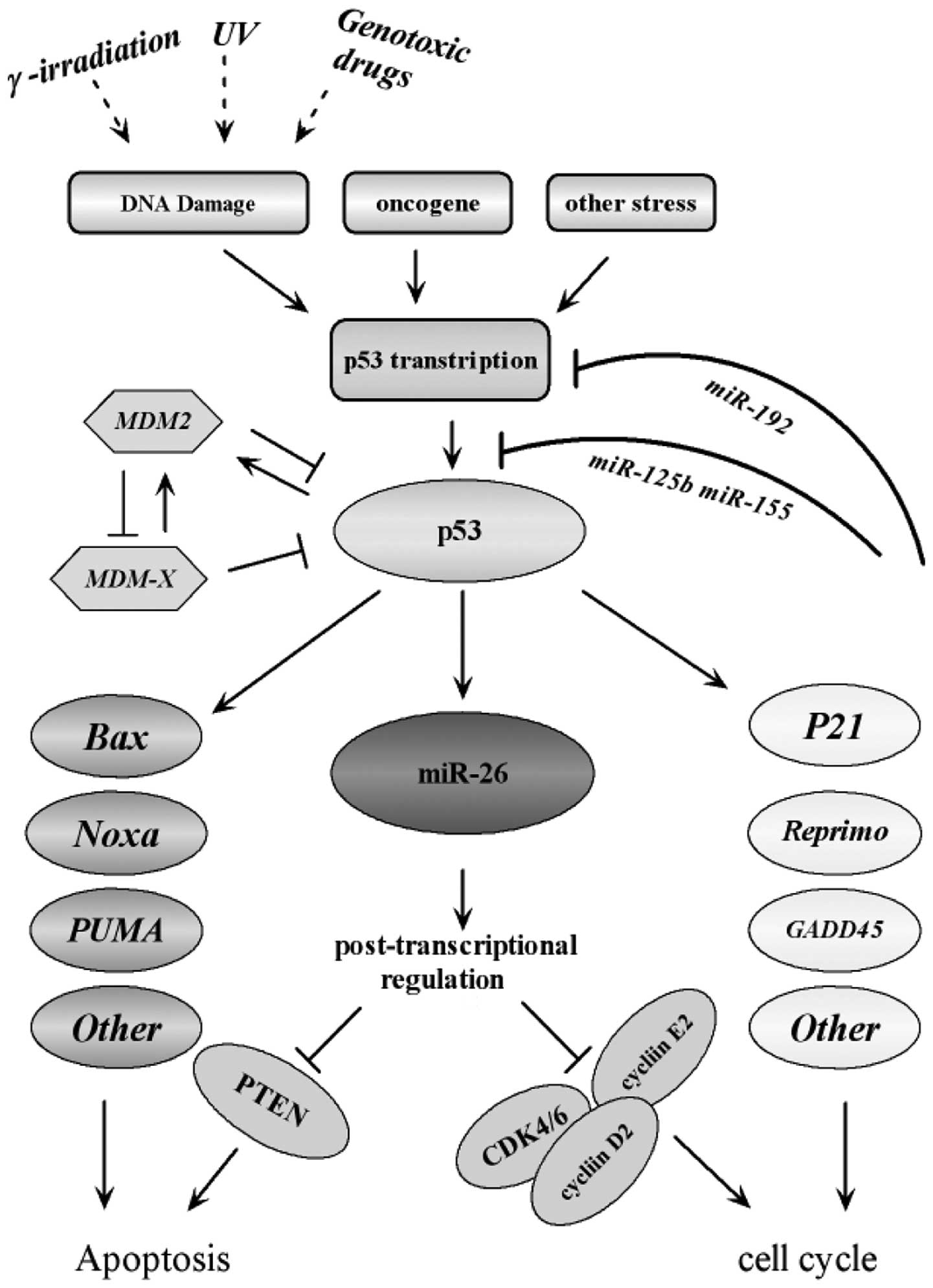
|
1
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
2
|
O’Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005.PubMed/NCBI
|
|
3
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qi LQ, Bart J, Tan LP, et al: Expression
of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial
atypia of the breast in relation to ductal carcinoma in situ and
invasive carcinoma. BMC Cancer. 9:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pineau P, Volinia S, McJunkin K, et al:
miR-221 overexpression contributes to liver tumorigenesis. Proc
Natl Acad Sci USA. 107:264–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Voorhoeve PM, Le Sage C, Schrier M, et al:
A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in
testicular germ cell tumors. Cell. 124:1169–1181. 2006. View Article : Google Scholar
|
|
7
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Chaerkady R, Beer MA, Mendell JT
and Pandey A: Identification of miR-21 targets in breast cancer
cells using a quantitative proteomic approach. Proteomics.
9:1374–1384. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamamichi N, Shimomura R, Inada KI, et al:
Locked nucleic acid in situ hybridization analysis of miR-21
expression during colorectal cancer development. Clin Cancer Res.
15:4009–4016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gaur A, Jewell DA, Liang Y, et al:
Characterization of microRNA expression levels and their biological
correlates in human cancer cell lines. Cancer Res. 67:2456–2468.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji JF, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szumiel I: Intrinsic radiation
sensitivity: cellular signaling is the key. Radiat Res.
169:249–258. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
An IS, An S, Kang SM, et al: Titrated
extract of Centella asiatica provides a UVB protective effect by
altering microRNA expression profiles in human dermal fibroblasts.
Intl J Mol Med. 30:1194–1202. 2012.
|
|
16
|
Chen CF, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaudhry MA: Real-time PCR analysis of
micro-RNA expression in ionizing radiation-treated cells. Cancer
Biother Radiopharm. 24:49–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo L, Huang ZX, Chen XW, et al:
Differential expression profiles of microRNAs in NIH3T3 cells in
response to UVB irradiation. Photochem Photobiol. 85:765–773. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(−Delta Delta C) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
20
|
Alexiou P, Maragkakis M, Papadopoulos GL,
Simmosis VA, Zhang L and Hatzigeorgiou AG: The DIANA-mirExTra web
server: from gene expression data to microRNA function. PLoS One.
5:e91712010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murakami Y, Yasuda T, Saigo K, et al:
Comprehensive analysis of microRNA expression patterns in
hepatocellular carcinoma and non-tumorous tissues. Oncogene.
25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Löbrich M and Jeggo PA: The impact of a
negligent G2/M checkpoint on genomic instability and cancer
induction. Nat Rev Cancer. 7:861–869. 2007.PubMed/NCBI
|
|
24
|
Spitz DR, Azzam EI, Li JJ and Gius D:
Metabolic oxidation/reduction reactions and cellular responses to
ionizing radiation: a unifying concept in stress response biology.
Cancer Metastasis Rev. 23:311–322. 2004. View Article : Google Scholar
|
|
25
|
Luo H, Zou J, Dong Z, Zeng Q, Wu D and Liu
L: Up-regulated miR-17 promotes cell proliferation, tumour growth
and cell cycle progression by targeting the RND3 tumour suppressor
gene in colorectal carcinoma. Biochem J. 442:311–321. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Creevey L, Ryan J, Harvey H, Bray IM,
Meehan M, Khan AR and Stallings RL: MicroRNA-497 increases
apoptosis in MYCN amplified neuroblastoma cells by targeting the
key cell cycle regulator WEE1. Mol Cancer. 12:232013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Varnholt H: The role of microRNAs in
primary liver cancer. Ann Hepat. 7:104–113. 2008.
|
|
29
|
He L, He XY, Lowe SW and Hannon GJ:
microRNAs join the p53 network--another piece in the
tumour-suppression puzzle. Nat Rev Cancer. 7:819–822. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rossi JJ: New hope for a microRNA therapy
for liver cancer. Cell. 137:990–992. 2009. View Article : Google Scholar : PubMed/NCBI
|