Introduction
The ‘fetal origins of adult chronic diseases’ theory
has been proposed in modern developmental biology. This theory
suggests that several adult chronic diseases, including coronary
heart disease, type II diabetes and hypertension, originate during
developmental plasticity in the embryonic and infant stages
(1–3). Therefore, external stimuli during
pregnancy may affect embryonic development and play a key role in
adult chronic diseases (4). One of
these diseases is essential hypertension (EH), which is a common
cardiovascular illness. Although its etiology has not been fully
elucidated, it has been shown that the occurrence of EH is
associated with the interaction between congenital genetic
susceptibility and acquired environmental factors. Previous studies
have mainly focused on the identification of relevant susceptible
genes, including the use of candidate gene approaches and
genome-wide scanning (4). To date,
>100 genes have been studied, including those involved in the
renin-angiotensin-aldosterone system, adrenergic receptors of the
sympathetic nervous system, endothelial functional proteins,
G-protein signaling proteins, cytokines and ion channels (5–7).
Although a number of these genes have been shown to be associated
with EH, they do not fully explain its occurrence.
Previous studies have shown that inflammation is
closely associated with EH (8,9). In
2002, Parissis et al(10)demonstrated that the expression of
inflammatory cytokines in the peripheral blood of EH patients was
increased, and that the increased inflammatory cytokine levels
during hypertension were mainly related to peripheral vascular
inflammation and endothelial cell dysfunction. Studies by
Samuelsson et al(11,12)
reported that following the stimulation of pregnant rats with IL-6,
the offspring exhibited elevated blood pressure levels, obesity and
enhanced activity of the hypothalamic-pituitary-adrenal axis. Since
hypertension was caused by one single cytokine in these studies,
this suggests that inflammation may be one of the direct causes of
EH. However, although the level of inflammatory cytokine IL-6 is
significantly increased during inflammation, IL-6 levels alone do
not reflect the immune-inflammatory status of the body (13).
Based on the available literature and our
experimental studies, we hypothesized that prenatal maternal
inflammation is important in EH pathogenesis. In previous studies
conducted by our group, lipopolysaccharide (LPS) from the cell wall
of Gram-negative bacteria and zymosan (Zym), both capable of
initiating systemic and non-specific inflammatory immune responses,
were used as immuno-inflammatory stimuli. Either LPS or Zym was
intraperitoneally injected into female rats on gestational days 8,
10 and 12. The 6-week-old offspring of these rats displayed
significantly higher blood pressure levels compared with the
respective controls, and at 24 weeks, a hypertension level of 140
mmHg was observed. Furthermore, the body weights of offspring in
the experimental group were significantly increased compared with
that of the controls (14–17).
In the present study, the Affymetrix
GeneChip® Rat Genome 230 2.0 Array was used to
investigate expression profile changes in the embryos after
pregnant female rats were intraperitoneally injected with LPS or
Zym. Additionally, fluorescent quantitative (q)PCR was used to
validate the differentially expressed genes determined using the
microarray analysis. The results of this study shed light on
embryonic development following immune-inflammatory stimulation
during pregnancy, providing valuable information for future studies
investigating the association between hypertension and
inflammation.
Materials and methods
Animals
Pregnant Sprague-Dawley rats (200–250 g) were
purchased from the Experimental Animal Center of the Third Military
Medical University (Chongqing, China). The rats were raised under
the following conditions: 23–25°C, 60% relative humidity, 12-h
light/dark cycle, periodic air changes (every 15 min) and ad
libitum access to food and water. This study was performed in
strict accordance with recommendations in the Guide for the Care
and Use of Laboratory Animals published by the National Institutes
of Health (NIH Publication no. 85-23, revised 1996; http://www.nap.edu/openbook.php?record_id=5140). The
protocol was approved by the Ethics Committee for Animal
Experimentation of the Third Military Medical University. All the
surgical procedures were performed under ether anesthesia and all
efforts were made to minimize suffering.
Model preparation
Fifteen pregnant rats were randomly divided into
three groups (5 animals/group); the control, LPS and Zym groups and
animals in these groups were intraperitoneally injected with 1
ml/kg sterile saline, 0.79 mg/kg LPS or 8 mg/kg Zym, respectively.
The solutions were administered between 8:00 and 9:00 a.m. on
gestational days 8, 10 and 12. Twelve hours after the final
injection, the rats were anesthetized and caesarean sections were
performed immediately under sterile conditions in order to obtain
the embryos. One embryo was removed from each pregnant rat and
placed into an RNAlater solution, which was incubated at 4°C
overnight and then stored at −80°C (Qiagen, Mainz, Germany).
Extraction of total RNA
Each whole embryo was homogenized and the total RNA
was extracted using the Qiagen RNA extraction kit (Qiagen). Total
RNA then was purified using the Qiagen RNeasy Mini Kit (total RNA
yield, >45 μg; Qiagen).
Microarray hybridization
The One-Cycle cDNA Synthesis kit (Affymetrix Inc.,
Santa Clara, CA, USA) was used to generate first- and second-strand
cDNA from a sample of total RNA (1–8 μg). The synthesized cDNA was
purified using the cDNA Cleanup Spin Column (Affymetrix Inc.). The
cRNA was then synthesized and purified using the GeneChip IVT
Labeling kit, then fragmented to a size range of 35–200 bp for
microarray hybridization. The microarray hybridization was
performed by Gene Tech Co., Ltd. (Shanghai, China). The
differentially expressed genes were determined based on the
following two criteria: i) signal ratios of >2.0 and <0.5
were considered to be upregulated and downregulated genes,
respectively; and ii) at least one signal of the two signal
detection values was required to be significantly higher than the
backgroud value.
Data analysis
Gene expression changes following LPS and Zym
stimulation were subjected to pathway analysis using the GenMAPP
and MAPPFinder programs (http://www.genmapp.org). Functional analysis was
performed based on the biological function classification in the
Gene Ontology (GO) database (http://www.geneontology.org). More specifically, the
analysis was performed using three major characteristics;
biological processes, molecular functions and cellular components.
GenMAPP classifies signaling pathways into four major groups;
cellular processes, metabolic processes, molecular functions and
physiological processes. All the expression profile data from the
LPS vs. control and Zym vs. control experiments were first compiled
using the document format required by GenMAPP. Subsequently, the
two data sets were imported into the Expression Dataset Manager of
the GenMAPP software. In the color sets, the colors were used to
indicate the up- and downregulated genes, as well as the
fold-changes for gene expression.
qPCR
Twenty genes of interest were selected to be
examined using qPCR. Total RNA extraction and cDNA generation were
performed according to the manufacturer’s instructions (Takara
Biomedical Technology, Beijing, China). Detection was performed
using the SYBR® Green PCR Master mix (Takara Biomedical
Technology). The double-standard curve method was adopted to
perform qPCR analysis for the target genes and a housekeeping gene
(β-actin) for each sample. The amplified products were subjected to
melting curve analysis to verify their specificity. For each
sample, the concentration of a target gene was divided by that of
the housekeeping gene to estimate the relative content of the
target gene in the sample.
Results
Microarray analysis
To determine the differentially expressed genes,
signal ratios of >2.0 and <0.5 were used as the cut-off
values for upregulated and downregulated genes, respectively. Rat
embryos in the LPS group produced 183 upregulated and 270
downregulated genes, while rat embryos in the Zym group generated
144 upregulated and 417 downregulated genes, both compared with the
control group. Results of our previous studies demonstrated that
the prenatal administration of either LPS or Zym caused elevated
blood pressure levels in rat offspring. Consequently, this study
aimed to identify genes that changed in the same direction in both
the LPS and Zym groups. A total of 50 upregulated and 173
downregulated genes occurred in both the LPS and Zym groups. These
genes included a number of transcripts with unknown functions and
several external sequence tags (ESTs). With regard to genes with a
known function and that were shared by the two groups, 10 were
upregulated and 85 were downregulated.
Biological function analysis
Functional analysis was classified based on
biological function annotations in the GO database, and was mainly
performed using three major characteristics (biological processes,
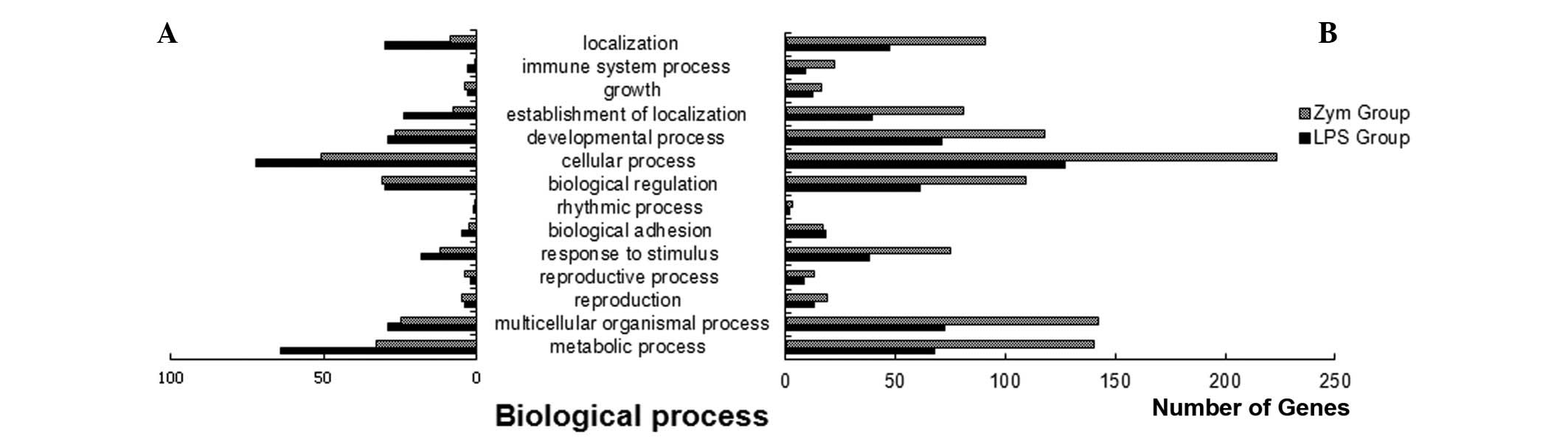
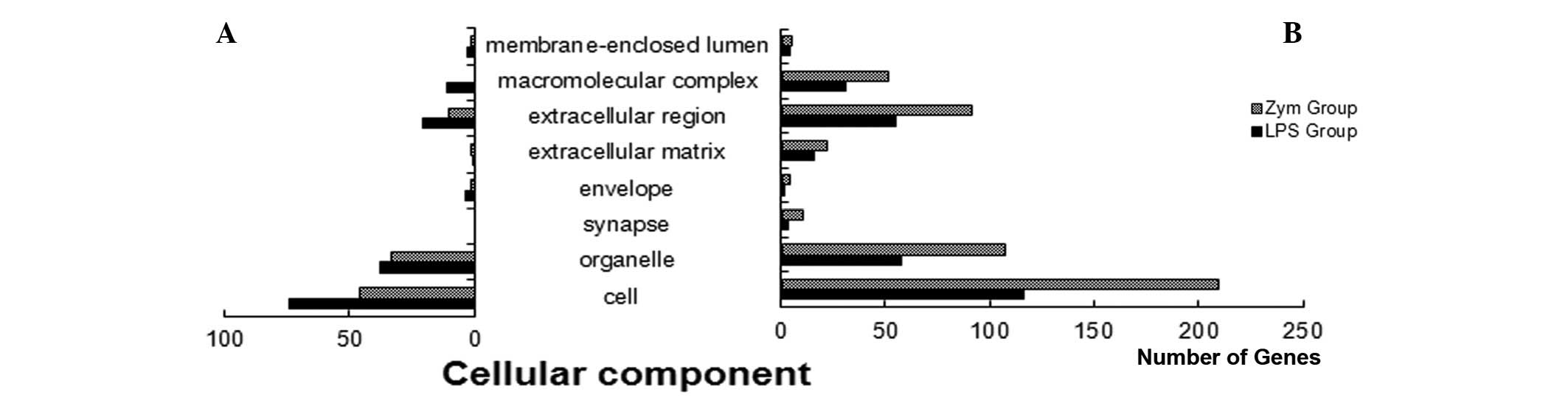
molecular functions and cellular components). The biological
process analysis showed that there were fewer upregulated than
downregulated genes, and that there were even fewer upregulated and
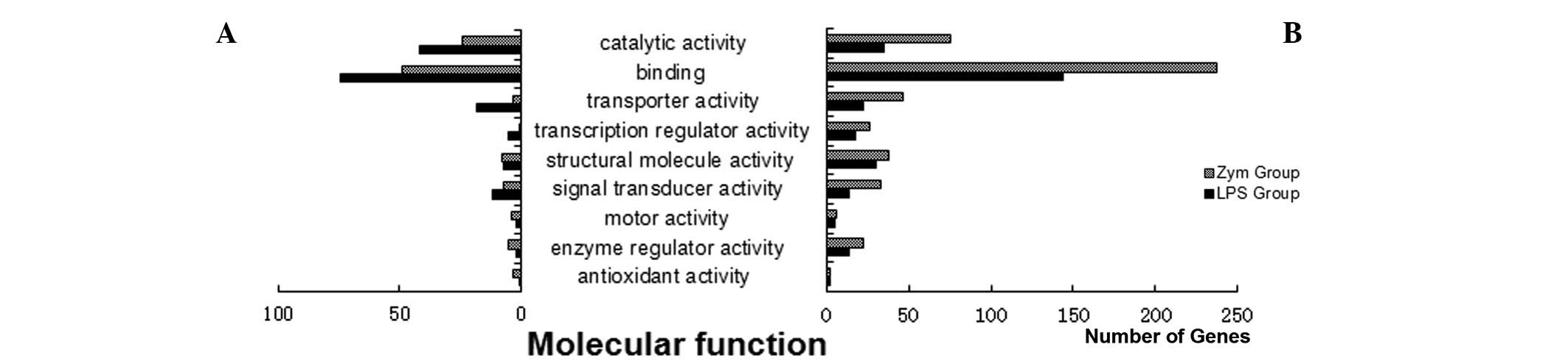
more downregulated genes in the Zym group. The molecular function
analysis demonstrated that the differentially expressed genes were
involved in a variety of biological pathways. The pathways with
more differentially expressed genes included genes involved in
cellular processes, development, metabolism and biological
regulation (Fig. 1). The cellular
component analysis indicated that the experimental groups had fewer
upregulated than downregulated genes (Fig. 2). The focus of the present study
was the analysis of molecular function, which, as previously
stated, involved more downregulated than upregulated genes, and
this pattern was more apparent in the Zym group. Additionally,
detailed functional analysis showed that the differentially
expressed genes consisted primarily of binding molecules, proteins
with catalytic activity, transporters, signal transduction
molecules and transcriptional regulators (Fig. 3) (18–20).
GenMAPP signaling analysis
The Gene Microarray Pathway Profile 2.1 (GenMAPP
2.1) software was used for signaling pathway analysis. The database
(Biological Pathway Map) was composed primarily by the KEGG pathway
database (http://www.genome.jp/kegg), which is
a database provided by a number of biologists and is maintained by
a group of bioinformaticians (21–23).
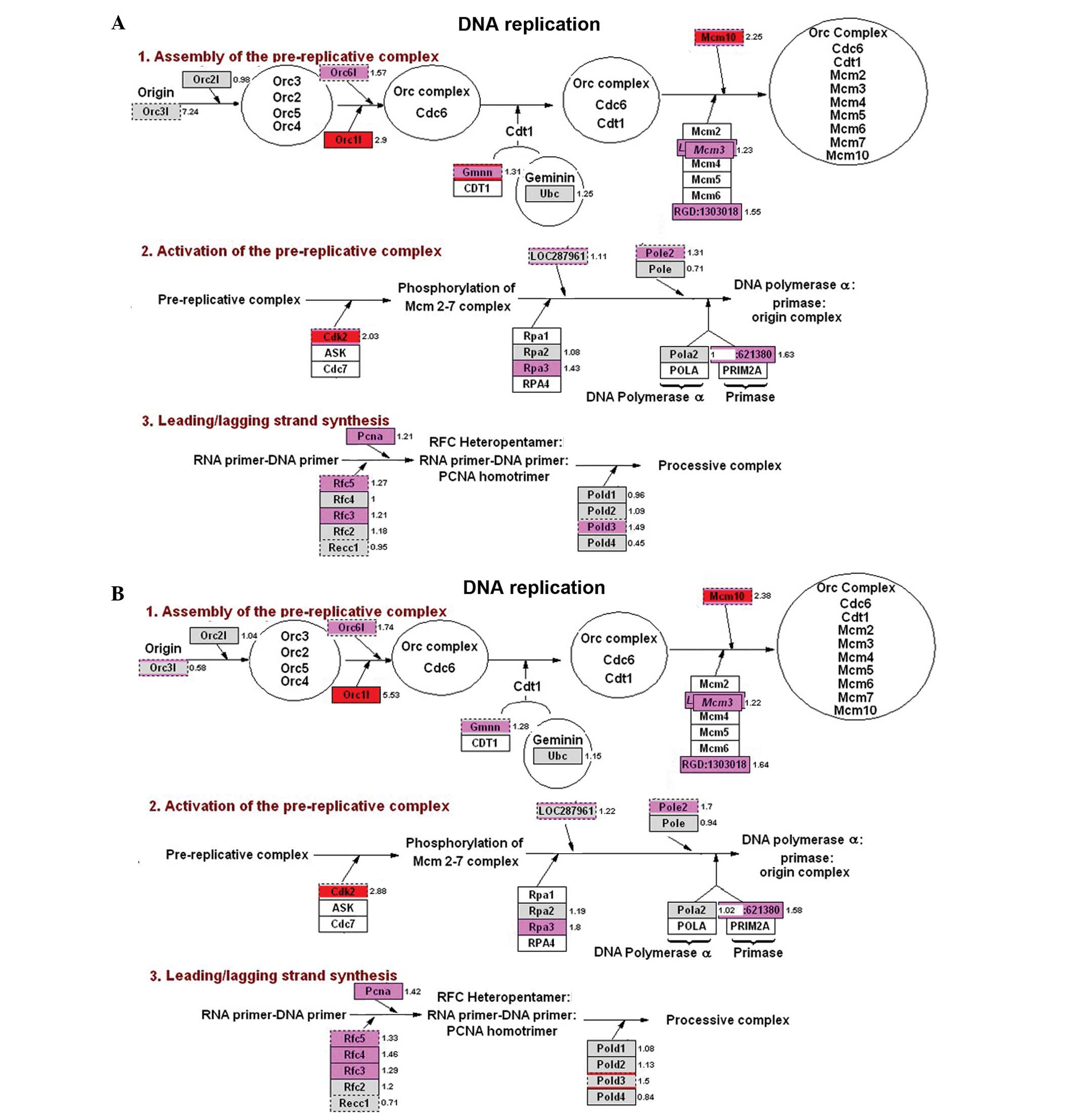
The analysis of genes associated with DNA
replication displayed numerous pink and red expression levels,
indicating that the corresponding genes were highly expressed in
the experimental groups. Additionally, there was high consistency
between the LPS and Zym groups. The upregulated genes included
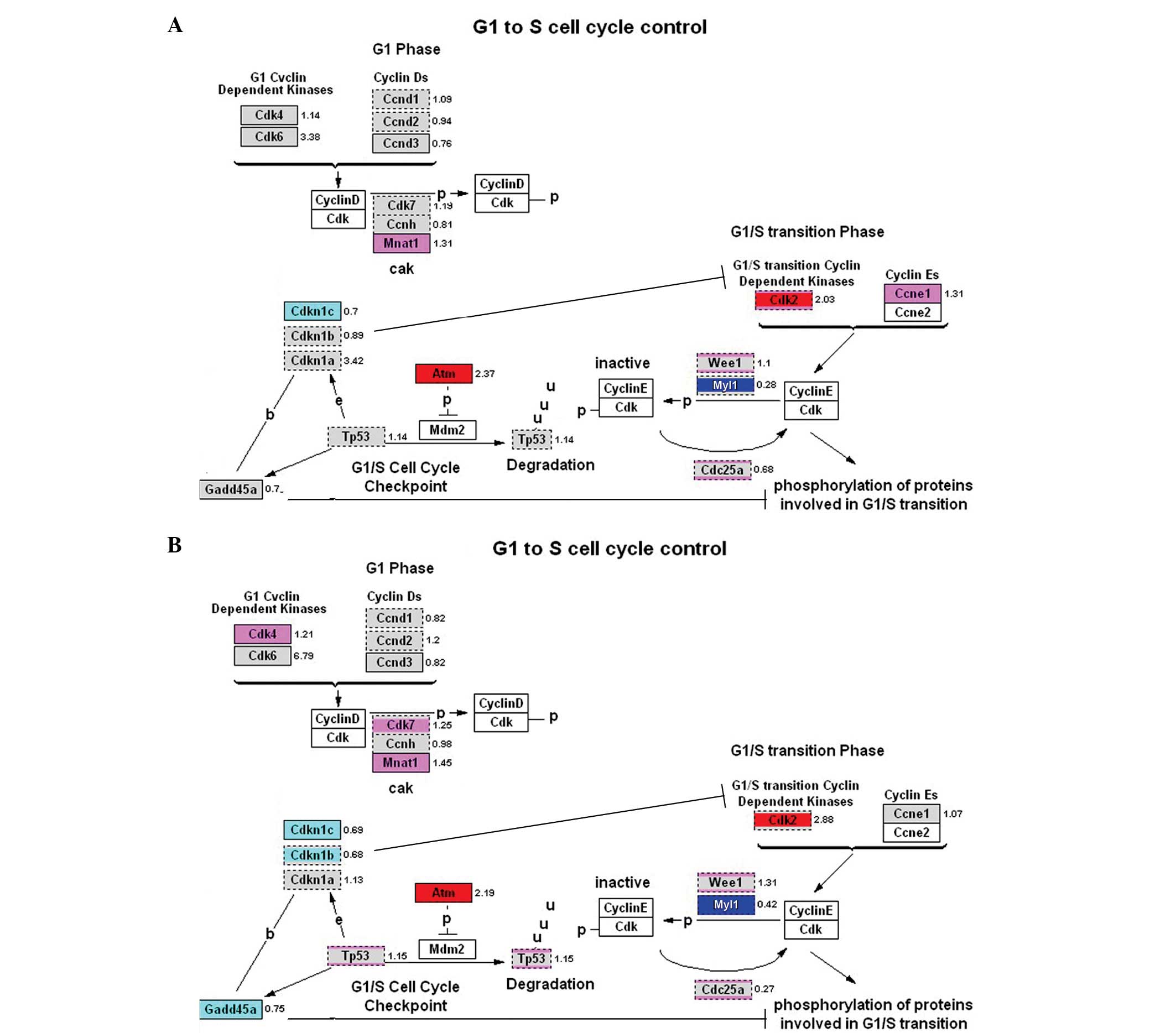
Orc1l, Cdk2, Mcm10, Gmnn, Rfc5, Rpa3 and PCNA (Fig. 4). Analysis of the G1/S regulatory
pathways showed that there was a similar pattern during DNA
synthesis following cell division. In particular, the upregulated
genes included Atm, Mcm family genes and Cdk2. Furthermore, Myt1
was downregulated (Fig. 5). Genes
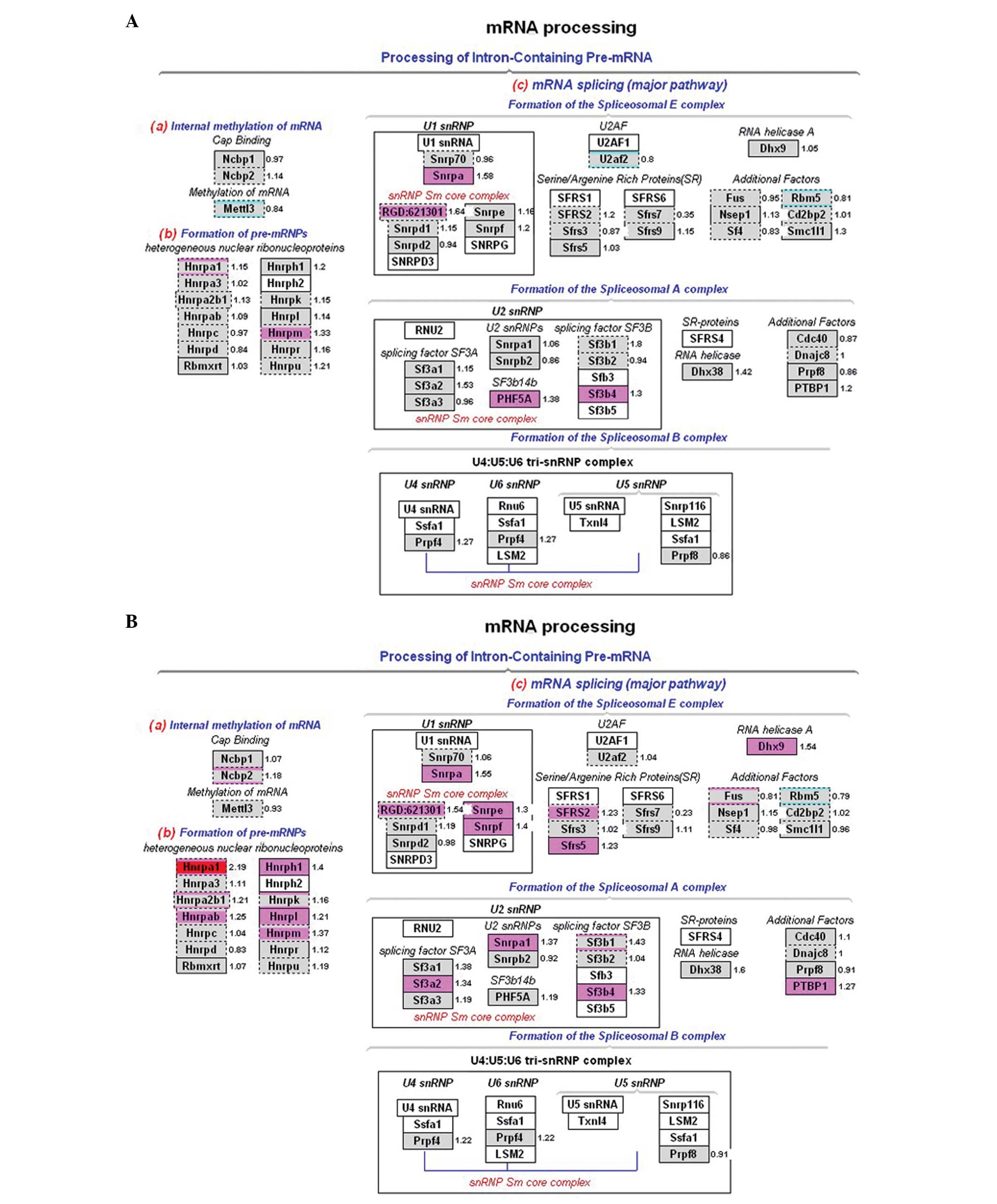
involved in mRNA processing also exhibited an upregulated pattern
that was consistent in both experimental groups. The upregulated
genes, including Hnrpa1, Snrpa and Sf3b4, were mainly involved in
mRNA splicing (Fig. 6). Several of
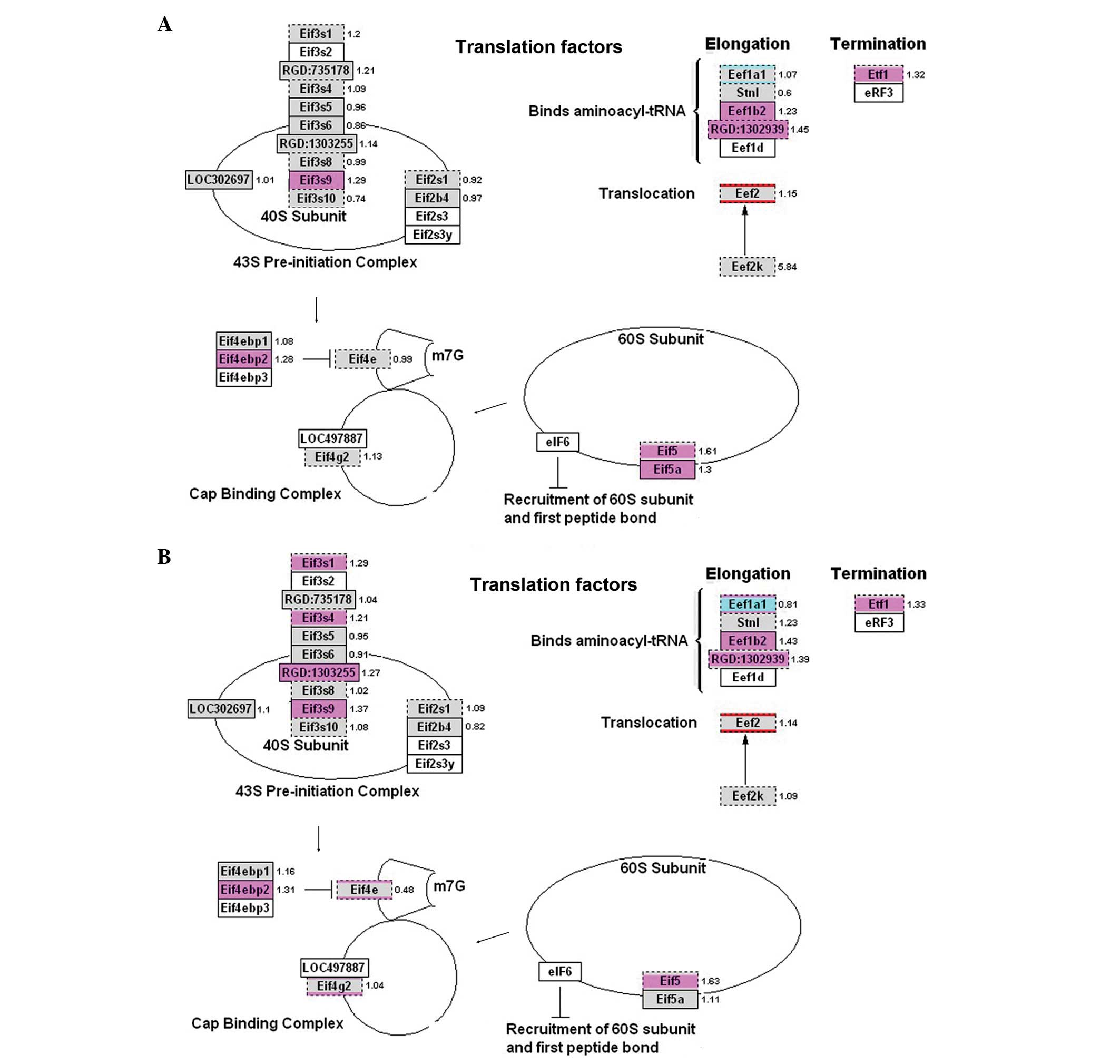
the translation factors were also upregulated in the experimental
groups; however, none of these genes exhibited a >2-fold
increase in expression. Nevertheless, these results indicated that
the upregulated genes were dominant in the entire process (Fig. 7).
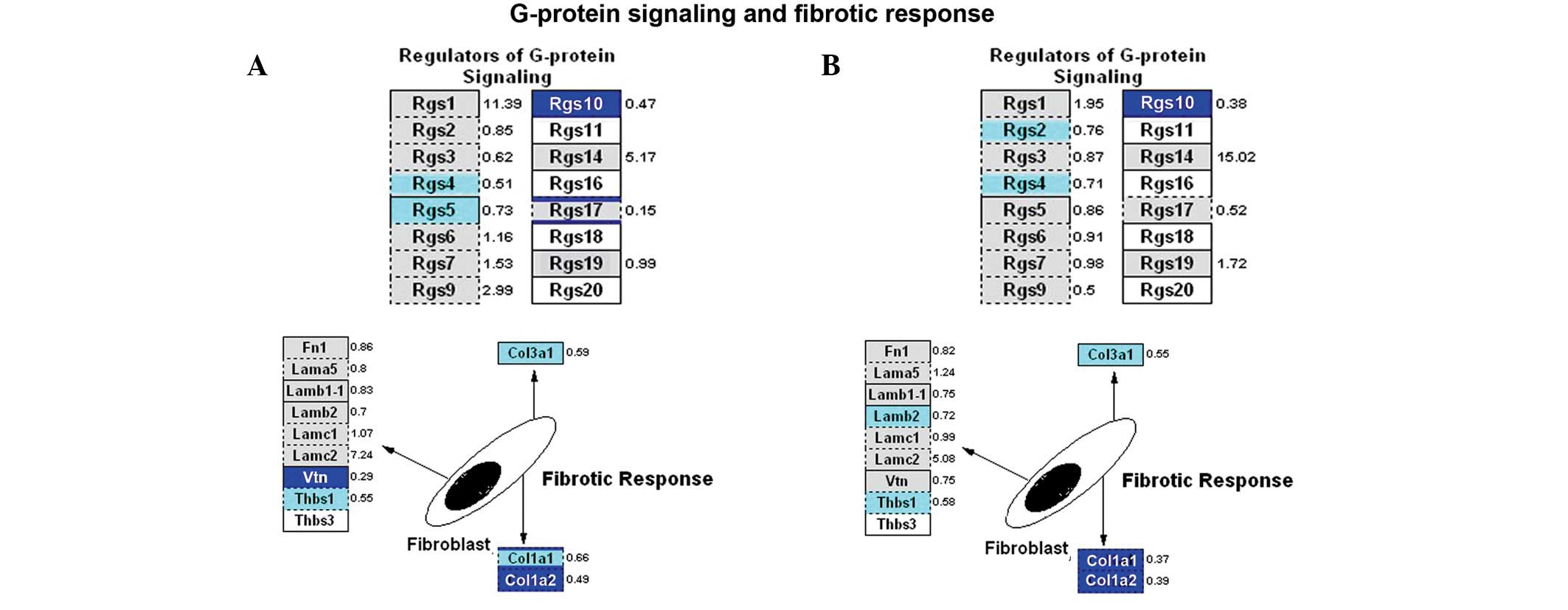
Expression of the majority of downregulated genes
was observed in calcium regulation pathways, including those
involved in G-protein signaling, CaM kinase, PKC pathways and
fibrotic response. Downregulated genes that occurred in both
experimental groups included Rgs10, Rgs4, Col3a1, Col1a1 and Col1a2
(Fig. 8).
Discussion
GO analysis was initially performed in order to
analyze the data in this study. GO is a collection of databases
established by the Gene Ontology Consortium, which aims to generate
a gene product classification standard to annotate gene and protein
functions for various species. Currently, GO contains >12
databases (18). GO is primarily
classified into three features; biological processes (e.g., orderly
integration of molecular functions leading to increased biological
activities), molecular functions (e.g., functions of individual
gene products) and cellular components (e.g., mainly referring to
cellular and subcellular structures, locations and macromolecular
complexes) (19,20).
In the present study, the distribution of
differentially expressed genes, in terms of their biological
processes, was found to vary greatly. Pathways with increased
numbers of differentially expressed genes were mainly involved in
cellular processes, development, metabolism and biological
regulation. Following the preliminary analysis, classification of
the differential expression data into one category was difficult;
therefore, our initial GO analysis focused on molecular function.
The categories for the differentially expressed genes were
determined to be binding molecules, proteins with catalytic
activity, transporters, signal transduction molecules and
transcriptional modulators, which included cytokines and signal
transduction proteins, transcription factors, enzymes directly
involved in metabolism and troponin. The binding molecules mainly
included genes with protein products that are responsible for
nucleic acid and protein binding. The nucleic acid binding
molecules were mainly regulators of transcription and translation,
while the protein binding molecules were predominantly receptor
binding proteins and signaling proteins. There were more
downregulated than upregulated genes. This downregulation of
embryonic gene expression may be important in the development of
rat offspring. Although their function is unclear, these genes may
play distinct roles during embryonic development.
The GenMAPP software was used to analyze the
signaling pathways, and the visualization method was used to
present information regarding the up- or downregulation of factors
from certain pathways and their fold-change in gene expression.
Using the GenMAPP software, the differentially expressed genes from
biological processes of interest were analyzed (21,22).
Our analyses showed that in DNA synthesis, G1/S regulation, RNA
processing and translational factors, the differentially expressed
genes were mostly upregulated and were highly consistent between
the LPS and Zym groups. This finding suggests that inflammatory
stimulation is highly associated with the upregulation of genes
involved in DNA replication, transcription and translation in rat
embryos. Among the most important genes, Orc1l, Cdk2, Atm, Mcm10,
Gmnn, Rfc5, Rpa3 and PCNA are all involved in DNA replication;
Hnrpa1, Snrpa and Sf3b4 in mRNA processing; and Eif3s9, Eif4ebp2,
Eif5, Eef1b2 and Etf1 in translation. Differentially expressed
genes were mainly downregulated in the inflammatory response
pathways. For example, Rgs10, Rgs4, Col3a1, Col1a1 and Col1a2 were
downregulated in both experimental groups. It remains unclear
whether the downregulation of inflammatory response pathways is
associated with feedback inhibition resulting from maternal
inflammatory responses.
Future studies investigating gene expression and
gene and protein function will allow the underlying molecular
mechanisms of offspring hypertension resulting from inflammatory
stimulation during pregnancy to be further elucidated. However, it
is currently possible to identify a number of specific inflammatory
factors that are important in the model of inflammation-induced
hypertension in rodent offspring.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81072630, 30973523 and
30900527).
References
|
1
|
McMillen IC and Robinson JS: Developmental
origins of the metabolic syndrome: prediction, plasticity, and
programming. Physiol Rev. 85:571–633. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barker DJ: The developmental origins of
chronic adult disease. Acta Paediatr Suppl. 93:26–33. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gluckman PD, Lillycrop KA, Vickers MH,
Pleasants AB, Phillips ES, et al: Metabolic plasticity during
mammalian development is directionally dependent on early
nutritional status. Proc Natl Acad Sci USA. 104:12796–12800. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson AD, Newton CC, Chasman DI, Ehret
GB, Johnson T, et al; Cohorts for Heart and Aging Research in
Genomic Epidemiology Consortium; Global BPgen Consortium; Women’s
Genome Health Study. Association of hypertension drug target genes
with blood pressure and hypertension in 86,588 individuals.
Hypertension. 57:903–910. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lima SG, Hatagima A and Silva NL:
Renin-angiotensin system: is it possible to identify hypertension
susceptibility genes? Arq Bras Cardiol. 89:427–433. 2007.PubMed/NCBI
|
|
6
|
Marcano AC, Onipinla AK, Caulfield MJ and
Munroe PB: Recent advances in the identification of genes for human
hypertension. Expert Rev Cardiovasc Ther. 3:733–741. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McBride MW, Graham D, Delles C and
Dominiczak AF: Functional genomics in hypertension. Curr Opin
Nephrol Hypertens. 15:145–151. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bautista LE, Vera LM, Arenas IA and
Gamarra G: Independent association between inflammatory markers
(C-reactive protein, interleukin-6, and TNF-α) and essential
hypertension. J Hum Hypertens. 19:149–154. 2005.
|
|
9
|
Sesso HD, Buring JE, Rifai N, Blake GJ,
Gaziano JM and Ridker PM: C-reactive protein and the risk of
developing hypertension. JAMA. 290:2945–2951. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parissis JT, Korovesis S, Giazitzoglou E,
Kalivas P and Katritsis D: Plasma profiles of peripheral
monocyte-related inflammatory markers in patients with arterial
hypertension. Correlations with plasma endothelin-1. Int J Cardiol.
83:13–21. 2002. View Article : Google Scholar
|
|
11
|
Samuelsson AM, Ohrn I, Dahlgren J,
Eriksson E, Angelin B, et al: Prenatal exposure to interleukin-6
results in hypertension and increased
hypothalamic-pituitary-adrenal axis activity in adult rats.
Endocrinology. 145:4897–4911. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Samuelsson AM, Jennische E, Hansson HA and
Holmäng A: Prenatal exposure to interleukin-6 results in
inflammatory neurodegeneration in hippocampus with NMDA/GABA(A)
dysregulation and impaired spatial learning. Am J Physiol Regul
Integr Comp Physiol. 290:R1345–R1356. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pauletto P and Rattazzi M: Inflammation
and hypertension: the search for a link. Nephrol Dial Transplant.
21:850–853. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei YL, Li XH and Zhou JZ: Prenatal
exposure to lipopolysaccharide results in increases in blood
pressure and body weight in rats. Acta Pharmacol Sin. 28:651–656.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao W, Wei Y, Yu C, Zhou J, Li S, et al:
Prenatal exposure to zymosan results in hypertension in adult
offspring rats. Clin Exp Pharmacol Physiol. 35:1413–1418.
2008.PubMed/NCBI
|
|
16
|
Hao XQ, Zhang HG, Yuan ZB, Yang DL, Hao
LY, et al: Prenatal exposure to lipopolysaccharide alters the
intrarenal renin-angiotensin system and renal damage in offspring
rats. Hypertens Res. 33:76–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao XQ, Zhang HG, Li SH, Jia Y, Liu Y, et
al: Prenatal exposure to inflammation induced by zymosan results in
activation of intrarenal renin-angiotensin system in adult
offspring rats. Inflammation. 33:408–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, et al: Gene ontology: tool for the unification of
biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou X and Su Z: EasyGO: Gene
Ontology-based annotation and functional enrichment analysis tool
for agronomical species. BMC Genomics. 8:2462007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alam-Faruque Y, Huntley RP, Khodiyar VK,
Camon EB, Dimmer EC, et al: The impact of focused Gene Ontology
curation of specific mammalian systems. PLoS One. 6:e275412011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dahlquist KD, Salomonis N, Vranizan K,
Lawlor SC and Conklin BR: GenMAPP, a new tool for viewing and
analyzing microarray data on biological pathways. Nat Genet.
31:19–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Salomonis N, Hanspers K, Zambon AC,
Vranizan K, Lawlor SC, et al: GenMAPP 2: new features and resources
for pathway analysis. BMC Bioinformatics. 8:2172007.PubMed/NCBI
|
|
23
|
Doniger SW, Salomonis N, Dahlquist KD,
Vranizan K, Lawlor SC and Conklin BR: MAPPFinder: using Gene
Ontology and GenMAPP to create a global gene-expression profile
from microarray data. Genome Biol. 4:R72003. View Article : Google Scholar : PubMed/NCBI
|