Introduction
microRNAs (miRNAs) are noncoding RNA molecules that
act as post-transcriptional regulators of specific messenger RNA
transcripts (mRNAs), resulting in targeted degradation and
suppression of gene expression. miRNAs play major roles in the
normal developmental processes (1,2), and
their dysregulation significantly contributes to various aspects of
tumorigenesis in the majority of types of cancer, negatively
regulating tumor suppressor genes and oncogenes (3). Although the regulator in control of
gene expression has been predicted to regulate ~30% of all human
genes, an increasing number of miRNAs assigned to their target
mRNAs with specific functions must be excluded.
Laryngeal squamous cell carcinoma (LSCC) is one of
the most common types of head and neck squamous carcinoma (HNSCC),
and HNSCC is the sixth most frequent type of cancer in the world
(4–6). At present, the main treatment
strategy for LSCC is surgery or total laryngectomy followed by
radiotherapy; however, the prognosis remains poor. Thus, to further
improve the survival and remission rates, the carcinogenic
mechanisms of LSCC requires further elucidation.
The majority of evidence suggested that miR-34a and
miR-34c is aberrantly expressed in the majority of types of cancer,
which lead to significant malignancies, including growth and
invasion alteration (7,8). However, the mechanism of miRNAs
functioning as tumor suppressor or oncogenes in the process of
tumor development is complex and varied, and may involve molecular
and network regulatory pathways. Thus, the association between
miR-34a and miR-34c and LSCC requires further investigation,
particularly, their functional targets. In the current study,
miR-34a and miR-34c were observed to be downregulated in eight
human laryngeal cancer tissues compared with the relative adjacent
normal tissue. Overexpression of miR-34a or miR-34c in the
laryngeal cancer cell line Hep-2 may suppress cell growth with MTT
and colony formation assay. Co-expression of miR-34a and miR-34c
may restrict cell movement with wound healing assay. Furthermore,
UDP-N-acetyl-α-D-galactosamine:polypeptide-N-acetylgalactosaminyltransferase
7 (GALNT7) was identified as a novel functional target of miR-34a
and miR-34c in the Hep-2 laryngeal carcinoma cell line.
Understanding the modulatory pathways of miR-34a and miR-34c may
aid in characterizing the progression of laryngeal carcinoma.
Materials and methods
Laryngeal carcinoma samples, cell lines,
transfection and RNA extraction
Human laryngeal carcinoma samples were obtained from
The First People’s Hospital of Jining (Jining, Shandong, China)
with patients’ informed consent and approval by the Ethics
Committee of of the First People’s Hospital of Jining. The Hep-2
laryngeal carcinoma cell line was cultured at 37°C with 5%
CO2 in RPMI-1640 media (Gibco-BRL, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (FBS). Hep-2 cells
were transfected with Lipofectamine 2000 reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA). Enriched small RNAs were
extracted using the mirVana™ miRNA Isolation kit (Ambion Inc.,
Austin, TX, USA). Total RNA was extracted using the TRIzol reagent
(Invitrogen Life Technologies).
Cell growth assay
Cells were seeded in 96-well plates at 8,000
cells/well and transfected the following day. An MTT assay was used
to determine relative cell growth 24, 48 and 72 h following
transfection. A MTT solution of 20 μl was added into 100 μl culture
media and cells were incubated for a further 4 h at 37°C. Next, the
optical density was measured at 570 nm (A570).
Colony formation assay
The cells were seeded into 12-well plates at a
density of 200 cells/well following transfection. The medium was
changed every three days. Approximately 10 days later, the majority
of the cell clones contained >50 cells. The clones were washed
with 1× phosphate-buffered saline (PBS) and stained with crystal
violet for ~5 min. Finally, images were captured of the clones and
clones were counted. The colony formation rate = (no. of
clones)/(no. of seeded cells) × 100%.
Wound healing assay
Cells were transfected and cultured to >90%
confluency in 24-well dishes. A sterile 200 μl pipette tip was used
to scratch three separate wounds through the cells moving
perpendicular to the line drawn in the step above. The medium was
removed and replaced with fresh medium. Images were captured
immediately above and below each line to ensure that the line
appeared in each image. Two straight lines were created close to
the border of the wound, and the width of the wound was defined as
the vertical dimension of the two lines. The wound border was
measured in nine random fields.
Quantitative polymerase chain reaction
(qPCR)
The stem-loop qPCR method (9) was used to detect the miR-34a and
miR-34c levels in Hep-2 cells and laryngeal carcinoma tissue. The
detection of GALNT7 mRNA levels was in accordance with a previous
study (10). The SYBR-Green Mix
Taq™ kit (Takara Bio, Inc., Shiga, Japan) was used to trace the
amplified DNA.
Western blot analysis
The Hep-2 cells were seeded into a 6-well plate at a
density of 3×105 cells/well and the cells were
transfected when the cell density reached ~80% confluency in the
second day. At 48 h following transfection, the cells were lysed
using RIPA buffer for 30 min at 4°C. The protein concentration was
measured by the BCA method and 20 μg protein was loaded into
SDS-PAGE for analysis. The first antibody was rabbit polyclonal
anti-human GALNT7 antibody (1:1,000; Abcam, Cambridge, MA, USA) and
rabbit monoclonal anti-human GAPDH antibody (1:1,000; Abcam). The
second antibody was goat anti-rabbit IgG conjugated with
horseradish peroxidase (1:1,000; Abcam). The bound antibodies were
detected with the use of ECL Plus Western Blotting Detection system
(GE Healthcare, Piscataway, NJ, USA) and the chemiluminescent
signals were detected with the use of high-performance
chemiluminescence film (GE Healthcare).
Luciferase reporter assay
Plasmids containing GALNT7-3′UTR and GALNT7-3′UTR
mutations were constructed via technical support from Dajin Co.
(Guangdong, China). 3′-UTR sequence of GALNT7 was predicted by six
miRNA target prediction algorithms to interact with miR-34a,
miR-34c and a mutated sequence of the 3′-UTR sequence was inserted
into pGL3 vectors (Promega, Madison, WI, USA). Following
transfection of miR-34a or miR-34c for 24 h, Hep-2 cells were
transfected with pGL3/GALNT7-3′UTR and pGL3/GALNT7-3′UTR mutant
plasmids. After 48 h, luciferase activity of Hep-2 cells was
measured 96 h after transfection using the Dual-Luciferase reporter
assay system (Promega).
Statistical analysis
All the data are shown as the mean ± SD, and the
difference between groups was determined by a two-tailed Student’s
t-test. P<0.05 and P<0.01 were considered to indicate a
statistically significant difference.
Results
Downregulation of miR-34a and miR-34c in
laryngeal carcinoma tissue
A previous study showed that miR-34c was
downregulated in the laryngeal cancer tissue compared with normal
laryngeal tissue (11) using miRNA
profiling. Cai et al (8)
reported that miR-34c is capable of suppressing growth and invasion
of human laryngeal carcinoma cells by targeting c-Met. However, the
precise expression level of miR-34a in laryngeal carcinoma tissue
remains unclear. Thus, qPCR between 8 pairs of laryngeal carcinoma
and adjacent non-tumor tissue was performed. As shown in Fig. 1, miR-34a and miR-34c expression was
decreased compared with the adjacent non-tumor tissue. These data
suggested that miR-34a and miR-34c were downregulated in laryngeal
carcinoma, which implied that the two of these miRNAs play tumor
suppressor roles in laryngeal carcinoma development.
miR-34a and miR-34c inhibit the growth of
Hep-2 cells in vitro
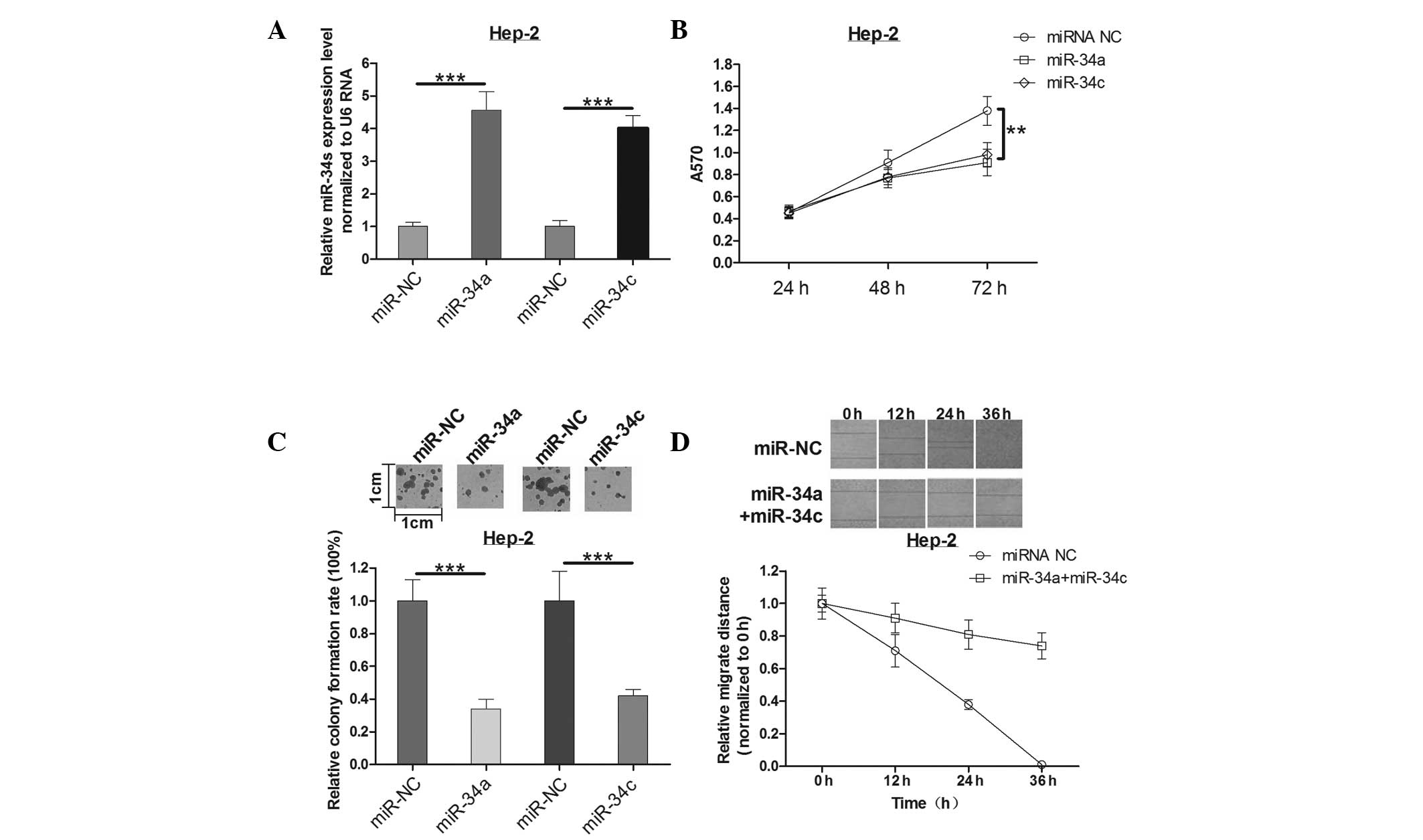
MTT and colony formation assays were used to
determine the effects of miR-34a and miR-34c in Hep-2 cells. The
expression of miR-34a and miR-34c mimics was identified by qPCR
(Fig. 2A). The mean A570 values in
each group (Fig. 2B) suggest that
transfection of Hep-2 cells with miR-34a or miR-34c caused
suppression of Hep-2 cell proliferation at 72 h post-infection when
compared with controls. As expected, miR-34a or miR-34c had a
similar effect on the colony formation ability of Hep-2 cells as
that on the cell viability. Overexpression of miR-34a reduced the
colony formation ability by ~64%, while miR-34c led to a 58%
reduction (Fig. 2C). This implied
that miR-34a and miR-34c suppresses cell growth significantly in
human laryngeal carcinoma.
Combination expression of miR-34a and
miR-34c induces decreased cell motility of Hep-2 cells
To further investigate the effect of miR-34a and
miR-34c on cell motility, a wound healing assay was measured
post-infection for 0, 12, 24 and 36 h in Hep-2 cells. Notably, a
slight decrease (not statistically significant, data not shown) in
cell motility of Hep-2 cells either transfected with miR-34a or
miR-34c alone was observed. However, as time increased, the
difference of migration distance was gradually increased in
co-expression of miR-34a and miR-34c groups compared with the
control group (Fig. 2D). These
results indicated that the combination of miR-34a and miR-34c plays
an important role in Hep-2 cell motility, which may be explained by
a synergism mechanism in biology. Thus, it was concluded that the
combination of miR-34a and miR-34c could decrease cell motility of
Hep-2 cells in human laryngeal carcinoma.
Identification of GALNT7 as a target of
miR-34a and miR-34c
As miR-34a and miR-34c have pivotal functions in
LSCC, the question as to how miRNA exerts its role in LSCC requires
investigation. In the current study, the TargetScan algorithm was
used to identify the target genes of miR-34a and miR-34c. A list of
functional genes were identified, including MYCN, Cyclin D1, B-cell
lymphoma 2 (BCL-2), E2F1, E2F3, CDC25A and c-Met, which have been
reported to be direct targets of miR-34a or miR-34c in various
types of cancers. Although a number of target genes of miR-34a and
miR-34c were validated, it has been proposed that a single miRNA
may target several genes and multiple miRNAs may target a single
gene in a comprehensive manner (12,13).
Thus, the functional target of miR-34a and miR-34c in laryngeal
carcinoma requires further investigation. Of the identified genes,
GALNT7 was selected, which is reported to play important roles in
the pathogenesis of cervical cancer and the behavior of melanoma
cells (10,14).
To confirm whether the 3′UTR of GALNT7 was a
functional target of miR-34a and miR-34c in LSCC, multiple miRNA
prediction algorithm screen methods were prepared. As shown in
Fig. 3A, the table suggests that
the six programs, TargetScan, miRDB, RNA22, microRNA.org, DIANA LAB
and PicTar, predicted GALNT7 as a potential target of miR-34a, and
each yielded a high score, respectively. Furthermore, four
programs, TargetScan, miRDB, RNA22 and microRNA.org, predicted that
miR-34c may directly target GALNT7. Next, whether miR-34a and
miR-34c directly target GALNT7 in laryngeal carcinoma was
investigated using a Luciferase reporter system. The alignment of
miR-34a and miR-34c with the GALNT7 3′UTR insert is presented in
Fig. 3B, including the seed
sequence 459–465. Thus, a mutated 3′UTR vector was constructed with
four nucleotides deleted in the seed sequence (Fig. 3C). When miR-34a was overexpressed
by miR-34a mimics, the Luciferase expression level was
significantly lower compared with the miR-negative control group.
However, the Luciferase intensity, with the mutated 3′UTR, was not
affected by miR-34a (Fig. 3D). The
same effect was observed when cells were transfected with miR-34c
in Hep-2 cells (Fig. 3E). Thus, it
was concluded that GALNT7 is a direct target of miR-34a and miR-34c
in Hep-2 cells.
miR-34a and miR-34c downregulate GALNT7
expression in Hep-2 cells
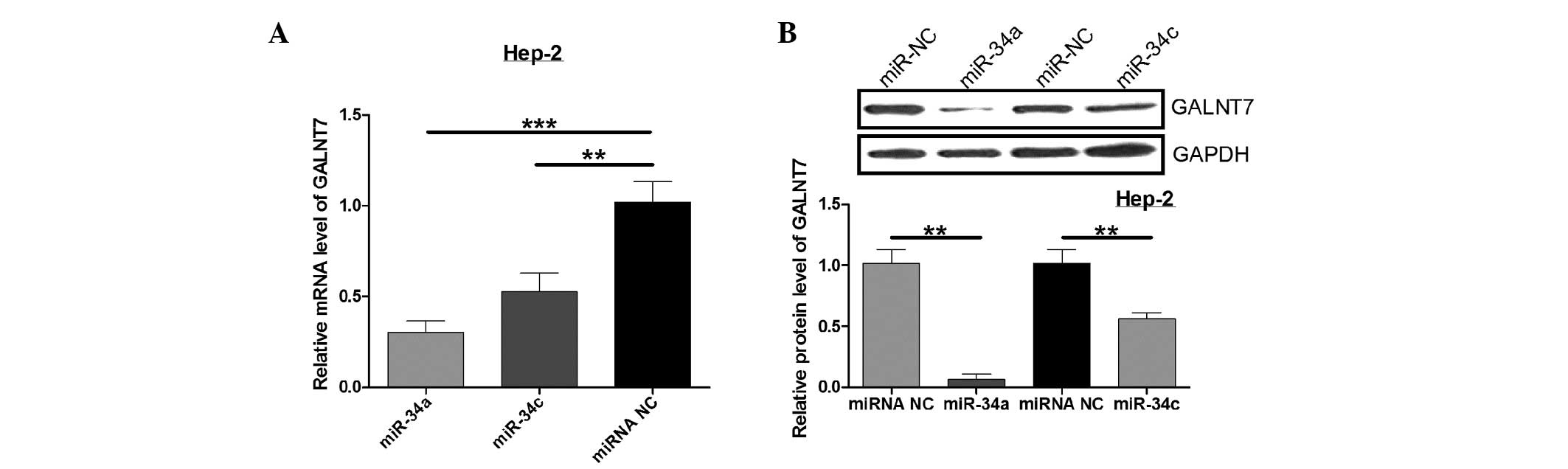
Next, qPCR and western blot analysis were performed
to determine the regulation of endogenous GALNT7 by miR-34a and
miR-34c. Compared with the control, miR-34a and miR-34c
significantly decrease GALNT7 mRNA expression, while the GALNT7
protein level was also significantly downregulated when Hep-2 cells
were cotransfected with miR-34a or miR-34c (P<0.05; Fig. 4). These results indicate that
miR-34a and miR-34c negatively regulate GALNT7 expression in the
Hep-2 laryngeal carcinoma cell line.
Discussion
Over the past number of years, a large number of
miRNAs have been reported to play important roles in regulating
cell biology processes, including tumorigenesis, and reduce its
target gene expression by mRNA degradation or mRNA cleavage
(15,16). The miRNAs have been confirmed as
tumorigenic and tumor suppressing genes, and include miR-17-92 and
let-7 (17,18).
Among these functional miRNAs, miR-34a and miR-34c
were firmly established to exhibit tumor suppressive effects in
multiple types of cancer, including leukemias, hepatocellular
carcinoma, pancreatic and colon cancer (19–22).
In the present study, miR-34a and miR-34c were observed to be
downregulated in human laryngeal carcinoma compared with the
adjacent normal tissue. Next, transfection of Hep-2 cells with
miR-34a and miR-34c was shown to significantly induce growth and
migration inhibition by MTT and colony formation assays. These
results confirmed the tumor suppressor role of miR-34a and miR-34c
in laryngeal carcinoma. Notably, when Hep-2 cells were transfected
with miR-34a and miR-34c alone, there were little changes with the
movement ability of Hep-2 cells using a wound healing assay.
However, co-expression of miR-34a and miR-34c significantly reduces
Hep-2 cell motility. Therefore, miR-34a and miR-34c were
hypothesized to affect the motility of Hep-2 cells and may
cooperate with each other by targeting cell metastasis-associated
genes. Thus, the functional targets of the miR-34 family in
laryngeal carcinoma require further investigation.
At present, miR-34a and miR-34c have multiple
experimentally validated targets involved with cellular
proliferation and apoptosis, including MYCN, BCL2, SIRT1, SFRP1,
CAMTA1, NOTCH1, JAG1, CCND1, CDK6 and E2F3 (21,23–29).
In the current study, the GALNT7 was identified as a novel target
using 6 algorithms, TargetScan, miroRNA.org, PicTar, miRDB, DIANA
LAB and RNA22. These were selected because greater specificity in
miRNA prediction may be attained using the consensus of multiple
algorithms. Notably, the Luciferase report assay confirmed that
miR-34a and miR-34c may directly target GALNT7-3′UTR with the seed
sequence of 459–465 sites. The western blot analysis and qPCR assay
demonstrated that the miR-34a and miR-34c also regulate endogenous
GALNT7 expression in Hep-2 cells in mRNA and protein levels.
GALNT7, a member of glycosyltransferases, catalyzes
the transfer of GalNAc to serine and threonine residues on target
proteins, thus, initiating mucin-type O-linked glycosylation in the
Golgi apparatus (30). Previous
studies have reported that glycosylation may play a role in
carcinogenesis and metastasis in a number of types of common
cancers (31). A recent study
revealed that ectopic expression of miR-30b/30d promoted the
metastatic behavior of melanoma cells, and in vitro and
in vivo data implicated GALNT7 inhibition as a key
contributor of the prometastatic effects of miR-30d. This
miR-30d/GALNT7 axis may be involved in the regulation of the
synthesis of the immunosuppressive cytokine, IL-10, and reduced
immune cell activation and recruitment (14). Other miRNAs were also found to be
upstream regulators of GALNT7, including miR-378 (32). In the current study, miR-34a and
miR-34c were identified to regulate GALNT7 expression and directly
target its 3′UTR in Hep-2 cells. To the best of our knowledge,
there may be many more miRNAs that regulate GALNT7 in tumor models,
thus, future studies aim to determine the number of miRNAs which
directly target GALNT7, and which may support the key contribution
to regulate GALNT7 repression in Hep-2 cells.
In conclusion, the expression of miR-34a and miR-34c
in eight pairs of laryngeal carcinoma were evaluated, and the two
were observed to be downregulated in laryngeal carcinoma tissue
compared with their adjacent normal tissue. Overexpression of
miR-34a and miR-34c alone suppressed cell growth of Hep-2 cells,
and co-expressed miR-34a and miR-34c are capable of reducing the
movement ability of Hep-2 cells. The phenotypic changes of Hep-2
cells by miR-34a and miR-34c potentially occur through inhibition
of GALNT7. These data suggest that the association between miR-34a
and miR-34c with its target novel GALNT7 may aid in understanding
the molecular mechanism of the tumorigenesis of laryngeal
carcinoma.
References
|
1
|
Breving K and Esquela-Kerscher A: The
complexities of microRNA regulation: mirandering around the rules.
Int J Biochem Cell Biol. 42:1316–1329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim VN: Small RNAs: classification,
biogenesis, and function. Mol Cells. 19:1–15. 2005.PubMed/NCBI
|
|
3
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
4
|
Mao L, Hong WK and Papadimitrakopoulou VA:
Focus on head and neck cancer. Cancer Cell. 5:311–316. 2004.
View Article : Google Scholar
|
|
5
|
Licitra L, Bernier J, Grandi C, et al:
Cancer of the larynx. Crit Rev Oncol Hematol. 47:65–80. 2003.
View Article : Google Scholar
|
|
6
|
Vokes EE, Weichselbaum RR, Lippman SM and
Hong WK: Head and neck cancer. N Engl J Med. 328:184–194. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tivnan A, Tracey L, Buckley PG, Alcock LC,
Davidoff AM and Stallings RL: MicroRNA-34a is a potent tumor
suppressor molecule in vivo in neuroblastoma. BMC Cancer.
11:332011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai KM, Bao XL, Kong XH, et al:
Hsa-miR-34c suppresses growth and invasion of human laryngeal
carcinoma cells via targeting c-Met. Int J Mol Med. 25:565–571.
2010.PubMed/NCBI
|
|
9
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng RQ, Wan HY, Li HF, Liu M, Li X and
Tang H: MicroRNA-214 suppresses growth and invasiveness of cervical
cancer cells by targeting
UDP-N-acetyl-α-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 7. J Biol Chem. 287:14301–14309.
2012.PubMed/NCBI
|
|
11
|
Liu M, Wu H, Liu T, et al: Regulation of
the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma.
Cell Res. 19:828–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peter ME: Targeting of mRNAs by multiple
miRNAs: the next step. Oncogene. 29:2161–2164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hobert O: miRNAs play a tune. Cell.
131:22–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gaziel-Sovran A, Segura MF, Di Micco R, et
al: miR-30b/30d regulation of GalNAc transferases enhances invasion
and immunosuppression during metastasis. Cancer Cell. 20:104–118.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayashita Y, Osada H, Tatematsu Y, et al:
A polycistronic microRNA cluster, miR-17-92, is overexpressed in
human lung cancers and enhances cell proliferation. Cancer Res.
65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji Q, Hao X, Zhang M, et al: MicroRNA
miR-34 inhibits human pancreatic cancer tumor-initiating cells.
PLoS One. 4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pang RT, Leung CO, Ye TM, et al:
MicroRNA-34a suppresses invasion through downregulation of Notch1
and Jagged1 in cervical carcinoma and choriocarcinoma cells.
Carcinogenesis. 31:1037–1044. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li N, Fu H, Tie Y, et al: miR-34a inhibits
migration and invasion by down-regulation of c-Met expression in
human hepatocellular carcinoma cells. Cancer Lett. 275:44–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Welch C, Chen Y and Stallings RL:
MicroRNA-34a functions as a potential tumor suppressor by inducing
apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei JS, Song YK, Durinck S, et al: The
MYCN oncogene is a direct target of miR-34a. Oncogene.
27:5204–5213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun F, Fu H, Liu Q, et al: Downregulation
of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett.
582:1564–1568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bommer GT, Gerin I, Feng Y, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu H, Brannon AR, Reddy AR, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: with
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luan S, Sun L and Huang F: MicroRNA-34a: a
novel tumor suppressor in p53-mutant glioma cell line U251. Arch
Med Res. 41:67–74. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ten Hagen KG, Fritz TA and Tabak LA: All
in the family: the UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferases. Glycobiology. 13:1R–16R.
2003.PubMed/NCBI
|
|
31
|
Casey RC, Oegema TR Jr, Skubitz KM,
Pambuccian SE, Grindle SM and Skubitz AP: Cell membrane
glycosylation mediates the adhesion, migration, and invasion of
ovarian carcinoma cells. Clin Exp Metastasis. 20:143–152. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kahai S, Lee SC, Lee DY, et al: MicroRNA
miR-378 regulates nephronectin expression modulating osteoblast
differentiation by targeting GalNT-7. PLoS One. 4:e75352009.
View Article : Google Scholar : PubMed/NCBI
|