Introduction
Regular joint loading is required for the
maintenance of healthy cartilage tissue. The ability of cartilage
to withstand these stress loads predominantly depends on
extracellular matrix (ECM), proteoglycans and collagens.
Physiological mechanical stress is essential for chondrocyte
metabolism, and a pre-requisite for maintaining the normal
composition and structure of joint cartilage. Conversely, excessive
mechanical stress is recognized as a definite degradation factor
for cartilage ECM and as a key factor in osteoarthritis (OA).
OA is a predominant and much neglected cause of
disability in the aged. During the development of this
multifactorial degenerative disease, joint pain and progressive
limitation of motion are commonly accompanied by the gradual loss
of joint cartilage, sclerotic and cystic changes in local bone, and
osteophyte formation. Although the increased synthetic activity
within OA cartilage has been previously demonstrated, the ability
of chondrocytes to synthesize novel matrix is exceeded by the rate
of cartilage degradation (1).
Previous studies have suggested that the elevated proteolytic
activities mediated by several proteinases may be involved in the
degradation of the cartilage ECM (2). Among these enzymes, the plasminogen
activators (PAs) and the matrix metalloproteinases (MMPs) are the
most strongly implicated in cartilage degradation (3). Following the demonstration of the
involvement of PAs in the endogenous activation of latent
collagenases (4), intra-articular
PA has been suggested as a possible activator of cartilage
destruction in rheumatoid arthritis and OA. Several studies have
demonstrated that the PA levels in OA patients were positively
correlated with the degradation of cartilage (5–7).
Furthermore, the activity of PA was also correlated with the levels
of active MMP1 and MMP3, which were directly involved in the
degradation of cartilage proteoglycans and collagens during the
pathological processes of OA (8–10).
However, these studies only determined the involvement of PA in the
cartilage destruction of patients with OA, and did not demonstrate
that mechanical load results in OA by affecting the expression of
PA. In the temporomandibular joint (TMJ), mandibular motions may
result in diversely static and/or dynamic loading during talking
and chewing. Cyclic mechanical stress is suggested to be the
predominant factor resulting in the etiology of TMJ OA among
various types of mechanical stress (11). However, whether cyclic mechanical
stress induces the PA-mediated degradation of cartilage remains
unclear.
The aim of the present study was to investigate the
effects of cyclic mechanical stress on PA activity and expression
in cultured mandibular condylar chondrocytes (MCCs). Furthermore,
based on the suggestion that PA may be intensified and activated by
its cell-bound receptor (uPAR) and inactivated by its specific
inhibitor (PAI-1), it was also investigated whether other
predominant components of the PA system, uPAR and PAI-1, are
involved in the regulation of PA activity during mechanical
loading. The results demonstrated that the PA system increased the
activity of the MCCs as their response to excessive mechanical
stress was dependent upon the mechanical induction of uPA, tPA and
uPAR. Thus, PA functions as the active enzyme in the process of
mechanical stress responsiveness, largely via uPAR not PAI-1.
Materials and methods
Cell isolation and culture
Mandibular condylar cartilage was isolated from
Sprague-Dawley rats (age, 1–2 weeks) and was subsequently minced.
Following digestion with 0.25% trypsinase and 0.2% collagenase, the
primary MCCs were rinsed with phosphate-buffered saline three times
and prepared as a single-cell suspension in a growth medium of
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal calf serum, 25 mM HEPES, 1% L-glutamine and 100 mg/ml
kanamycin. MCCs were separated from the debris by filtration
through a 40-μm mesh nylon sieve. The cells were then washed,
suspended in DMEM and counted. Viable cells were seeded at a
density of 2×105 cells/cm2 in a humidified
atmosphere at 37°C and 5% CO2. Following 5–7 days in
culture, third-generation MCCs were validated by type II collagen
immunohistochemical staining as described previously (12). The cells were then utilized for the
subsequent experiments. This study was approved by the
Institutional Animal Care and Use Committee of Sichuan University
(Chengdu, Sichuan, China).
Lentiviral short hairpin RNA (shRNA)
vectors and transfection
In order to knockout uPAR expression, RNAi-Ready
Plvx-shRNA2 vector (Clontech Laboratories, Inc., Palo Alto, CA,
USA) was used. This vector was provided prelinearized for ligation
with a double-stranded oligonucleotide encoding a shRNA. In
addition to expressing shRNAs, plvx-shRNA2 also expressed the
fluorescent protein ZsGreen 1 and an ampicillin resistance gene for
the selection of stable transfectants. Two sequences of the human
uPAR gene Plaur (accession no. NM 017350) were selected as targets
for RNA interference, including shuPAR 1 (start 89:
CGGTGCATACAGTGCGAAA) and shuPAR 2 (start 174: GGGAATGGGAAGACGCCGA)
along with shLuciferase (start 1594: CTGTCTGGCACAAGAAAGTGT). For
the majority of the experiments, shuPAR 1 was used. To produce
recombinant lentivirus for target cell infection, Lenti-X plasmid
vectors were cotransfected into Lenti-X 293T cells, along with a
Lenti-X HTX Packaging mix (Clontech Laboratories, Inc.). Empty
vector (EV) expressing enhanced green fluorescent protein was used
as a control for infection efficiency. At 48 h after transfection,
supernatants containing the lentivirus were collected and frozen at
−70°C. Cells were infected with twice-diluted supernatant and 4
μg/ml polybrene for 8 h prior to being washed. Lentiviral vector
expressing enhanced green fluorescent protein served as a control
for infection efficiency; 90–95% of the cells were fluorescent for
4–6 days following infection and only a fraction of cells (<10%)
were destroyed during the 2-day selection process.
Mechanical stress on the cell
culture
When the cells had reached confluence, culture
plates were transferred to bending dishes filled with 25 ml fresh
medium. The cells were then subjected to cyclic uniaxial mechanical
compressive stress using the four-point bending system (patent no.
01129166.4 and 01256849.x), which consisted of a
computer-controlled, servomotor-driven, linear actuator assembly
with an interface controller that controlled vertical displacement
and actuator ram speed (displacement rate). Culture plates were
loaded with or without cyclic mechanical compressive stress of
2000, 4000, 6000 and 8000 μ strain at 0.5 Hz for 6, 12 and 24 h,
separately.
Determination of the rates of DNA,
proteoglycan and collagen synthesis
For determining the rate of DNA synthesis, MCC
cultures (on day 7) were exposed to 5 μCi/ml [3H] thymidine in DMEM
containing 0.5% fetal bovine serum (FBS) for the final 4 h of the
application of varying mechanical stress. The radioactive counts of
the cell layer were measured by a scintillation counter. To
determine the rate of proteoglycan and collagen syntheses, the MCC
cultures were exposed to 2.5 μCi/ml [35S] sulfate or 10 μCi/ml
[2,3-3H] proline in DMEM containing 0.5% FBS for the final 4 h of
the application of various mechanical stressors. The rates of
proteoglycan and collagen synthesis were determined by measuring
the incorporation of [35S] sulfate or [2,3-3H] proline into the
materials precipitated with cetylpyridinium chloride (Nacalai
Tesque Inc., Kyoto, Japan) in a scintillation counter.
qPCR
qPCR was used to measure the mRNA expression levels
of uPA, tPA, uPAR and PAI-1 in MCCs subjected to different methods
of mechanical loading. Total RNA was isolated from cultured rat
MCCs, which had or had not been subjected to cyclic mechanical
stress, with TRIzol reagent (Invitrogen Life Technologies Inc.,
Carlsbad, CA, USA) while being treated with RNase-free DNase I
(Takara Bio, Inc., Shiga, Japan) to avoid genomic DNA
contamination. A single-stranded cDNA was synthesized from 1 μg
total RNA using a RevertAid First-Strand cDNA Synthesis kit
(Fermentas, Vilnius, Lithuania) with a random hexamer primer. The
genes were amplified in a 25-μl reaction mixture containing
PreDeveloped TaqMan Assay Reagents (Applied Biosystems, Foster
City, CA, USA) using ABI PRISM 7700 (Applied Biosystems). The
conditions comprised an initial denaturation step at 94°C for 1
min, followed by 45 cycles at 94°C for 10 sec, 55°C for 30 sec and
72°C for 1 min, and finally an extension step at 72°C for 5 min. As
an internal control of each sample, the glyceraldehyde
3-phosphatase dehydrogenase (GAPDH) gene was used for
standardization. Quantitative results of real-time PCR were
assessed with a cycle threshold (Ct) value, which identified
a cycle when the fluorescence of a given sample became
significantly different from the base signal. The sequences of the
primers and probes for uPA, tPA, uPAR, PAI-1, MMP-2, MMP-9 and
GAPDH, are listed in Table I. The
relative expression levels of the genes were calculated using the
ΔΔCt method comparing the results with those of the 0 h control
groups.
 | Table IOligonucleotide primers and probes for
qPCR. |
Table I
Oligonucleotide primers and probes for
qPCR.
| Gene | Forward primer
5′→3′ | Reverse primer
5′→3′ | TaqMan probe
5′→3′ |
|---|
| uPA | TCACTGGCTT
CGGACAAGAGA | TCCAATGTGG
GACTGAATCCA | TGCTCGGGAG
ATTCAGGAGGACCTCTTA |
| tPA | GGCCAAATGC
CATCAAGCT | CGTGGTATAC
TTCCCTGCCTTAAA | TACTGCAGAA
ACCCAGACCGAGACGTG |
| uPAR | GTCCTGTTGG
TCTTCTCCTTGTG | CACGGTGCTT
CGGGAATG | TCACCACCTC
CAGCTCCTCGGC |
| PAI-1 | CCTCGGTGCT
GGCTATGCT | GTGCCCCTCT
CACTGATATTGAA | ACCACAGCAG
GGAAAACCCGGC |
| GAPDH | CAAGTTCAAC
GGCACAGTCAA | TGGTGAAGAC
GCCAGTAGACTC | TCTTCCAGGA
GCGAGATCCCGCTAAC |
Western blot analysis
Total proteins were extracted from MCCs that had or
had not been subjected to mechanical stress load and were
quantified using a Bicinchoninic Protein Assay kit (Pierce
Biotechnology Inc., Rockford, IL, USA). Total protein (25 μg) from
each sample was separated by 10% sodium dodecyl sulfate
polyacrylamide gel and transferred electrophoretically onto
polyvinylidene difluoride (PVDF) membranes (Millipore, Boston, MA,
USA) using a Bio-Rad Mini Trans-Blot system (Bio-Rad Laboratories,
Hercules, CA, USA). Membranes were blocked with 5% non-fat milk in
Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h and
were incubated with primary antibodies against uPA (1:400), tPA
(1:400), uPAR (1:800), PAI-1 (1:500) and GAPDH (1:1,000) for 4 h at
room temperature. Immunoreactive bands were visualized using
horseradish peroxidase-conjugated secondary antibody (1:5,000
diluted in TBST with 2% bovine serum albumin) following 1 h of
incubation. The bands were scanned using a densitometer (GS-700;
Bio-Rad Laboratories), and quantification was performed using
Quantity One 4.6.3 software (Bio-Rad Laboratories). All antibodies
were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA,
US).
Determination of PA activity
The conversion of plasminogen (PLG) to plasmin by PA
in cell lysates was determined by the hydrolysis of the chromogenic
substrate H-D-Val-Leu-Lys-pNA (S-2251, KabiVitrum, Stockholm,
Sweden) as described previously. Briefly, cells were lysed by
repeated freezing and thawing, and were then mixed with PLG (1.3
mM; Sigma-Aldrich, St. Louis, MO, USA), S-2251 (0.7 mM) and TBST
(50 mM Tris and 0.05% Tween 80, pH 8.0). The optimal density was
measured at 450 nm in a microtest plate spectrophotometer. The PA
activity was quantified with a calibration curve using human
urokinase (Sigma-Aldrich) as a standard and was presented in units
according to international standards.
Statistical analysis
Results are expressed as the mean ± SD. Statistical
analysis was performed using analysis of variance followed by
Scheffe’s test for multiple comparisons and an independent
Student’s t-test for comparisons of two groups of data. P<0.05
was considered to indicate a statistically significant
difference.
Results
Effects of mechanical stress on cell
appearance and mechanical stress condition selection
To mimic the natural living status and select
appropriate mechanical stress conditions, cyclic mechanical
stresses of 2000, 4000, 6000 and 8000 μ strain at 0.5 Hz were
adopted for this investigation. By day 7, the cultures reached
confluence (Fig. 1A) and type II
collagen staining was positive in the cytoplasm (Fig. 1B). The cell appearance, including
morphology and adherence, was unchanged in the cultures with 2000 μ
cyclic mechanical strain during the experimental period (Fig. 1C). Subsequent to 6 h of loading,
cyclic mechanical strains of 4000 and 6000 μ strain resulted in a
marked change in cell appearance of the MCCs, which manifested in
the change in morphology from star-shaped to shuttle- or
spindle-shaped. Furthermore, the long axes of the MCCs was oriented
parallel to the direction of mechanical stress (Fig. 1D and 1E). Increasing the loading
magnitude at 8,000 μ strain of stress, markedly decreased cell
adhesion, resulting in the detachment of a number of MCCs from the
culture plate during stress loading (Fig. 1F). These results indicated that an
abnormally high magnitude of mechanical stress (8000 μ strain) as
an in vitro regulatory factor for mechanical loading may be
unsuitable for this study. The appropriate stress magnitude was
thus defined at 2000, 4000 and 6000 μ strain.
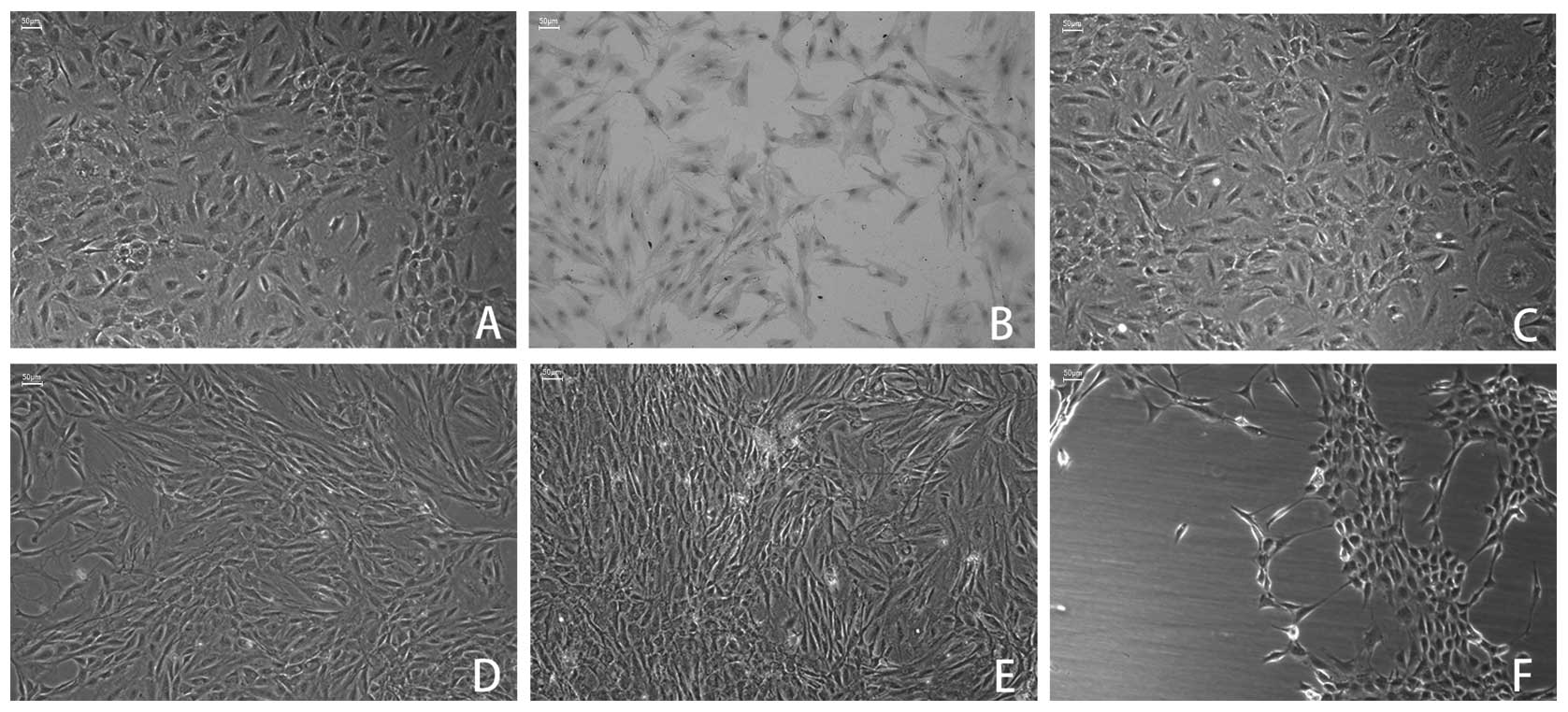 | Figure 1Effects of cyclic mechanical stress on
MCC cell appearance. (A) Third-generation MCCs were polygon-shaped,
expressed a vivid typical ‘pavement stone’ appearance. (B) Positive
immunoperoxidase staining of type II collagen was observed in the
cytoplasm of MCCs. (C) No obvious arrangements of MCC cell
appearance at cyclic mechanical stress of 2,000 μ strain. (D and E)
Following 6 h of loading, MCCs rearranged at cyclic mechanical
stress of 4,000 and 6,000 μ strain and the cell morphology changed
from star-shaped to shuttle- or spindle-shaped, and the cells long
axes were oriented parallel to the direction of stress loading. (F)
Large numbers of MCCs were detached from the plate following 6 h of
8,000 μ strain of mechanical stress. Magnification, ×100; scale
bar, 50 μm. MCC, mandibular condylar chondrocyte. |
Furthermore, to investigate the effects of
mechanical stimulation on chondrocyte metabolism, the synthesis of
DNA, collagen and proteoglycans was measured by [3H] thymidine,
[35S] sulfate and [2,3-3H] proline incorporation into MCCs at
different mechanical stress levels. Compared with the unloaded
control, cyclic mechanical stress of 2000 μ strain significantly
increased the DNA synthesis of MCCs, whereas cyclic mechanical
stress of 4000 or 6000 μ strain decreased the rate of DNA synthesis
(Fig. 2A). Similar observations
were identified in the synthesis of collagen and proteoglycans.
Cyclic mechanical stress of 2000 μ strain also increased the
synthesis of collagen and proteoglycans in MCCs, whereas cyclic
mechanical stresses of 4000 and 6000 μ strain inhibited the MCC
collagen and proteoglycan synthesis during loading periods
(Fig. 2B and 2C). Based on these
data, the cyclic mechanical stress of 4000 and 6000 μ strain were
defined as excessive stress stimuli, whereas cyclic mechanical
stress of 2000 μ strain was defined as a moderate stress stimuli,
as described previously (14–16).
Effect of mechanical stress on PA
activity
The cell-associated PA activity on cell surface and
soluble PA activity was determined in the culture supernatant of
MCCs with and without subjection to mechanical stress. Compared
with the unloaded control, the PA activity on the cell surface was
gradually increased following loading of 4000 and 6000 μ strain;
however, did not significantly change under 2000 μ strain (Fig. 3A). The soluble PA activity showed
no significant change at 2000 and 4000 μ strain loading; however
showed a significant increase at 6000 μ strain loading (Fig. 3B). The cell-associated PA activity
at 6000 μ strain was significantly higher compared with that of the
4000 μ strain (Fig. 3C).
Effect of mechanical stress on the mRNA
expression of uPA, tPA, uPAR and PAI-1
To determine whether mechanical stress induced gene
expression of the predominant components of PAs, uPA, tPA, uPAR and
PAI-1, qPCR was performed using primer pairs with a probe for each.
Compared with the unloaded control, cyclic mechanical stress at
2000 μ strain exhibited no effect on the uPA mRNA levels. However,
cyclic mechanical stress at 4000 and 6000 μ strain significantly
increased the uPA mRNA levels during the different loading periods
(Fig. 4A). Cyclic mechanical
stress at 2000, 4000 and 6000 μ strain significantly increased the
tPA mRNA levels and these elevated mRNA levels were approximately
equivalent among different loading periods (Fig. 4B). Cyclic mechanical stress also
resulted in an increase in uPAR mRNA expression at 4000 and 6000 μ
strain and the increased mRNA level simultaneously peaked following
12 h of loading. However, cyclic mechanical stress at 2000 μ strain
exhibited no effect on uPAR mRNA expression (Fig. 4C). Cyclic mechanical stress at 2000
μ strain increased the PAI-1 mRNA expression as early as 6 h
following the application of mechanical loading. Conversely, cyclic
mechanical stress at 6000 μ strain decreased the PAI-1 mRNA
expression following 6 h of loading. Cyclic mechanical stress at
4000 μ strain showed no effect on the expression of PAI-1 mRNA
(Fig. 4D).
 | Figure 4Effect of cyclic mechanical stress on
the expression of mRNA. The mRNA level of (A) uPA, (B) tPA, (C)
uPAR and (D) PAI-1 were determined using qPCR. Rat MCCs were
exposed to cyclic mechanical stress of 2000, 4000 and 6000 μ strain
for 6, 12 and 24 h, respectively. Values were normalized with the
expression level of glyceraldehyde 3-phosphate dehydrogenase and
expressed as the fold-increase of mRNA expression relative to that
in 0 h group control cultures. *P<0.05 and
**P<0.01, vs. unloaded control. Mean ± SD; n=5. uPA,
urokinase-type plasminogen activator; tPA, tissue-type PA; uPAR,
uPA receptor; PAI-1, PA inhibitor 1; MCC, mandibular condylar
chondrocyte. |
Effect of mechanical stress on the
protein expression of uPA, tPA, uPAR and PAI-1
To investigate the involvement of PAs, the proteins
from MCCs, with and without exposure to cyclic mechanical stress,
were analyzed by western blot analysis. Compared with the unloaded
control, the expression of uPA protein was significantly increased
following the application of 4000 or 6000 μ strain of mechanical
loading. The increased uPA protein expression between 4000 and 6000
μ strain of mechanical stress showed no significant difference. No
significant change in the uPA protein expression was identified
following the application of mechanical stress at 2000 μ strain
(Fig. 5A). The expression of tPA
protein was significantly increased following the application of
loading at 2000, 4000 and 6000 μ strain and these elevated tPA
protein levels were approximately equivalent among the different
loading magnitudes (Fig. 5B). The
expression of uPAR was unchanged following mechanical stress of
2000 μ strain. In contrast, at cyclic mechanical stress of 4000 and
6000 μ strain, the uPAR protein expression significantly increased
following the application of mechanical loading. Furthermore, the
increased uPAR protein levels at 6000 μ strain magnitude were
higher than that of the 4000 μ strain (Fig. 5C). The expression of PAI-1 protein
gradually increased subsequent to the application of mechanical
loading of 2000 μ strain; however, decrease following the
application of mechanical loading at 6000 μ strain (Fig. 5D). No significant change in the
PAI-1 expression was observed following the application of
mechanical loading of 4000 μ strain. These results suggested that
the expression of proteins of the PA system was involved in the
regulation of tje effects of mechanical stress on PA activity.
 | Figure 5Effect of cyclic mechanical stress on
the expression of protein. Rat MCCs were exposed to cyclic
mechanical stress of 2000 and 4000 μ strain for 3, 6, 12 and 24 h.
The protein level of (A) uPA, (B) tPA, (C) uPAR and (D) PAI-1 were
determined by western blot analysis. All values were normalized
with the expression level of glyceraldehyde 3-phosphate
dehydrogenase and expressed as the fold-increase of protein
expression relative to that in 0 h group control cultures.
*P<0.05 and **P<0.01, vs. unloaded
control were indicated. Mean ± SD; n=3. uPA, urokinase-type
plasminogen activator; tPA, tissue-type PA; uPAR, uPA receptor;
PAI-1, PA inhibitor 1; MCC, mandibular condylar chondrocyte. |
Knockdown of uPAR expression by RNAi
inhibits PA activity
As the activity of PA was regulated by its receptor
(uPAR) and inhibitor (PAI-1), the potential of shRNA-mediated
downregulation of uPAR was determined in the regulation of PA
activity on the cell surface of MCCs with and without exposure to
cyclic mechanical stress for 24 h. Following knockdown of uPAR, the
uPAR expression was significantly reduced to 42% of that of the
controls (non-transfected MCCs) at 4000 μ strain mechanical loading
and to 38% at 6000 μ strain (Fig.
6A). Compared with the unloaded control group, knockdown of
uPAR significantly decreased the expression of uPAR and the
cell-associated PA activity, but it exhibited no effect on the
expression of uPA, tPA and PAI 1, or the soluble PA activity in the
culture supernatant (data not shown). The cell-associated PA
activity in shuPAR-transfected MCCs demonstrated no significant
difference at 4000 and 6000 μ strain mechanical loading (Fig. 6B). These results suggested that the
mechanical induction of the PA activity at the cell surface may be
substantially regulated by uPAR (Fig.
6B). Moreover, analysis of the cell-associated PA activity on
shu-PAR-transfected MCCs at 4000 and 6000 μ strain mechanical
loading showed that PA functions as the active enzyme and is not
regulated mainly by its specific inhibitor, PAI 1, alone (Fig. 6B).
Discussion
Several factors (including the position or form of
the mandibular condyle and other elements of the joint) affect the
load on the condyle, which renders it difficult to determine the
actual strain in the TMJ. For this reason, the effect of stress
applied to MCCs was determined and the magnitude of the applied
mechanical stress was obtained and assessed with reference to the
in vitro data of previous studies (12,14,17).
In the present study, rat MCCs were analyzed in a four-point
bending system to investigate the effect of cyclic mechanical
tension stress on the cell viability and metabolism. In this
system, it was demonstrated that mechanical stress of 8000 μ strain
resulted in a decrease in cell viability; whereas, following
continuous exposure to mechanical stress at 2,000, 4,000 and 6,000
μ strain, MCCs exhibited no obvious changes in cell viability.
These results indicated that the abnormally high magnitude of
mechanical stress affected the cell viability and was cytotoxic to
MCCs. Notably, cyclic mechanical stress at 4,000 and 6,000 μ strain
suppressed the synthesis of the intracellular DNA, proteoglycan and
collagen, whereas reducing the magnitude of loading to 2000 μ
strain, promoted their synthesis. Fujisawa et al(14) and De Witt et al(17) also demonstrated that moderate
mechanical stress exhibited anabolic effects on chondrocytes,
whereas high magnitude mechanical stress markedly suppressed the
intracellular anabolism of chondrocytes. Based on these results,
cyclic mechanical stresses of 4000 and 6000 μ strain were defined
as appropriately excessive mechanical loadings and imitated the
mechanical response to different mechanical stimuli.
The possibility that mechanical stimulation may be
involved in the regulation of PA activity was initially suggested
following an in vitro study, which demonstrated that
excessive shear stress induced the tPA expression in endothelial
cells (18). However, whether the
mechanical stimulation affected the PA expression and activity in
cartilage chondrocytes remains unclear, particularly on MCCs. In
the present study, using a direct in vitro cell loading
method, a marked increase in PA activity in MCCs in response to the
excessive mechanical stress was observed. Furthermore, the elevated
PA activity identified following excessive mechanical stress
appeared to be associated with an increase in PA and a decrease in
PAI-1 expression; however, it may be correlated with mechanical
induction of uPAR. No change was identified in the PA activity when
MCCs were subjected to moderate mechanical stress.
Due to the rupture of surface cartilage following
compressive overload in OA pathology (19,20),
it is suggested that excessive mechanical stress stimulates the
chondrocytes to synthesize PA. This may accelerate the activation
of plasminogen and subsequently promote cartilage degradation by
providing a proteolytic environment. Furthermore, the in
situ presence of enhanced PA gene expression has been
demonstrated in previous studies, particularly in the clefts and
fissures of the cartilage defects with TMJ disk displacement and in
knee OA cartilage lesions following anterior cruciate ligament
injury. The presence of PA was also concurrent with the increased
activity of plasmin and collagenase (21–23).
Moreover, Yamaguchi et al(24) demonstrated that mechanical stress
induced tPA expression and enhanced plasmin activity in human
periodontal ligament cells. In this study, the positive correlation
between the enhanced plasmin activity and the increased PAs (uPA
and tPA) expression indicated that the mechanical induction of PA
was involved in the stress-mediated regulatory mechanism of PA
activity for excessive mechanical stress responsiveness.
In the present study, the excessive mechanical
stimuli induced the mRNA and protein expression of uPAR in MCCs. As
demonstrated previously, uPA involved in tissue remodeling was
localized and/or modulated on the cell and/or matrix surface, where
uPA catalyzed and accelerated the conversion of inactive
plasminogen to plasmin through binding with high-affinity cell
and/or matrix binding sites (21).
uPAR, as a specific receptor of uPA, binds and localizes
uPA/pro-uPA to the cell surface through its N-terminal domain 1
structure. Receptor binding not only focalizes the PA activity on
the cell surface, but also increases the activation rate of PA
(25,26). Thus, if an increased number of cell
membrane uPAR receptors are expressed and highly bound, this would
suggest that increased active PA would be available at the cell
surface (27), whereas the absence
of extracellular uPA binding sites may reduce activation of the
inactive PA zymogen. In OA, the presence of uPAR in cartilage
lesions was often accompanied by an increased fibrinolytic
activity, whereas reducing the number of uPAR on chondrocyte
surfaces with a diacetylrhein agent resulted in a decrease in the
fibrinolytic activity (28,29).
The present study indicated that, in response to excessive
mechanical stress, the stress-mediated PA activity appeared to be
modulated primarily by mechanical induction of uPAR in MCCs. This
may also be implicated directly in the degradation of cartilage
matrix, for example by its involvement in the degeneration of
annulus tissue (30).
PAI-1, as a negative regulator of the PA system, is
able to bind to uPA bound receptors at the cell surface, where
PAI-1 accumulates in a complex form of PAI-1-uPA-uPAR and undergoes
internalization followed by degradation (27). Therefore, it may lead to an
imbalance between the quantity of the enzymes and the level of the
physiological inhibitor. Yeh et al(31) showed that mechanical stimuli
upregulated the PAI-1 gene expression in chondrocytes exposed to
moderate stress levels. Consistent with this study, a stimulatory
effect of mechanical stress was also observed on PAI-1 expression
accompanied by an increase in the tPA expression under moderate
mechanical stress. Furthermore, the PAs activity remained stable.
This indicated that PAI-1 was pivotal in balancing the PAs activity
when MCCs were subjected to moderate mechanical stress. Notably, it
was identified that, in the presence of mechanical induction of
uPA, tPA and uPAR with excessive mechanical responsiveness, the
expression of PAI-1 was decreased or remained unchanged. As a
result, the equilibrium of PA/PAI-1 may be disturbed in the MCC
response to excessive mechanical stimuli, which may subsequently
result in an enhancement in the PA activity. This may due to the
fact that, the relative activity levels of the PA/PAI-1 often
change in OA cartilage lesions in response to excessive stress,
where the cartilage chondrocytes bear abnormal loading (22,29).
In conclusion, this study demonstrated that moderate
mechanical stress may only induce physiological remodeling of
cartilage matrix by simultaneously inducing an increase of tPA and
PAI-1 expression. In contrast, excessive mechanical stress resulted
in an enhancement in the PA activity by inducing the upregulation
of uPA, tPA and uPAR, as well as changes in the PA/PAI-1 repair.
This may provide a pericellular proteolytic environment, which
subsequently affects the cartilage degradation in TMJ OA.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 30300391, 81172580 and
81272961) and the Fundamental Research Funds for the Central
Universities of China (2011).
References
|
1
|
Sandell LJ and Aigner T: Articular
cartilage and changes in arthritis. An introduction: cell biology
of osteoarthritis. Arthritis Res. 3:107–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldring MB: The role of the chondrocyte
in osteoarthritis. Arthritis Rheum. 43:1916–1926. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saksela O and Rifkin DB: Cell-associated
plasminogen activation: regulation and physiological functions.
Annu Rev Cell Biol. 4:93–126. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Werb Z, Mainardi CL, Vater CA and Harris
ED Jr: Endogenous activiation of latent collagenase by rheumatoid
synovial cells. Evidence for a role of plasminogen activator. N
Engl J Med. 296:1017–1023. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh YS, Yang SF, Lue KH, Chu SC and Lu
KH: Effects of different molecular weight hyaluronan products on
the expression of urokinase plasminogen activator and inhibitor and
gelatinases during the early stage of osteoarthritis. J Orthop Res.
26:475–484. 2008. View Article : Google Scholar
|
|
6
|
Nonaka T, Kikuchi H, Ikeda T, Okamoto Y,
Hamanishi C and Tanaka S: Hyaluronic acid inhibits the expression
of u-PA, PAI-1, and u-PAR in human synovial fibroblasts of
osteoarthritis and rheumatoid arthritis. J Rheumatol. 27:997–1004.
2000.PubMed/NCBI
|
|
7
|
Sakamaki H, Ogura N, Kujiraoka H, Akiba M,
Abiko Y and Nagura H: Activities of plasminogen activator, plasmin
and kallikrein in synovial fluid from patients with
temporomandibular joint disorders. Int J Oral Maxillofac Surg.
30:323–328. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milner JM, Elliott SF and Cawston TE:
Activation of procollagenases is a key control point in cartilage
collagen degradation: interaction of serine and metalloproteinase
pathways. Arthritis Rheum. 44:2084–2096. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pap G, Eberhardt R, Röcken C, Nebelung W,
Neumann HW and Roessner A: Expression of stromelysin and urokinase
type plasminogen activator protein in resection specimens and
biopsies at different stages of osteoarthritis of the knee. Pathol
Res Pract. 196:219–226. 2000. View Article : Google Scholar
|
|
10
|
Walter H, Kawashima A, Nebelung W, Neumann
W and Roessner A: Immunohistochemical analysis of several
proteolytic enzymes as parameters of cartilage degradation. Pathol
Res Pract. 194:73–81. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuroda S, Tanimoto K, Izawa T, Fujihara S,
Koolstra JH and Tanaka E: Biomechanical and biochemical
characteristics of the mandibular condylar cartilage.
Osteoarthritis Cartilage. 17:1408–1415. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li H, Yang HS, Wu TJ, et al: Proteomic
analysis of early-response to mechanical stress in neonatal rat
mandibular condylar chondrocytes. J Cell Physiol. 223:610–622.
2010.PubMed/NCBI
|
|
13
|
Sadowski T and Steinmeyer J: Differential
effects of nonsteroidal antiinflammatory drugs on the IL-1 altered
expression of plasminogen activators and plasminogen activator
inhibitor-1 by articular chondrocytes. Inflamm Res. 51:427–433.
2002. View Article : Google Scholar
|
|
14
|
Fujisawa T, Hattori T, Takahashi K, Kuboki
T, Yamashita A and Takigawa M: Cyclic mechanical stress induces
extracellular matrix degradation in cultured chondrocytes via gene
expression of matrix metalloproteinases and interleukin-1. J
Biochem. 125:966–975. 1999. View Article : Google Scholar
|
|
15
|
Hall AC, Urban JP and Gehl KA: The effects
of hydrostatic pressure on matrix synthesis in articular cartilage.
J Orthop Res. 9:1–10. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi K, Kubo T, Kobayashi K, et al:
Hydrostatic pressure influences mRNA expression of transforming
growth factor-beta 1 and heat shock protein 70 in chondrocyte-like
cell line. J Orthop Res. 15:150–158. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
De Witt MT, Handley CJ, Oakes BW and
Lowther DA: In vitro response of chondrocytes to mechanical
loading. The effect of short term mechanical tension. Connect
Tissue Res. 12:97–109. 1984.PubMed/NCBI
|
|
18
|
Diamond SL, Eskin SG and McIntire LV:
Fluid flow stimulates tissue plasminogen activator secretion by
cultured human endothelial cells. Science. 243:1483–1485. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Flachsmann R, Broom ND and Hardy AE:
Deformation and rupture of the articular surface under dynamic and
static compression. J Orthop Res. 19:1131–1139. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morel V, Berutto C and Quinn TM: Effects
of damage in the articular surface on the cartilage response to
injurious compression in vitro. J Biomech. 39:924–930. 2006.
View Article : Google Scholar
|
|
21
|
Appella E, Robinson EA, Ullrich SJ, et al:
The receptor-binding sequence of urokinase. A biological function
for the growth-factor module of proteases. J Biol Chem.
262:4437–4440. 1987.
|
|
22
|
Martel-Pelletier J, Faure MP, McCollum R,
Mineau F, Cloutier JM and Pelletier JP: Plasmin, plasminogen
activators and inhibitor in human osteoarthritic cartilage. J
Rheumatol. 18:1863–1871. 1991.PubMed/NCBI
|
|
23
|
Pelletier JP, Mineau F, Faure MP and
Martel-Pelletier J: Imbalance between the mechanisms of activation
and inhibition of metalloproteases in the early lesions of
experimental osteoarthritis. Arthritis Rheum. 33:1466–1476. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi M, Shimizu N, Ozawa Y, et al:
Effect of tension-force on plasminogen activator activity from
human periodontal ligament cells. J Periodontal Res. 32:308–314.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ellis V, Scully MF and Kakkar VV:
Plasminogen activation initiated by single-chain urokinase-type
plasminogen activator. Potentiation by U937 monocytes. J Biol Chem.
264:2185–2188. 1989.PubMed/NCBI
|
|
26
|
Vassalli JD, Baccino D and Belin D: A
cellular binding site for the Mr 55,000 form of the human
plasminogen activator, urokinase. J Cell Biol. 100:86–92. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Plow EF, Herren T, Redlitz A, Miles LA and
Hoover-Plow JL: The cell biology of the plasminogen system. FASEB
J. 9:939–945. 1995.PubMed/NCBI
|
|
28
|
Del Rosso M, Fibbi G, Magnelli L, et al:
Modulation of urokinase receptors on human synovial cells and
osteoarthritic chondrocytes by diacetylrhein. Int J Tissue React.
12:91–100. 1990.PubMed/NCBI
|
|
29
|
Fibbi G, Pucci M, Serni U, Cerinic MM and
Del Rosso M: Antisense targeting of the urokinase receptor blocks
urokinase-dependent proliferation, chemoinvasion, and chemotaxis of
human synovial cells and chondrocytes in vitro. Proc Assoc Am
Physicians. 110:340–350. 1998.
|
|
30
|
Ford JL and Downes S: Cellularity of human
annulus tissue: an investigation into the cellularity of tissue of
different pathologies. Histopathology. 41:531–537. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yeh CC, Chang HI, Chiang JK, et al:
Regulation of plasminogen activator inhibitor 1 expression in human
osteoarthritic chondrocytes by fluid shear stress: role of protein
kinase Calpha. Arthritis Rheum. 60:2350–2361. 2009. View Article : Google Scholar
|




















