Introduction
The methylation of deoxycytidine nucleotides
distributed in CpG islands is well known as an epigenetic
regulation mechanism for genomic function. Alteration of the DNA
methylation pattern has been identified to be closely associated
with carcinogenesis (1,2). Aberrant DNA hypermethylation at
promoter sequences leads to silencing of certain critical genes,
including the tumor suppressors, thus contributing to cancer
development (3,4). A number of studies have focused
extensively on the identification of DNA methylation patterns as
biomarkers for diagnosing cancer (5–7).
A global change in DNA methylation on a genome-wide
scale is able to be analyzed by DNA microarrays and high-throughput
DNA sequencing, which may not be accessible to a number of
institutions, particularly those in developing countries (8,9).
Additionally, DNA methylation at local genes may be analyzed by
methods based on the PCR approach, which is routinely used in every
laboratory that works with DNA (10). The majority of the PCR-based
methods use genomic DNA templates that have been treated with
sodium bisulfite. This chemical converts unmethylated cytosine, but
not methylated cytosine, to uracil residues (11). Specific primers were designed on
the basis of sequences that contain an adequate number of CpG
islands, thus the primers distinguish methylated from unmethylated
templates (12). The
methylation-specific PCR (MSP) is suitable and sensitive for the
detection of the CpG methylation status at any CpG islands
(10). Since the MSP primer sets
are specifically designed for the DNA whose composition was changed
following bisulfite conversion, a trace of unmodified DNA (native
DNA), due to uncompleted conversion in principle, is not amplified
during the PCR reactions (12,13).
Thereby, the majority of the control tests (positive or negative
controls) that are used to validate the MSP results for the DNA
methylation patterns in different types of cancers have used only
bisulfite-treated DNA and not untreated DNA extracted from
different cell lines (cancer or non-cancer) or from patient’s
specimens (14).
In the present study, the false-positive effect
caused by a trace of unmodified DNA on the MSP results was
reported, using previously published primer sets to identify the
methylation of the breast cancer 1 (BRCA1) and estrogen
receptor (ER) genes in Vietnamese females with breast
cancer. New primer sets and the set-up of additional standard
controls for eliminating false-positive results were designed in
order to improve the accurate positivity of the MSP method.
Materials and methods
Tissue samples
A total of 60 specimens of primary breast cancer
were collected from patients undergoing surgical resection at the
Department of Pathology, National Cancer Hospital K, Hanoi, the
largest cancer hospital in Vietnam. Informed consent was obtained
from patients in written form (ICF-ATF-FP-005-VN), and the study
was approved by the guidelines of the local ethical committee in
Vietnam (2205/QĐ-KHCN; Vietnam National University, Hanoi,
Vietnam).
Genomic DNA extraction and bisulfite
modification
Genomic DNA was extracted using a QIAamp DNA Mini
kit (Qiagen, Valencia, CA, USA) and treated with sodium bisulfite
using an EpiTect Bisulfite kit (Qiagen). During the modification,
the unmethylated cytosines of the genomic DNA were converted to
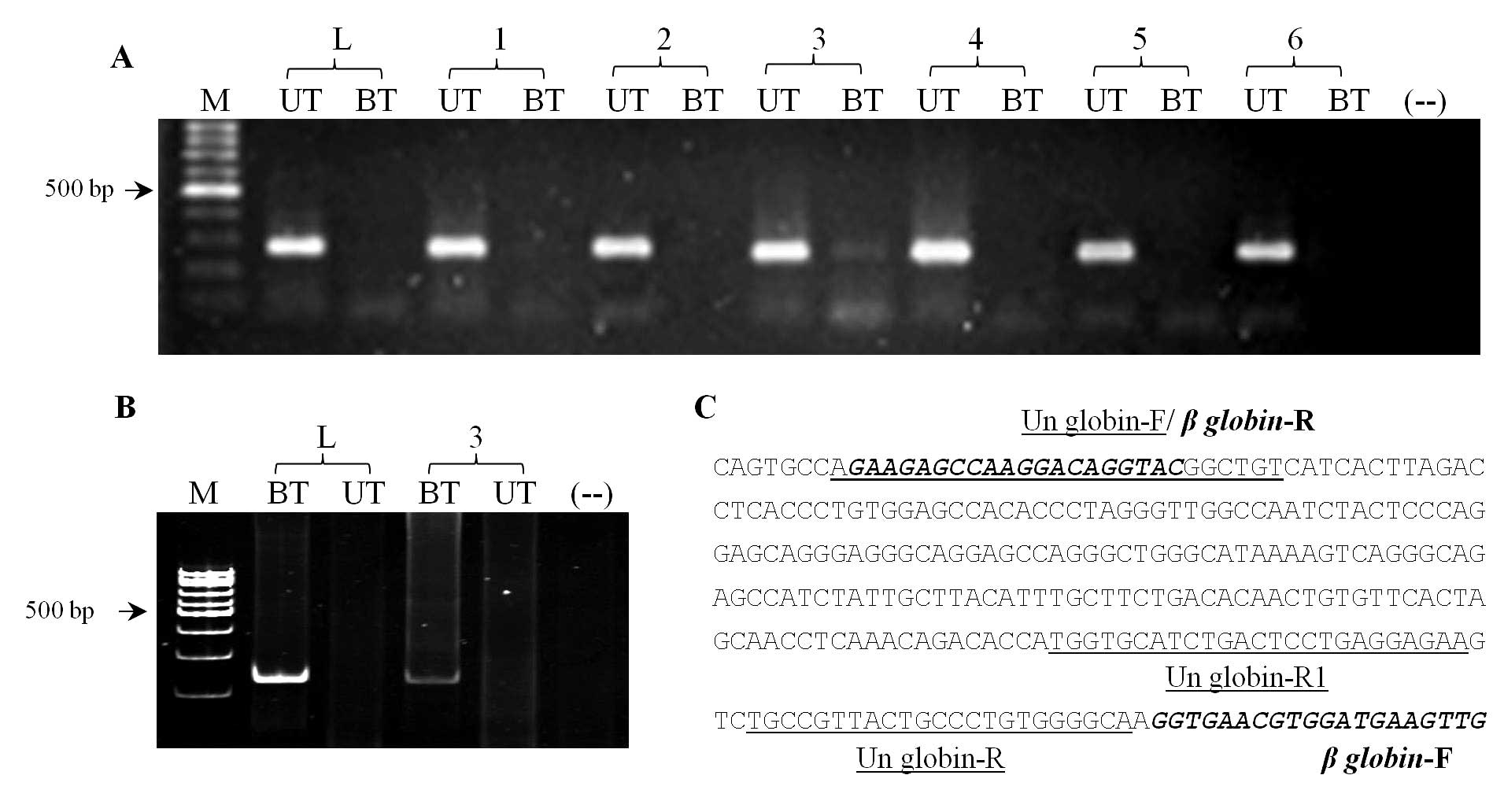
uracils, but the methylated cytosines remained unchanged (11). PCR that used β-globin-F/R primer
for the native DNA and Un-globin-F, -R and -R1 for treated DNA
(Fig. 1) was performed to
determine the efficiency of bisulfite conversion.
MSP
The methylation status of BRCA1 and ER
was evaluated using two primer sets for the MSP. The first set
included BRCA1 and ER primers that were originally
designed and reported by Esteller et al (15) and Lapidus et al (16), respectively. The second set
included new primers that were designed using the free online tool
from MethPrimer (http://www.urogene.org/methprimer/index1.html). The
primer sequences and amplicon lengths are shown in Table I. PCR amplification with the first
primer set was performed as described previously (15,16).
Bisulfite-treated DNA was subjected to a single round of PCR with
the new EM-F and ER4-R ER primers. Two rounds of PCR, the
first round with the BM-F/BRCA-R and the second round with
BM-F/BM-R primers, were performed to detect BRCA1
methylation. The 25 μl of the PCR reaction contained 0.3 μmol/l
primers, 100 μmol/l dNTPs, 2.0 U JumpStart Taq polymerase
(Sigma-Aldrich, St. Louis, MO, USA) and 1–2 μl of bisulfite-treated
DNA. The PCR conditions were follows: 94°C for 1 min, 40 cycles of
(94°C for 30 sec, 65°C for 10 sec and 72°C for 10 sec), and 72°C
for 5 min. The second 25 μl nested PCR reaction contained 1 μl of
the first PCR product and was performed with the conditions as
follows: 94°C for 1 min, 40 cycles of (94°C for 30 sec, 68°C for 10
sec and 72°C for 10 sec) and 72°C for 5 min. Two rounds of PCR were
performed with the new primer sets specific to unmethylated
BRCA1 and ER. The PCR products were subjected to
electrophoresis on a 12% polyacrylamide gel. All the PCR reactions
were replicated at least three times.
 | Table IMSP primers for analysis of
BRCA1 and ER gene methylation. |
Table I
MSP primers for analysis of
BRCA1 and ER gene methylation.
| Gene name | Primers | Sequence
(5′-3′) | Size, bp | First author
(ref.) |
|---|
| BRCA1 | BRCA-F |
TCGTGGTAACGGAAAAGCGC | 75 | Esteller et
al (15) |
| BRCA-R |
AAATCTCAACGAACTCACGCCG | | |
| BRCA-Un F |
TTGGTTTTTGTGGTAATGGAAAAGTG | 86 | Esteller et
al (15) |
| BRCA-Un R |
CAAAAAATCTCAACAAACTCACACCA | | |
| BM-F |
GGGTAGATTGGGTGGTTAATT | Round 1: 200 | Present study |
| BRCA-R |
AAATCTCAACGAACTCACGCCG | | |
| BM-F |
GGGTAGATTGGGTGGTTAATT | Round 2: 195 | Present study |
| BM-R |
TACACGAACTCACGCCGCGCAA | | |
| BU-F |
TTAATTTAGAGTTTTGAGAGAT | Round 1: 191 | Present study |
| BRCA-Un R |
CAAAAAATCTCAACAAACTCACACCA | | |
| BRCA-Un F |
TTGGTTTTTGTGGTAATGGAAAAGTG | Round 2: 76 | Present study |
| BU-R |
CAACAAACTCACACCACACAA | | |
| ER | ER4-F |
CGAGTTGGAGTTTTTGAATCGTTC | 151 | Lapidus et
al (16) |
| ER4-R |
CTACGCGTTAACGACGACCG | | |
| ER4-Un F |
ATGAGTTGGAGTTTTTGAATTGTTT | 158 | Lapidus et
al (16) |
| ER4-Un R |
ATAAACCTACACATTAACAACAACCA | | |
| EM-F |
GATACGGTTTGTATTTTGTTCG | 247 | Present study |
| ER4-R |
CTACGCGTTAACGACGACCG | | |
| EU4-F |
GTGGGGATATGGTTTGTATTTTGTTTG | Round 1: 258 | Present study |
| ER4-Un R |
ATAAACCTACACATTAACAACAACCA | | |
| ER4-Un F |
ATGAGTTGGAGTTTTTGAATTGTTT | Round 2: 154 | Present study |
| EU4-R |
ACCTACACATTAACAACAACCACAACA | | |
DNA that was extracted from the lymphocytes of the
healthy volunteers and then treated with bisulfite was used as a
positive control for BRCA1 and ER unmethylation. A
mixture of plasmid DNA containing methylated BRCA1 or
ER sequences and DNA extracted from normal lymphocytes was
used as a positive control for BRCA1 and ER
methylation. Water without a DNA template was included in each PCR
reaction as a control for any contamination. The methylation status
was confirmed by sequencing the cloned MSP products for a subset of
samples from each assay.
Results
The full conversion of genomic DNA that was
extracted from the primary breast cancer specimens was verified by
PCR with a β-globin primer set (Fig.
1). Using primers designed from native DNA sequences, the
majority of the PCR products were revealed to be amplified from
untreated and not bisulfite-converted DNA (Fig. 1A). By contrast, the PCR products
amplified by primers designed for unmethylated globin sequences
were detected from the bisulfite-treated DNA, but not the native
DNA (Fig. 1B). Negligible PCR
products were amplified from several treated DNA samples possibly
due to an incomplete conversion. Incompletely and completely
converted DNA were applied separately to MSP with the first
BRCA1 and ER primer sets. Unexpectedly, in several
samples, methylation of BRCA1 and ER was detected
from the incompletely modified DNA and not from the fully modified
DNA (Fig. 2A and C). It was likely
that the primer sets specifically designed for methylated
BRCA1 and ER wrongly amplified the native DNA
template that was not modified, and this template remained through
the bisulfite reaction.
 | Figure 2Representative analysis of MSP
products amplified by (A and B) the first primer sets of
BRCA1 and (C and D) ER. UnFT, incompletely converted
DNA; FT, completely converted DNA; mx, mixture of untreated and
completely converted DNA; UT, untreated DNA; BT, bisulfite-treated
DNA without verifying the efficiency of full conversion; S1, S3,
S11 and S47, different samples of breast cancer tissue; M, DNA
ladder; (−−), negative control without DNA templates; MSP,
methylation-specific PCR; BRCA1, breast cancer 1; ER,
estrogen receptor. |
To confirm this hypothesis, untreated genomic
(native) DNA was subjected to MSP with the first BRCA1 and
ER primer sets, which were appropriate for detecting
methylation (Fig. 2B and D). PCR
products were amplified from untreated genomic DNA and from a
mixture of untreated genomic DNA and completely modified DNA. In
addition, the PCR products were also amplified from untreated DNA
by using the primer sets specifically designed for unmethylated
ER and BRCA1 (data not shown). The analysis indicated
a false-positive result that was due to a trace of native DNA not
being converted, but being retained through bisulfite
treatment.
Based on the primer design strategies for the MSP
method, new primers for BRCA1 and ER were designed. A
number of these primers were used in combination with the published
primers (Table I). PCR was
performed in which either untreated or bisulfite-treated genomic
DNA was used as a template. The methylation of BRCA1 and
ER was detected from the treated DNA, but not from the
untreated DNA (Fig. 3), and
unmethylation of BRCA1 and ER was also detected from
the treated DNA, but not from the untreated DNA (data not shown).
This indicates the precision and specificity of the new primer sets
in distinguishing methylated from unmethylated and untreated
sequences.
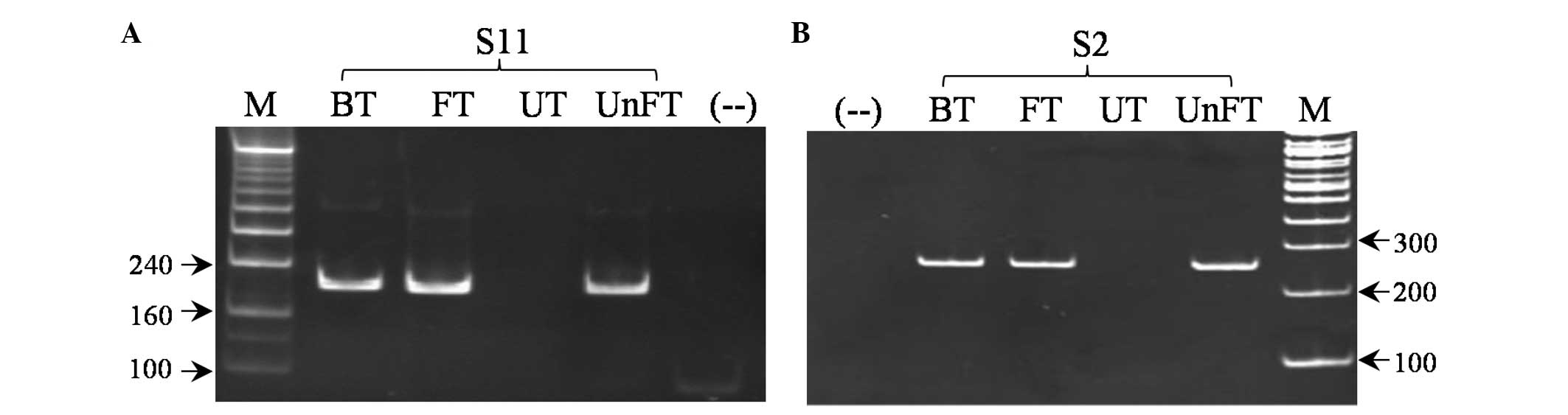 | Figure 3Representative analysis of MSP
products amplified by the new primer sets of (A) BRCA1 and
(B) ER. BT, bisulfite-treated DNA without verifying the
efficiency of full conversion; FT, completely converted DNA; UT,
untreated DNA; UnFT, incompletely converted DNA; S2 and S11, breast
cancer tissue samples; M, DNA ladder; (−−), negative control
without DNA templates; MSP, methylation-specific PCR; BRCA1,
breast cancer 1; ER, estrogen receptor. |
Genomic DNA extracted from 60 breast cancer
specimens was treated with bisulfite and subjected directly to MSP
without verifying the full conversion following treatment. The
number of cases of methylated BRCA1 and ER detected
by the first primer set was 25/60 and 47/60, respectively and that
detected by the second primer set was 7/60 and 21/60, respectively
(Figs. 4 and 5). When treated DNA whose full conversion
was examined through PCR with the β-globin primers and with the new
primers were used as templates for the two primer sets, the same
result (7/60 and 21/60 methylated DNA, respectively) was obtained.
Therefore, incompletely converted DNA resulted in 18 and 26 cases
of false-positive methylation of BRCA1 and ER,
respectively. Unmethylation of BRCA1 and ER, was
detected in the DNA of all 60 breast cancer patients.
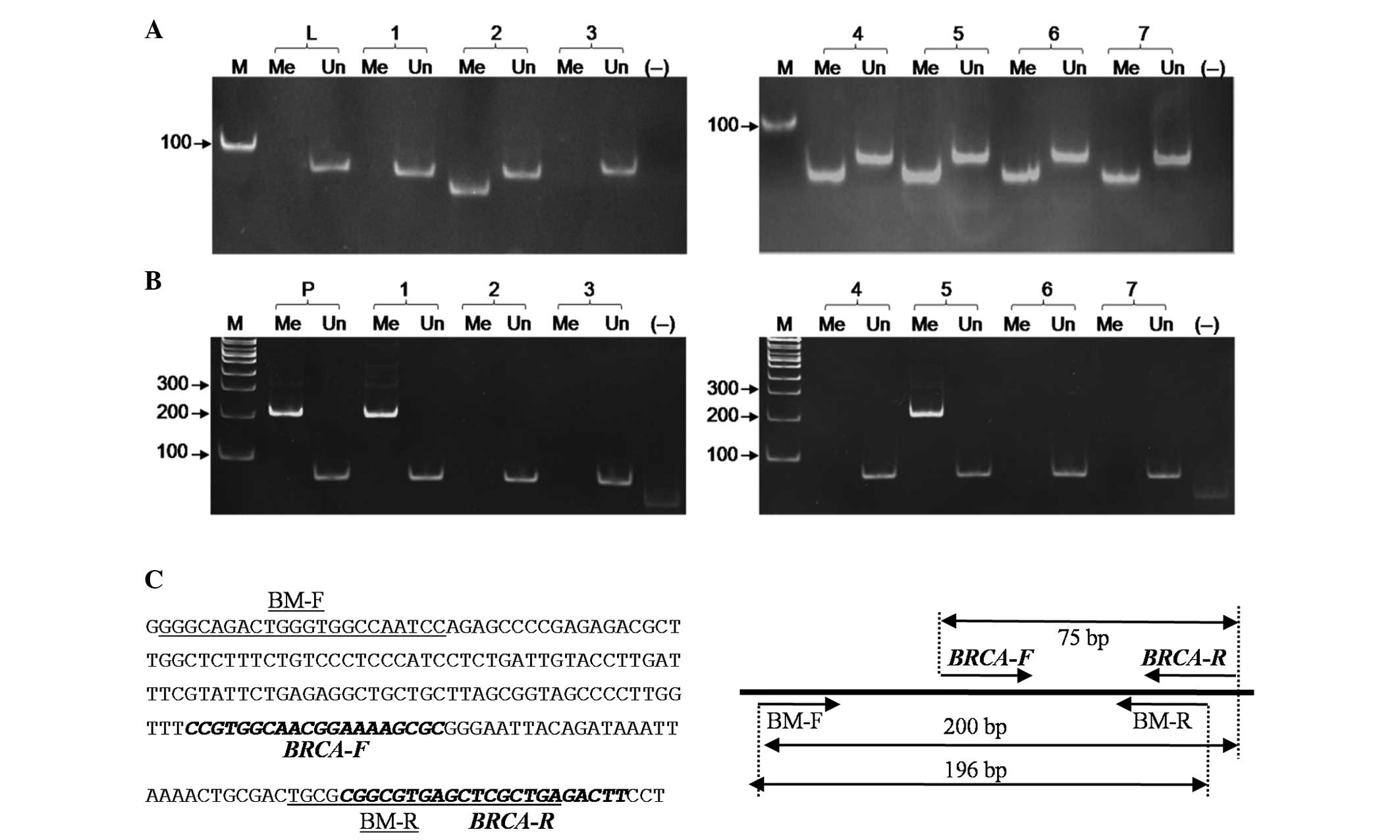 | Figure 4Representative analysis of
BRCA1-MSP products amplified by (A) the first primer set and
(B) the second primer set without verifying the efficiency of full
conversion of the DNA templates. 1–7, breast cancer samples; Me,
the presence of BRCA1 methylation; Un, the presence of BRCA1
unmethylation; L, lymphocytes of the healthy volunteer; P, plasmid
DNA, including BRCA1 methylated sequence mixed with DNA
extracted from lymphocytes of the healthy volunteer; M, DNA ladder;
(−−), negative control without DNA template. (C) The nucleotide
sequence of the 5′ region of BRCA1 (accession no.
NG-005905.2) and the location of the BRCA1-MSP primers listed in
Table I. BM indicated the primers
specific to methylated BRCA1. BRCA1, breast cancer; MSP,
methylation-specific PCR; F, forward; R, reverse. |
 | Figure 5Representative analysis of
ER-MSP products amplified by (A) the first primer set and
(B) the second primer set without verifying the efficiency of the
full conversion of the templates. 1–7, breast cancer samples; Me,
the presence of ER methylation; Un, the presence of
ER demethylation; L, lymphocytes of the healthy volunteer;
P, plasmid DNA, including ER methylated sequence mixed with
DNA extracted from lymphocytes of the healthy volunteer; M, DNA
ladder 100 bp; (−−), negative control without DNA template. (C) The
nucleotide sequence of the 5′ region of ER (accession no.
AL356311.6) and the location of the ER-MSP primers listed in
Table I. EM indicated the primers
specific to methylated ER. ER, estrogen receptor; MSP,
methylation-specific polymerase chain reaction. |
False priming events of the first primer set were
confirmed by cloning and sequencing the MSP products that were
amplified from untreated DNA templates (data not shown). The
nucleotide sequences amplified by the first primer set specific to
BRCA1 and ER methylation were revealed to be
identical to native sequences. In addition, three representatives
of the MSP products amplified from either incompletely converted or
fully converted DNA by the second BRCA1 and ER primer
set were also cloned and subsequently sequenced. The nucleotide
sequences revealed that all cytosine residues were converted to
thymidines in BRCA1 and ER unmethylated products, and
that all cytosines in the CpG sites remained as cytosines. The
cytosines that were not in the CpG sites were converted to
thymidines in the BRCA1 and ER methylated
products.
Discussion
Among the different types of markers that are
capable of distinguishing tumors from normal tissue, the DNA
methylation marker has become the most attractive due to its
sensitivity, specificity and applicability to a variety of clinical
specimens (12,17). MSP is the most widely used method
for the sensitive detection of DNA methylation (10). As this method requires common
equipment only, MSP may allow every laboratory to approach and
develop the DNA methylation marker for the purpose of diagnosis and
prognosis of cancers (5–7).
Using the MSP method, aberrant methylation at the 5′
region has been reported on a number of genes in different types of
cancer (18–20). The MSP result for one gene is
dependent on the analyzed sequence of the 5′ region and the type of
cancer. Thus, for a specific type of cancer, utilization of the
same panel of targeted genes and of the same region of the gene for
analysis of DNA methylation should be validated and reproduced to
increase the accuracy of DNA methylation markers in clinical
applications (14).
The BRCA1 and ER genes are the targets
of aberrant DNA methylation in breast tumors; thus, they are a
subject being studied extensively (21–24).
The BRCA1 gene encodes a multifunctional protein that is
involved in DNA repair, cell cycle control and chromatin remodeling
(25). The ER has a central
role in an important signaling pathway of mammary cells (26). The primers that were first designed
for analysis of BRCA1 (15)
and ER methylation (16) by
the MSP method have been subsequently applied to numerous studies
to detect the BRCA1 and ER methylation status in
different types of cancer, including breast cancer (27–30).
In the present study, these primers were also employed for the
analysis of the BRCA1 and ER methylation status in
females with breast cancer, using untreated and treated DNA as
templates. The results shown in Fig.
2 revealed that methylation of BRCA1 and ER was
detected in both types of DNA, and this indicates that these
primers did not discriminate between methylated and unconverted
sequences. The sequencing data confirmed that the first set of
BRCA1 and ER primers amplified the unconverted
sequences whose cytosine residues were retained. In replicated
experiments, the co-amplification of untreated sequences by only
the first primer set was confirmed by MSP and sequencing (data not
shown). The number of cases of methylated BRCA1 and
ER detected by the first primer set was 25/60 (41.7%) and
47/60 (78.3%), respectively, and that detected by the second primer
set was 7/60 (11.7%) and 21/60 (35.0%), respectively. A big
difference in the methylation levels (4-fold in BRCA1
methylation and 2-fold in ER methylation) was revealed
between the two primer sets. A significant difference in the DNA
methylation of the same gene(s) in one cancer type, for example,
eight-fold difference (5–40%) in the BRCA1 methylation in
breast cancer was reviewed by a number of different laboratories,
thus barriers in the performance of DNA methylation as cancer
biomarkers have been observed (14,31).
The results of the present study indicate that in numerous previous
studies, the significant difference in gene methylation analyzed by
the MSP in general, and in particular for BRCA1 and
ER methylation in breast cancer, was an overestimation that
resulted from the shortcomings of control tests for the accuracy of
MSP primers specific to the treated sequences only. An
overestimation may be prevented by the full conversion of the DNA
template, which may be verified through PCR with housekeeping gene
primers (Fig. 1) (32). However, such test controls are
required for each bisulfite-treated DNA template; thus, they are
laborious. The present study provided a simple control test that
eliminated the overestimation without verifying the full
conversion. Since the precision of the MSP primers was affirmed
through PCR with untreated DNA, a trace of uncompleted treated DNA
was not inferred from the MSP results (Fig. 3). Indeed, in the present study, the
BRCA1 and ER methylation levels detected by the new
primers, BM-F/BRCA-R and BM-F/BM-R, and EM-F and ER4-R (Table I) were four- and two-fold less than
that detected by the set of primers reported by Esteller et
al (15) and Lapidus et
al (16), respectively, and
much less than that detected by the first set of primers from
previous studies (26–56%), in which no control tests for the full
conversion through PCR were reported (22,33).
Thus, an accurate evaluation of the MSP primer specificity to
treated sequences only must avoid false-positive results.
MSP is a highly sensitive method; thus, different
approaches developed from or in combination with MSP, including
BS-MSP (Bisulfite conversion-Specific and Methylation-Specific
PCR), MEP (Methylation Enrichment Pyrosequencing) and ConLight MSP
(MSP, Conversion-specific hybridization and MethyLight), for
analysis of DNA methylation have been reported (34–36).
However, the precision of MSP primers specific to methylated
sequences only has not been verified in these methods to date.
Previous results have demonstrated that incomplete conversion may
typically be in the order of 2%, even when a commercial kit is used
(37). Considering the data of the
present study, it is proposed that all studies based on the MSP
approach should incorporate more steps in the control of the
specificity and precision of primers. By using untreated sequences
as the template for amplification with MSP primer sets,
overestimation of DNA methylation may be avoided. MSP is simple,
highly sensitive, extremely cost-effective and does not require any
special equipment; thus, MSP is the most widely used method for the
analysis of DNA methylation in the majority of laboratories,
particularly in those that are moderately equipped in developing
countries. The present study contributed to the standardization of
the MSP method and the validation of its precision. The study may
also promote the fast progression of the DNA methylation marker
towards its clinical application.
Acknowledgements
The present study was supported by the Ministry of
Science and Technology, Vietnam (nos. NAFOSTED106.06/2010.20 and
KC.04.05/11–15).
References
|
1
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dworkin AM, Huang TH and Toland AE:
Epigenetic alterations in the breast: Implications for breast
cancer detection, prognosis and treatment. Semin Cancer Biol.
19:165–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel A, Groopman JD and Umar A: DNA
methylation as a cancer-specific biomarker: from molecules to
populations. Ann NY Acad Sci. 983:286–297. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brooks J, Cairns P and Zeleniuch-Jacquotte
A: Promoter methylation and the detection of breast cancer. Cancer
Causes Control. 20:1539–1550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer - a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visvanathan K, Sukumar S and Davidson NE:
Epigenetic biomarkers and breast cancer: cause for optimism. Clin
Cancer Res. 12:6591–6593. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andrews J, Kennette W, Pilon J, Hodgson A,
Tuck AB, Chambers AF and Rodenhiser DI: Multi-platform whole-genome
microarray analyses refine the epigenetic signature of breast
cancer metastasis with gene expression and copy number. PLoS One.
5:e86652010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng S, Rubbi L, Jacobsen SE and
Pellegrini M: Determining DNA methylation profiles using
sequencing. Methods Mol Biol. 733:223–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kristensen LS and Hansen LL: PCR-based
methods for detecting single-locus DNA methylation biomarkers in
cancer diagnostics, prognostics, and response to treatment. Clin
Chem. 55:1471–1483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clark SJ, Harrison J, Paul CL and Frommer
M: High sensitivity mapping of methylated cytosines. Nucleic Acids
Res. 22:2990–2997. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hughes S and Jones JL: The use of multiple
displacement amplified DNA as a control for methylation specific
PCR, pyrosequencing, bisulfite sequencing and methylation-sensitive
restriction enzyme PCR. BMC Mol Biol. 8:912007. View Article : Google Scholar
|
|
14
|
Kagan J, Srivastava S, Barker PE, Belinsky
SA and Cairns P: Towards clinical application of methylated DNA
sequences as cancer biomarkers: A joint NCI’s EDRN and NIST
workshop on standards, methods, assays, reagents and tools. Cancer
Res. 67:4545–4549. 2007.PubMed/NCBI
|
|
15
|
Esteller M, Silva JM, Dominguez G, et al:
Promoter hypermethylation and BRCA1 inactivation in sporadic breast
and ovarian tumors. J Natl Cancer Inst. 92:564–569. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lapidus RG, Nass SJ, Butash KA, et al:
Mapping of ER gene CpG island methylation by methylation-specific
polymerase chain reaction. Cancer Res. 58:2515–2519.
1998.PubMed/NCBI
|
|
17
|
Cottrell SE and Laird PW: Sensitive
detection of DNA methylation. Ann NY Acad Sci. 983:120–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rabiau N, Thiam MO, Satih S, et al:
Methylation analysis of BRCA1, RASSF1, GSTP1 and EPHB2 promoters in
prostate biopsies according to different degrees of malignancy. In
Vivo. 23:387–391. 2009.PubMed/NCBI
|
|
19
|
Yamashita M, Toyota M, Suzuki H, et al:
DNA methylation of interferon regulatory factors in gastric cancer
and noncancerous gastric mucosae. Cancer Sci. 101:1708–1716. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou JL, Su HY, Chen LY, et al: Promoter
hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and
tumor suppressor, is associated with poor prognosis in human
ovarian cancer. Lab Invest. 90:414–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansmann T, Pliushch G, Leubner M, et al:
Constitutive promoter methylation of BRCA1 and RAD51C in patients
with familial ovarian cancer and early-onset sporadic breast
cancer. Hum Mol Genet. 21:4669–4679. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu NC, Huang YF, Yokoyama KK, Chu PY,
Chen FM and Hou MF: Methylation of BRCA1 promoter region is
associated with unfavorable prognosis in women with early-stage
breast cancer. PLoS One. 8:e562562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Archey WB, McEachern KA, Robson M, et al:
Increased CpG methylation of the estrogen receptor gene in
BRCA1-linked estrogen receptor-negative breast cancers. Oncogene.
21:7034–7041. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mann M, Cortez V and Vadlamudi RK:
Epigenetics of estrogen receptor signaling: Role in hormonal cancer
progression and therapy. Cancers (Basel). 3:1691–1707. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marquis ST, Rajan JV, Wynshaw-Boris A, et
al: The developmental pattern of BRCA1 expression implies a role in
differentiation of the breast and other tissues. Nat Genet.
11:17–26. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hayashi S, Niwa T and Yamaguchi Y:
Estrogen signaling pathway and its imaging in human breast cancer.
Cancer Sci. 100:1773–1778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tapia T, Smalley SV, Kohen P, et al:
Promoter hypermethylation of BRCA1 correlates with absence of
expression in hereditary breast cancer tumors. Epigenetics.
3:157–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YQ, Zhang JR, Li SD, He YY, Yang YX,
Liu XL and Wan XP: Aberrant methylation of breast and ovarian
cancer susceptibility gene 1 in chemosensitive human ovarian cancer
cells does not involve the phosphatidylinositol 3′-kinase-Akt
pathway. Cancer Sci. 101:1618–1623. 2010.PubMed/NCBI
|
|
29
|
Harder J, Engelstaedter V, Usadel H, et
al: CpG-island methylation of the ER promoter in colorectal cancer:
analysis of micrometastases in lymph nodes from UICC stage I and II
patients. Br J Cancer. 100:360–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei M, Xu J, Dignam J, et al: Estrogen
receptor alpha, BRCA1, and FANCF promoter methylation occur in
distinct subsets of sporadic breast cancers. Breast Cancer Res
Treat. 111:113–120. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Senturk E, Cohen S, Dottino PR and
Martignetti JA: A critical re-appraisal of BRCA1 methylation
studies in ovarian cancer. Gynecol Oncol. 119:376–383. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wilcox CB, Baysal BE, Gallion HH, Strange
MA and DeLoia JA: High-resolution methylation analysis of the BRCA1
promoter in ovarian tumors. Cancer Genet Cytogenet. 159:114–122.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Zhou J, Xu Y, et al: BRCA1
promoter methylation associated with poor survival in Chinese
patients with sporadic breast cancer. Cancer Sci. 100:1663–1667.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sasaki M, Anast J, Bassett W, Kawakami T,
Sakuragi N and Dahiya R: Bisulfite conversion-specific and
methylation-specific PCR: a sensitive technique for accurate
evaluation of CpG methylation. Biochem Biophys Res Commun.
309:305–309. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shaw RJ, Akufo-Tetteh EK, Risk JM, Field
JK and Liloglou T: Methylation enrichment pyrosequencing: combining
the specificity of MSP with validation by pyrosequencing. Nucleic
Acids Res. 34:e782006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rand K, Qu W, Ho T, Clark SJ and Molloy P:
Conversion-specific detection of DNA methylation using real-time
polymerase chain reaction (ConLight-MSP) to avoid false positives.
Methods. 27:114–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warnecke PM, Stirzaker C, Song J, Grunau
C, Melki JR and Clark SJ: Identification and resolution of
artifacts in bisulfite sequencing. Methods. 27:101–107. 2002.
View Article : Google Scholar : PubMed/NCBI
|