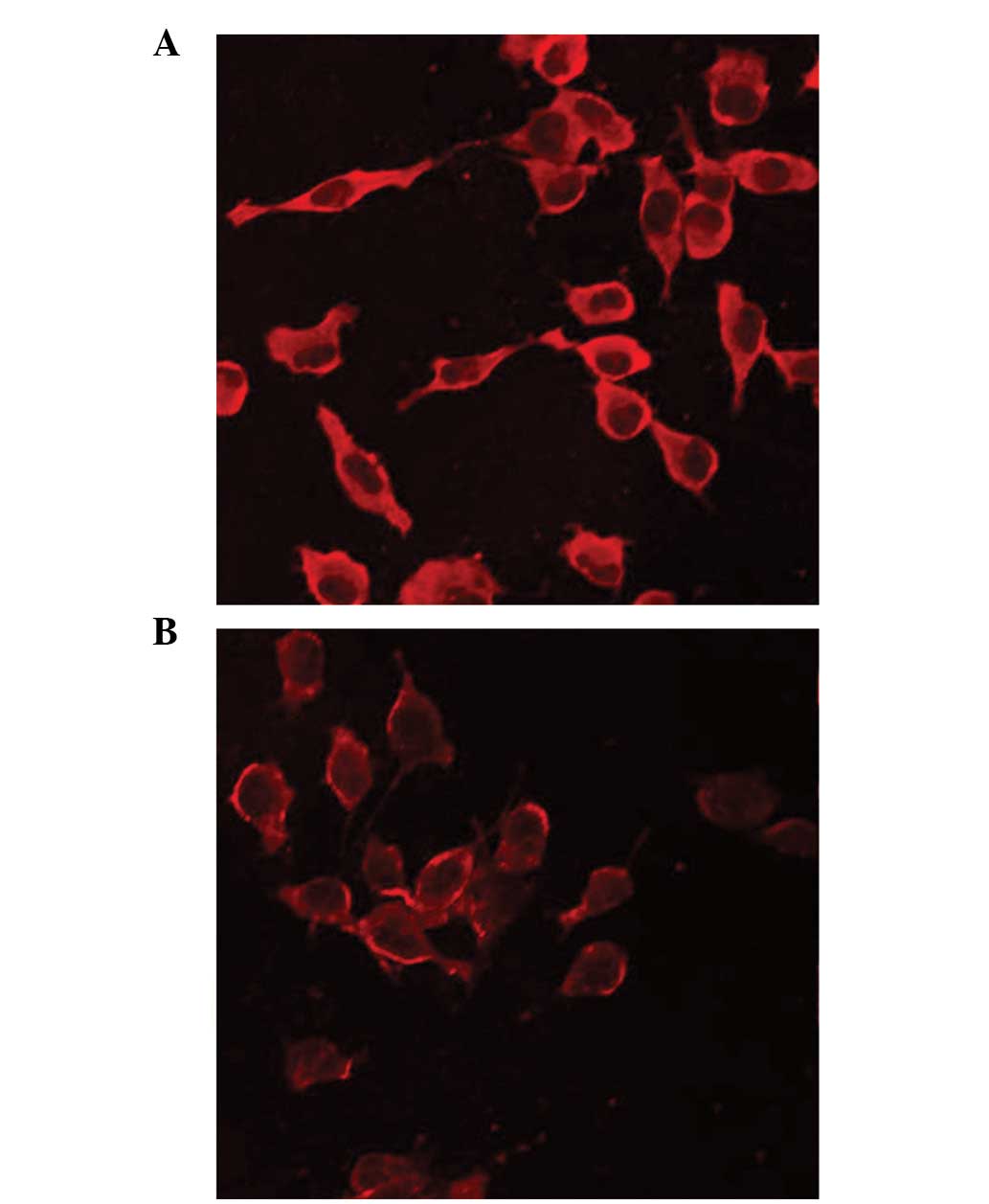
GBLP expression was decreased by
treatment with S(−)-PRO
Due to the fact that GBLP is important in several
signaling pathways and is under consideration as a new therapeutic
drug target for angiocardiopathy, we selected it for further
evaluation using western blot analysis and LSCM. We discovered that
the expression of GBLP was downregulated by treatment with S(−)-PRO
(P<0.05; Figs. 2 and 3), which is consistent with the results
observed in the 2-D gel.
Discussion
In the present study, we observed that 22 proteins
were differentially expressed between HUVECs treated with R(+)-PRO
and S(−)-PRO, of which 10 were successfully identified. Of these, 4
proteins were expressed more strongly in the HUVECs treated with
R(+)-PRO, while 6 were downregulated in the cells treated with
R(+)-PRO when compared with the cells treated with S(−)-PRO. The
majority of these differentially expressed proteins were involved
in signal transmission inside and outside the cells, energy
metabolism and stress response, indicating that the proteomic
differences between the two groups of cells was induced by 1 or
more proteins. Two of the differentially expressed proteins were
involved in energy (sugar, fat and amino acid) metabolism, TGM2 and
GCNT2. TGM2 is a key enzyme in the human glutathione cycle, which
affects the cellular metabolism in several ways, increases the
energy supply of the body and prevents angina. TGM2 may reduce
blood pressure and combat oxygenation (5–6).
TGM2 and GCNT2 were significantly upregulated in the HUVECs treated
with R(+)-PRO compared with those treated with S(−)-PRO, implying
that R(+)-PRO is a potential drug for treating angina and high
blood pressure.
GBLP is a key protein in numerous signaling pathways
(7–9). We deduced that GBLP is involved in
the chiral pharmacological mechanism of PRO, indicating that PRO
has multiple effects. Our results demonstrated that GBLP is
downregulated by treatment with S(−)-PRO compared with treatment
with R(+)-PRO. GBLP, also known as RACK1, belongs to a sub-protein
family of WD-40, and is a member of the RACK protein family. RACK1
is a receptor for active PKC and binding to it induces the
inversion of PKC, causing it to become active (10). RACK1 also combines with multiple
active proteins using different WD40 structure domain sites,
including the Src proto-carcinogenic protein (11), integrins, PDE4D5 (12), STAT and type I insulin-like growth
factor I (IGF-I) and its receptor (IGF-IR) (13,14).
Therefore, RACK1 may play various roles in multiple signaling
pathways and, at the same time, and function as multiple targets.
To date, studies on RACK1 have focused on its biological behavior
(15,16), and as a treatment for diabetes
(17) and other non-cardiovascular
diseases. However, certain studies have demonstrated that RACK1 may
participate in the occurrence and development of cardiovascular
diseases and may potentially be developed into a cardiovascular
drug. An experimental study conducted by Hermanto et
al(14) indicated that RACK1
is related to IGF-I and IGF-IR. IGF-I, IGF-IR and IGFBP1–6 are
found in several different types of tissue of the cardiovascular
system. IGF-I is thought to be the most important sex hormone
produced in the heart and may play a role in pathological and
physiological processes throughout the endocrine, autocrine and
paracrine systems. It is also thought to stimulate myocyte growth,
affect cardiac ion channels, strengthen myocardial contractility,
increase cardiac output and improve the heart ejection function.
Patterson et al(18)
observed that RACK1 adjusts the release of Ca2+, the
concentration of which plays a key role in cardiac excitability and
vascular telescopic state. In a study conducted by Bolger et
al(12), silencing RACK1
decreased the phosphorylation of β2-AR, indicating that RACK1 has a
function in signal transduction mediated by β2-AR. It is well known
that receptor phosphorylation induces arrhythmia by injuring the
β-receptor pathway and desensitization of the β-receptor and
arrhythmia may be treated by inhibiting the desensitization of the
β receptor. In our study, we observed that the expression of RACK1
in HUVECs treated with R(+)-PRO was downregulated compared with
those treated with S(−)-PRO, which explains why S(−)-PRO is a more
efficacious treatment for arrhythmia than R(+)-PRO.
As a representative nonselective β-AR antagonist
drug, PRO enantiomers are attracting increasing attention within
the research community and experiments exploring the
stereoselective mechanism in the ADME (absorption, distribution,
metabolism, excretion) drug process are still being performed.
References
|
1
|
Steiner S and Anderson NL: Pharmaceutical
proteomics. Ann NY Acad Sci. 919:48–51. 2000. View Article : Google Scholar
|
|
2
|
Graves PR, Kwiek JJ, Fadden P, et al:
Discovery of novel targets of quinoline drugs in the human purine
binding proteome. Mol Pharmacol. 62:1364–1372. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruneau JM, Maillet J, Tagat E, et al:
Drug induced protome changes in Candida albicans: comparison
of the effect of beta(1,3) glucan synthase inhibitors and two
triazoles, fluconazole and itraconazole. Proteomics. 3:325–336.
2003.PubMed/NCBI
|
|
4
|
You QD and Lin GQ: Chiral Drugs: Research
and Application. 34. Chemical Industry Press; Beijing: pp. 230–234.
2003
|
|
5
|
Lee DH, Jacobs DR Jr, Gross M, et al:
Gamma-glutamyltransferase is a predictor of incident diabetes and
hypertension: the Coronary Artery Risk Development in Young Adults
(CARDIA) Study. Clin Chem. 49:1358–1366. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee DH, Ha MH, Kim JH, et al:
Gamma-glutamyltransferase and diabetes - a 4 years follow-up study.
Diabetologia. 46:359–364. 2003.PubMed/NCBI
|
|
7
|
Dorsch M, Danial NN, Rothman PB and Goff
SP: A thrombopoietin receptor mutant deficient in Jak-STAT
activation mediates proliferation but not differentiation in UT-7
cells. Blood. 94:2676–2685. 1999.PubMed/NCBI
|
|
8
|
Guil S, de La Iglesia N, Fernández-Larrea
J, et al: Alternative splicing of the human proto-oncogene c-H-ras
renders a new Ras family protein that trafficks to cytoplasm and
nucleus. Cancer Res. 63:5178–5187. 2003.PubMed/NCBI
|
|
9
|
Kim IS, Ryang YS, Kim YS, et al:
Leukotactin-1-induced ERK activation is mediated via Gi/Go
protein/PLC/PKC delta/Ras cascades in HOS cells. Life Sci.
73:447–459. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mochly-Rosen D, Khaner H and Lopez J:
Identification of intracellular receptor proteins for activated
protein kinase C. Proc Natl Acad Sci USA. 88:3997–4000. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mamidipudi V, Dhillon NK, Parman T, Miller
LD, Lee KC and Cartwright CA: RACK1 inhibits colonic cell growth by
regulating Src activity at cell cycle checkpoints. Oncogene.
26:2914–2924. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolger GB, Baillie GS, Li X, et al:
Scanning peptide array analyses identify overlapping binding sites
for the signaling scaffold proteins, beta-arrestin and RACK1, in
cAMP-specific phosphodiesterase PDE4D5. Biochem J. 398:23–36. 2006.
View Article : Google Scholar
|
|
13
|
Kiely PA, O'Gorman D, Luong K, Ron D and
O'Connor R: Insulin-like growth factor I controls a mutually
exclusive association of RACK1 with protein phosphatase 2A and
beta1 integrin to promote cell migration. Mol Cell Biol.
26:4041–4051. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hermanto V, Zong CS, Li W and Wang LH:
RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting
protein, modulates IGF-I-dependent integrin signaling and promotes
cell spreading and contact with extracellular matrix. Mol Cell
Biol. 22:2345–2365. 2002. View Article : Google Scholar
|
|
15
|
Yaka R, Thornton C, Vagts AJ, Phamluong K,
Bonci A and Ron D: NMDA receptor function is regulated by the
inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci USA.
99:5710–5715. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Battaini F, Pascale A, Paoletti R and
Govoni S: The role of anchoring protein RACK1 in PKC activation in
the ageing rat brain. Trends Neurosci. 20:410–415. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiu Y, Mao T, Zhang Y, et al: A crucial
role for RACK1 in the regulation of glucose-stimulated IRE1alpha
activation in pancreatic beta cells. Sci Signal.
3:ra72010.PubMed/NCBI
|
|
18
|
Patterson RL, van Rossum DB, Barrow RK and
Snyder SH: RACK1 binds to inositol 1,4,5-trisphosphate receptors
and mediates Ca2+ release. Proc Natl Acad Sci USA.
101:2328–2332. 2004. View Article : Google Scholar : PubMed/NCBI
|