Introduction
Esophageal squamous cell carcinoma (ESCC) was once
the dominant type of esophageal malignancy in Western and Asian
countries and China has the highest incidence of esophageal
carcinoma in the world (1). The
molecular mechanisms leading to neoplastic progression in
esophageal carcinoma have not yet been fully understood.
A group of protein kinases known as cyclin-dependent
kinases (CDKs) regulate progression through the cell cycle. Two
classes of proteins, CDK inhibitors, negatively regulate the cell
cycle by binding to and inhibiting CDKs. INK4 proteins, p15, p16,
p18 and p19, specifically inhibit the CDK4/6 kinases, whereas
Cip/Kip proteins, p21cip1, p27kip1 and p57, target the majority of
cyclin-CDK complexes (2). The
p27kip1 protein promotes proliferation or blocks cell cycle
progression by inhibiting the activity of CDK, most notably CDK2.
p27kip1 binds and inhibits cyclin E- or cyclin A-associated CDKs,
and negatively regulates G1–G2 cell cycle progression (3). p27kip1 may be a potential tumor
suppressor gene, which is involved in the regulation of apoptosis
and cell motility (4). p27kip1
contributes to the repression of Sox2 during embryonic stem cell
differentiation (5). It is
hypothesized that combined deficiency in p21 and p27kip1 proteins
in mice is linked to more aggressive spontaneous tumorigenesis,
resulting in a decreased lifespan (6).
Alterations in the expression of p27kip1 cause
deregulation of cell growth and differentiation, and promote the
development of a variety of human tumors (7). A reduction in the level of p27kip1
protein contributes to tumor development by allowing an increase in
CDK2 activity and cell proliferation (8). Reduced p27kip1 expression has been
associated with the development of human epithelial tumors
originating from the majority of human organs, including the
esophagus (9). In gastric
adenocarcinoma, low p27kip1 protein expression is associated with
poorly differentiated and advanced tumors, and is a negative
prognostic factor of potential clinical value (10,11).
The developing data in human tumors suggest that p27kip1 may prove
to be a useful clinical tool even before the mechanisms of p27kip1
inactivation are completely understood.
The altered methylation of the promoter region in
tumor-associated genes may lead to transcriptional silencing, which
has increasingly been cited in a number of human cancers (12). However, promoter hypermethylation
has been reported as a common mechanism of p27kip1 inactivation
occurring at varying frequencies. The hypermethylation of p27 may
lead to the loss of p27kip1 mRNA transcription. The reduction or
loss of p27kip1 protein and mRNA is potentially involved in
hepatocarcinogenesis (13).
Silencing of the p27kip1 gene resulting from increased methylation
of the promoter region has also been observed in rodent pituitary
tumor cells (14). Previous
studies suggest that the evaluation of p27kip1 expression is likely
to aid in improving cancer diagnosis, prevention and treatment.
Therefore, clarification of the mechanisms and agents that regulate
p27kip1 expression and activity in tumor cells is an area of high
priority in cancer research (15,16).
The aim of the current study was to analyze the promoter
methylation status of the p27kip1 and mRNA expression in ESCC
patients in China.
Patients and methods
Patients and samples
Tumor samples from 50 esophageal cancer patients who
underwent surgical resection between 2004 and 2012 were
investigated, together with their adjacent non-tumor tissues. These
specimens were collected from Changzhou Tumor Hospital (Changzhou,
China). The present study was conducted in accordance with the
ethics standards of the Committee on Human Experimentation of
Soochow University (Changzhou, China). All samples were obtained
during treatment procedures under curative intent. The samples were
frozen in liquid nitrogen immediately following surgical resection
and were maintained in liquid nitrogen for long-term storage until
processing for RNA/DNA extraction.
Cell treatments
The ECa-9706 and TE-1 esophageal cancer cell lines
(Shanghai Institutes for Biological Sciences, Shanghai, China) were
maintained in the Clinical Oncology Laboratory of Changzhou Tumor
Hospital. Cells were cultured in RPMI medium (Invitrogen China
Ltd., Beijing, China) containing 10% fetal bovine serum. Cells were
cultured in a humidified 37°C incubator containing 5%
CO2. Cells were plated (3×105 cells/100 mm
dish) and treated 24 h later with 5×10−6 M
5-aza-2′-deoxycytidine (5-Aza-CdR) (Sigma-Aldrich, St. Louis, MO,
USA). The medium was changed 24 h following drug treatment, and RNA
and DNA were isolated six days after treatment.
Bisulfite modification and
methylation-specific polymerase chain reaction (MSP)
DNA was prepared from tissues and cells. Following
microdissection, the tissue samples were placed into Eppendorf
tubes and were incubated with proteinase K (Promega, Madison, WI,
USA) at 37°C overnight. The tissue was extracted twice in phenol
and twice in chloroform, followed by ethanol precipitation. Genomic
DNA (3 μg) from tissue and cells was denatured with 0.3 M NaOH at
37°C for 10 min. Next, freshly prepared (208 μl) 3 M sodium
bisulphate (pH 5.0) and 12 μl fresh 100 mM hydroquinone
(Sigma-Aldrich Shanghai Trading Co., Shanghai, China) solutions
were added. Bisulfite treatment, during which methylated DNA is
protected and unmethylated cytosine is converted to uracil, was
performed as described previously (17). Bisulphite-treated DNA was used for
amplification of the p27kip1 promoter. Primers (Sangon Biotech Co.,
Shanghai, China) specific for unmethylated p27kip1,
5′-ATGGAAGAGGTGAGTTAGT-3′ (sense) and 5′-AAAACCCCAATTAAAAACA-3′
(antisense); or methylated p27kip1, 5′-AAGAGGCGAGTTAGCGT-3′ (sense)
and 5′-AAAACGCCGCCGAACGA-3′ (antisense) were used, which amplify a
212 and 195 bp product, respectively (18). MSP was performed in a 25-μl
reaction volume with ~25 ng bisulfite-modified DNA. Reactions were
hot-started at 95°C for 5 min. This step was followed by 38 cycles
at 95°C for 45 sec, 57°C for 30 sec and 72°C for 30 sec, followed
by a 10-min extension at 72°C in DNA thermocycler (Agilent
Technologies, Inc., Santa Clara, CA, USA). The amplification
products were separated on a 2% agarose gel and visualized by
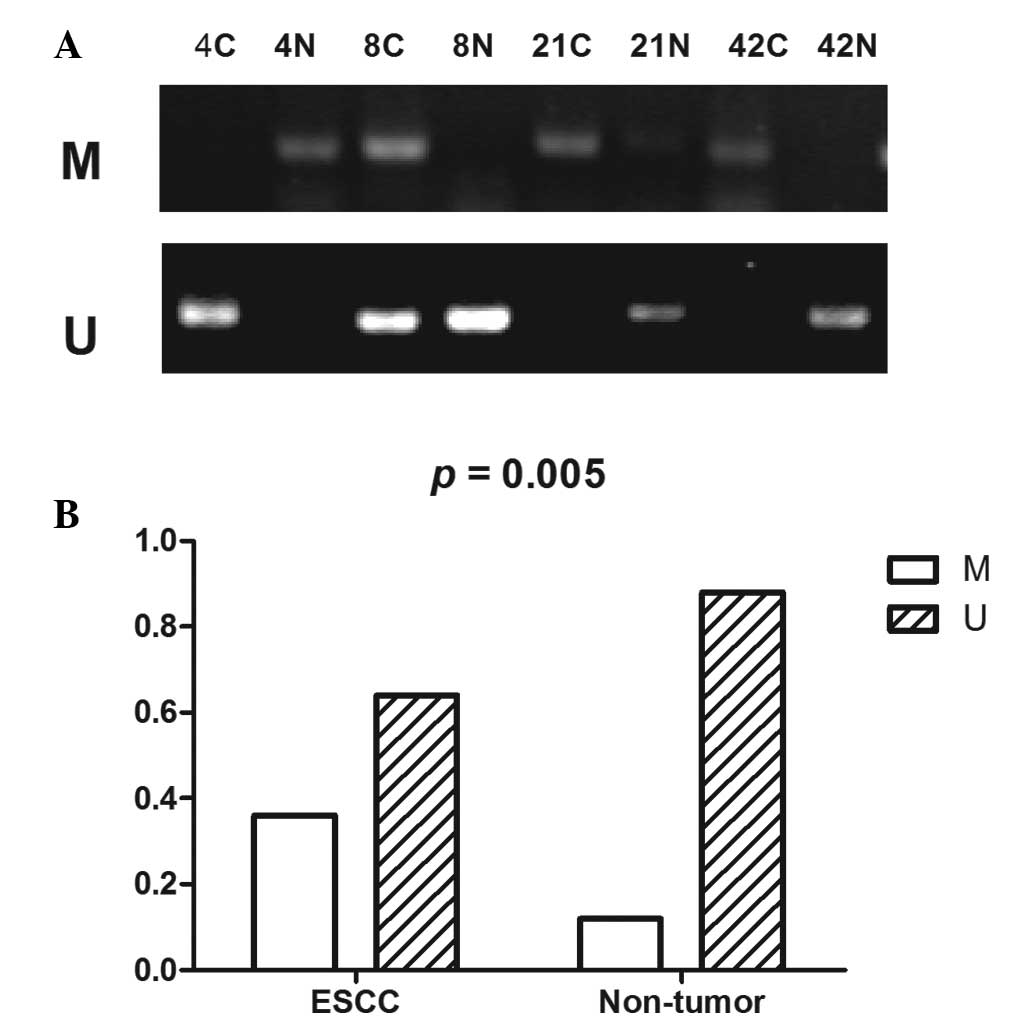
ethidium bromide staining and UV transillumination. Methylation (M)
was defined as M/(M + U) ≥ 0.5 and unmethylation (U) was defined as
M/(M + U) < 0.5.
Quantitative polymerase chain reaction
(qPCR) analysis for p27kip1
RNA was isolated from 50 ESCC tissues, adjacent
normal tissues and cultured cells. The first-strand cDNA was
synthesized from 2 μg total RNA. Primer sequences (Sangon Biotech
Co.) of p27kip1 for qPCR were 5′-TCCGGCTAACTCTGAGGACAC-3′ (sense)
and 5′-TGTTTTGAGTAGAAGAATCGTCGGT-3′ (antisense) for the
amplification of p27 mRNA (19).
qPCR was performed using the Mx3000P qPCR system (Stratagene, La
Jolla, CA, USA). The cDNA was then used for qPCR in a 20 μl SYBR
Premix Ex Taq (Takara Biotechnology Co., Ltd., Dailan, China). qPCR
for p27kip1 mRNA expression was performed under the following
conditions: 5 min at 95°C, 40 cycles of 30 sec at 95°C, 30 sec at
60°C and 1 min at 72°C. As an internal control for qPCR, β-actin
mRNA expression was amplified from the same cDNA samples. All
results were normalized to β-actin amplification. CT values for
triplicate reactions were averaged and relative p27kip1 expression
was determined with the comparative CT method, using average CT
values for p27kip1 and β-actin.
Statistical analysis
All data were generated without knowledge of the
clinical status of the samples analyzed with SPSS version 17.0
software (SPSS, Inc., Chicago, IL, USA). Comparisons were performed
with a t-test (unpaired or paired). Univariate analyses of the
interaction between p27kip1 methylation and clinical parameters
were performed with Pearson’s χ2 test or Fisher’s exact
test. All P-values presented were two-tailed. P<0.05 was
considered to indicate a statistically significant difference.
Results
Methylation status of the p27kip1
promoter region in esophageal cancer and its adjacent tissue
The methylation status of the p27kip1 promoter
region was analyzed by MSP. Methylation analysis of 50 ESCC tissues
and their adjacent non-tumor tissues was performed. The p27kip1
promoter was methylated in 36.0% (18/50) of esophageal cancers and
in 3.0% (6/50) of non-tumor samples. The difference in p27kip1
methylation between the tumor and non-tumor group was statistically
significant (P=0.005; Fig. 1).
Methylation status of the p27kip1
promoter with clinicopathologic parameters in ESCC
The association of p27kip1 methylation with
clinicopathologic parameters in esophageal cancer patients is
presented in Table I. The
methylation status of the p27kip1 promoter was associated with
tumor metastasis. Furthermore, the methylation of p27kip1 was
markedly increased in patients with metastasis (59.09%, 13/22)
compared with patients without metastasis (17.86%, 5/28). A
significant difference was identified between methylation of the
p27kip1 promoter and metastasis status in ESCC (P=0.002).
Methylation of p27kip1 was not associated with the remaining
clinicopathological parameters evaluated, including gender, tumor
differentiation and other features.
 | Table ICorrelation of p27kip1 methylation
with clinicopathological parameters of the ESCC patients. |
Table I
Correlation of p27kip1 methylation
with clinicopathological parameters of the ESCC patients.
| Variable | No. | M, n=18 | U, n=32 | P-valuea |
|---|
| Gender |
| Male | 43 | 14 | 29 | 0.209 |
| Female | 7 | 4 | 3 | |
| Depth of
invasion |
| T1–2 | 32 | 10 | 22 | 0.351 |
| T3–4 | 18 | 8 | 10 | |
| Lymph node
metastasis |
| N0–1 | 23 | 9 | 14 | 0.582 |
| N2–3 | 27 | 9 | 18 | |
| Distant
metastasis |
| M0 | 28 | 5 | 24 | 0.002 |
| M1 | 22 | 13 | 9 | |
| AJCC stage |
| I–II | 20 | 8 | 12 | 0.630 |
| III–IV | 30 | 10 | 20 | |
| Differentiation |
| G1 | 10 | 4 | 6 | 0.768 |
| G2–3 | 40 | 14 | 26 | |
Association of p27kip1 methylation with
mRNA expression in tissue
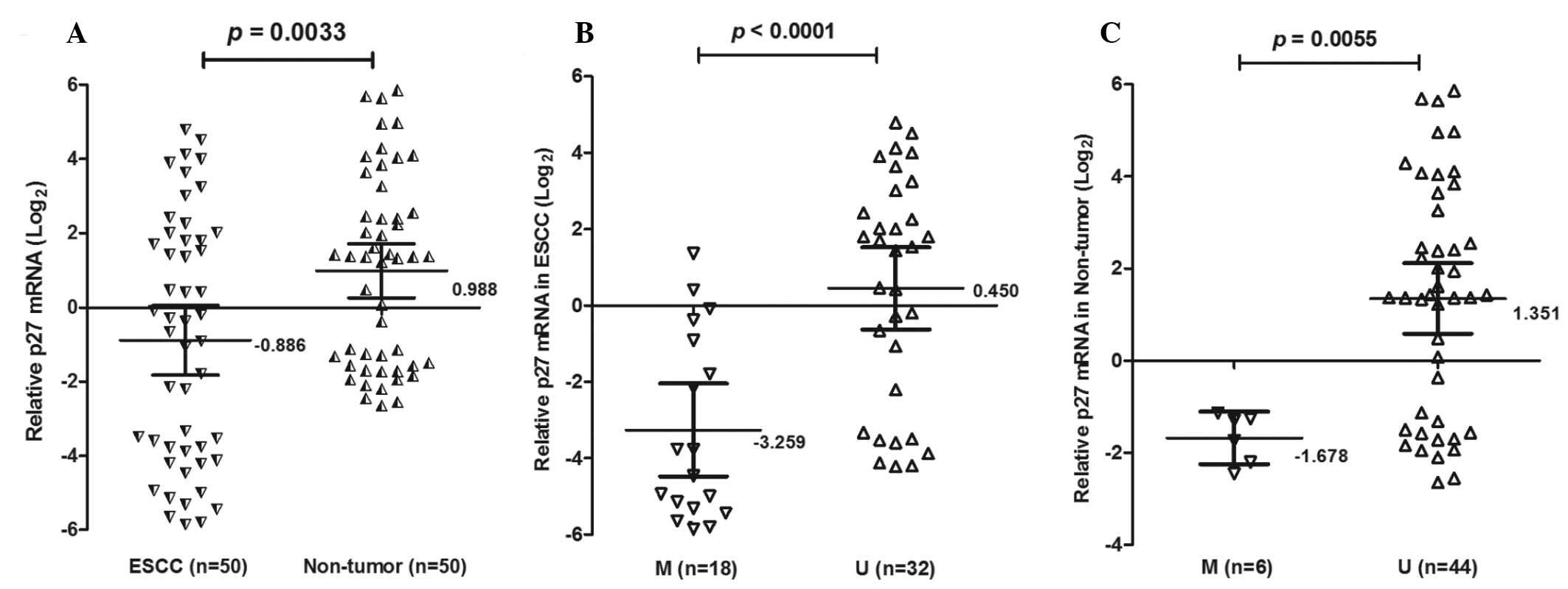
To determine whether loss of p27kip1 mRNA expression
was associated with promoter methylation, p27kip1 mRNA was analyzed
in 50 ESCC tissues using qPCR (Fig.
2). The expression of p27kip1 mRNA was lower (mean ± SD,
−0.886±3.298) in ESCC tissues compared with non-tumor tissues
(0.988±0.257; P=0.0033). Notably, the expression of p27kip1 mRNA
was decreased in ESCC patients with methylation (−3.259±2.439)
compared with unmethylation (0.449±2.970; P<0.0001).
Furthermore, p27kip1 mRNA was also decreased in non-tumor tissues
with methylation (01.678±0.545) compared with unmethylation
(1.351±2.523; P=0.0055). There was a statistically significant
association between the methylation status of the p27kip1 promoter
and p27kip1 mRNA expression (P<0.001) in cancer and non-tumor
samples.
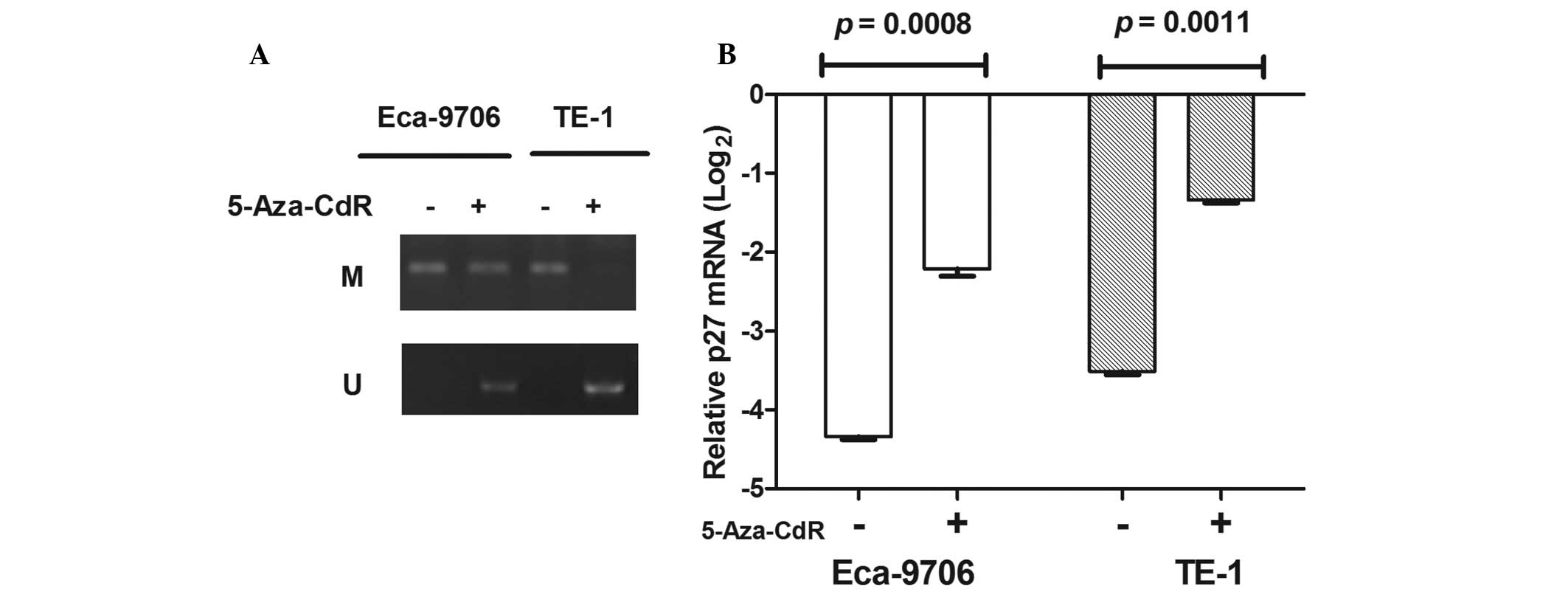
Demethylation by 5-Aza-CdR
The ECa-9706 and TE-1 esophageal cancer lines showed
methylation of the p27kip1 promoter (Fig. 3). To determine the effects of
5-Aza-CdR on p27kip1 gene expression, qPCR analyses were performed
using esophageal cancer lines treated with a final concentration of
5 μM p27kip1. However, following treatment with the demethylation
reagent, the methylation of the p27kip1 promoter was demethylated
(Fig. 3A). Following normalization
of mRNA levels to β-actin, the expression of the p27kip1 gene was
induced between −4.337±0.04 and −2.21±0.01 in the ECa-9706 cells
(P=0.0011); and between −3.513±0.07 and −1.337±0.07 in the TE-1
cells (P=0.0011; Fig. 3B). These
results suggested that the expression of p27kip1 may be activated
by 5-Aza-CdR.
Discussion
In the complex multistage process of esophageal
cancer, the accumulation of epigenetic alterations are required for
the emergence of a fully malignant tumor. Genes that inhibit cell
proliferation are ideal candidates for tumor suppressor genes,
which appear to be critical in the pathogenesis of a number of
human malignancies (20,21).
p27kip1 is an atypical tumor suppressor that
regulates G0 to S phase transitions by binding to and regulating
the activity of CDKs (22,23). Numerous antiproliferative signals
lead to p27kip1 accumulation, including mitogeny cytokine
withdrawal, cell-cell contact and agents such as cAMP and rapamycin
(24). A marked reduction in the
level of p27kip1 protein, or even a complete loss is observed in
~50% of all types of human cancer (25). Downregulation of p27kip1 expression
contributes to the increase in the percentage of cells entering S
phase, which may lead to increased radioresistance in the
established radioresistant cells (26). Although these studies are limited
in their ability to provide mechanistic information, they indicate
that low or absent p27kip1 protein in tumor cells is an important
clinical marker of disease progression in the majority of tumor
types (27).
In the present study, the frequency of
hypermethylation of the p27kip1 promoter was significantly higher
in esophageal cancer compared with the corresponding non-tumor
tissues (36.0 and 12.0%, respectively). In addition, there was a
significant association between the p27kip1 promoter methylation
status and its mRNA expression; samples with methylation exhibited
a lower p27kip1 mRNA expression, whereas samples without
methylation exhibited a higher expression (P<0.001). These
findings indicate that methylation of the p27kip1 promoter may
represent a common mechanism of inactivation of the tumor
suppressor gene in ESCC. In the current study, p27kip1 methylation
was observed to be associated with tumor metastasis in ESCC
patients (P=0.002). These findings suggest that p27kip1 methylation
may be a potential marker in association with the poor clinical
outcome of ESCC patients.
However, this cell cycle inhibitor has emerged to be
important in other cellular functions, including cell migration.
Understanding the diverse roles of p27kip1 protein during normal
cell cycle progression and tumor development may provide novel
insights into tumor prognosis and aid in the development of
therapeutics (28,29). 5-Aza-CdR is incorporated into DNA
during the replication process and binds DNMTs thereby causing
irreversible inhibition of their enzymatic activity. The present
study showed that the methylation inhibitor, 5-Aza-CdR, induced
p27kip1 mRNA expression in an esophageal cancer cell line, which
indicates that the epigenetic inhibitor may be useful in
considering the future clinical treatment in ESCC.
In conclusion, a high frequency methylation of the
p27kip1 tumor suppressor gene associated with tumor metastasis in
ESCC was reported. The inactivation of p27kip1 by promoter
methylation was a common phenomenon in ESCC. Additional studies are
required to clarify the promoter methylation and p27kip1 expression
in ESCC development.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (81372212 and 81071799); the Natural Science
Foundation of Jiangsu (BK2011251); Jiangsu Provincial Special
Program of Medical Science (BL2013012); the Health Talents Project
for Jiangsu, China (LJ201157, RC2011038 and BRA2011038); China
Postdoctoral Science Foundation Specific funded project
(201003380); and the Natural Science Foundation of Ningbo
(2011A610057).
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
MSP
|
methylation-specific PCR
|
|
5-Aza-CdR
|
5-aza-2′-deoxycytidine
|
References
|
1
|
Hongo M, Nagasaki Y and Shoji T:
Epidemiology of esophageal cancer: Orient to Occident. Effects of
chronology, geography and ethnicity. J Gastroenterol Hepatol.
24:729–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sherr CJ and Roberts JM: CDK inhibitors:
positive and negative regulators of G1-phase progression. Genes
Dev. 13:1501–1512. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bagui TK, Cui D, Roy S, et al: Inhibition
of p27Kip1 gene transcription by mitogens. Cell Cycle. 8:115–124.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li H, Collado M, Villasante A, et al:
p27(Kip1) directly represses Sox2 during embryonic stem cell
differentiation. Cell Stem Cell. 11:845–852. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garcia-Fernández RA, García-Palencia P,
Sánchez MA, et al: Combined loss of p21(waf1/cip1) and p27(kip1)
enhances tumorigenesis in mice. Lab Invest. 91:1634–1642.
2011.PubMed/NCBI
|
|
7
|
Clurman BE and Porter P: New insights into
the tumor suppression function of P27(kip1). Proc Natl Acad Sci
USA. 95:15158–15160. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Viglietto G, Motti ML and Fusco A:
Understanding p27(kip1) deregulation in cancer: down-regulation or
mislocalization. Cell Cycle. 1:394–400. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh SP, Lipman J, Goldman H, et al: Loss
or altered subcellular localization of p27 in Barrett’s associated
adenocarcinoma. Cancer Res. 58:1730–1735. 1998.
|
|
10
|
Nitti D, Belluco C, Mammano E, et al: Low
level of p27(Kip1) protein expression in gastric adenocarcinoma is
associated with disease progression and poor outcome. J Surg Oncol.
81:167–176. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng JY, Wang WZ, Li KZ, Guan WX and Yan
W: Effect of p27(KIP1) on cell cycle and apoptosis in gastric
cancer cells. World J Gastroenterol. 11:7072–7077. 2005.PubMed/NCBI
|
|
12
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lei PP, Zhang ZJ, Shen LJ, Li JY, Zou Q
and Zhang HX: Expression and hypermethylation of p27 kip1 in
hepatocarcinogenesis. World J Gastroenterol. 11:4587–4591.
2005.PubMed/NCBI
|
|
14
|
Qian X, Jin L, Kulig E and Lloyd RV: DNA
methylation regulates p27kip1 expression in rodent pituitary cell
lines. Am J Pathol. 153:1475–1482. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Serres MP, Kossatz U, Chi Y, Roberts JM,
Malek NP and Besson A: p27(Kip1) controls cytokinesis via the
regulation of citron kinase activation. J Clin Invest. 122:844–858.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jäkel H, Peschel I, Kunze C, Weinl C and
Hengst L: Regulation of p27 (Kip1) by mitogen-induced tyrosine
phosphorylation. Cell Cycle. 11:1910–1917. 2012.PubMed/NCBI
|
|
17
|
Zhang C, Ling Y, Xu Y, et al: The
silencing of RECK gene is associated with promoter hypermethylation
and poor survival in hepatocellular carcinoma. Int J Biol Sci.
8:451–458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling Y, Huang G, Fan L, et al: CpG island
methylator phenotype of cell-cycle regulators associated with TNM
stage and poor prognosis in patients with oesophageal squamous cell
carcinoma. J Clin Pathol. 64:246–251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim YK, Yu J, Han TS, et al: Functional
links between clustered microRNAs: suppression of cell-cycle
inhibitors by microRNA clusters in gastric cancer. Nucleic Acids
Res. 37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hitomi M, Yang K, Guo Y, Fretthold J,
Harwalkar J and Stacey DW: p27Kip1 and cyclin dependent kinase 2
regulate passage through the restriction point. Cell Cycle.
5:2281–2289. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vervoorts J and Lüscher B:
Post-translational regulation of the tumor suppressor p27(KIP1).
Cell Mol Life Sci. 65:3255–3264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hengst L and Reed SI: Inhibitors of the
Cip/Kip family. Curr Top Microbiol Immunol. 227:25–41.
1998.PubMed/NCBI
|
|
23
|
Blain SW and Massagué J: Breast cancer
banishes p27 from nucleus. Nat Med. 8:1076–1078. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sherr CJ and Roberts JM: Inhibitors of
mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149–1163.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Slingerland J and Pagano M: Regulation of
the cdk inhibitor p27 and its deregulation in cancer. J Cell
Physiol. 183:10–17. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tong Q, Zhang W, Jin S, Li S and Chen Z:
The relationship between p27(kip1) expression and the change of
radiosensitivity of esophageal carcinoma cells. Scand J
Gastroenterol. 46:173–176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sgambato A, Cittadini A, Faraglia B and
Weinstein IB: Multiple functions of p27(Kip1) and its alterations
in tumor cells: a review. J Cell Physiol. 183:18–27. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chu IM, Hengst L and Slingerland JM: The
Cdk inhibitor p27 in human cancer: prognostic potential and
relevance to anticancer therapy. Nat Rev Cancer. 8:253–267. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee J and Kim SS: The function of p27 KIP1
during tumor development. Exp Mol Med. 41:765–771. 2009. View Article : Google Scholar : PubMed/NCBI
|