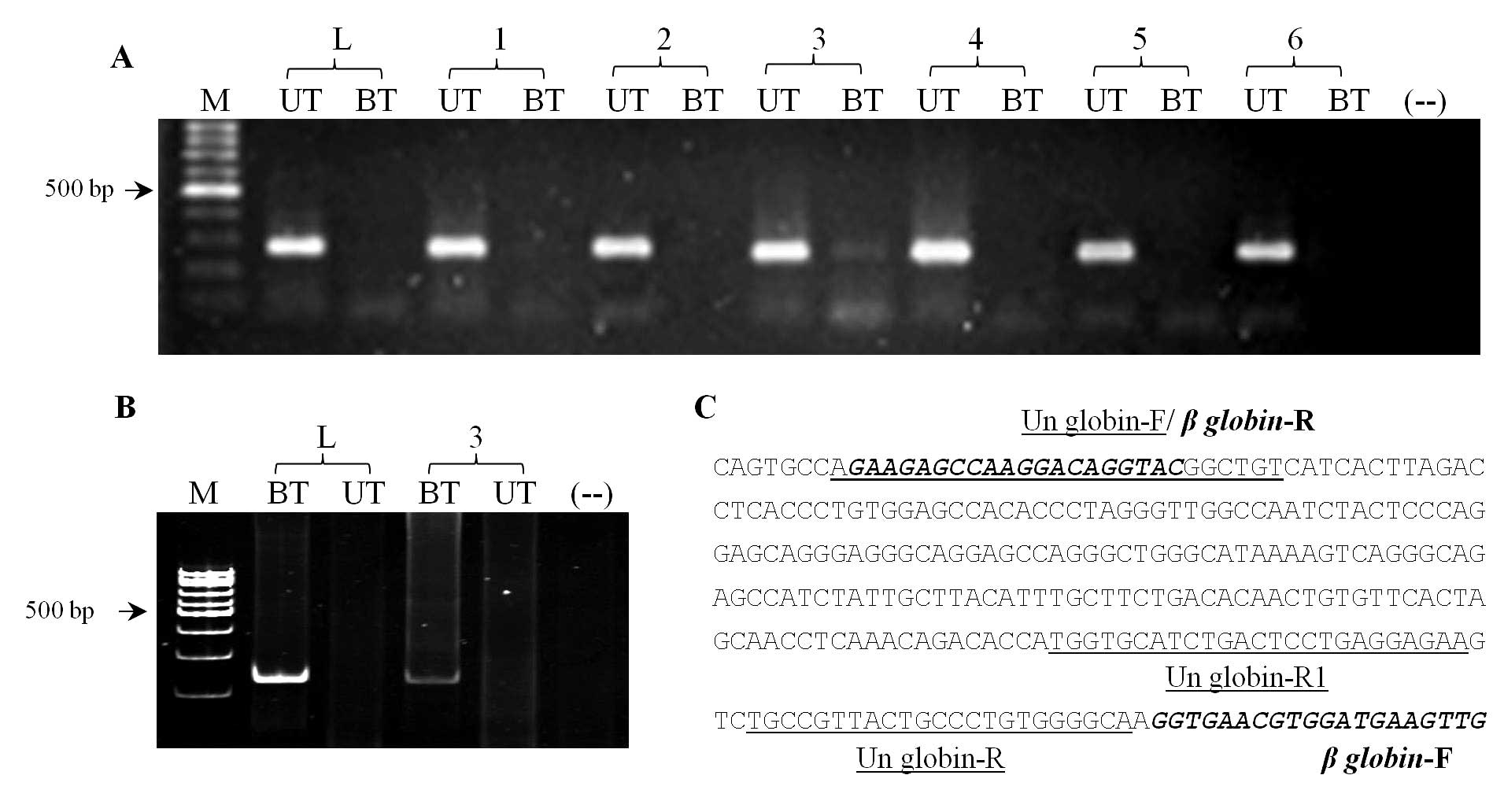
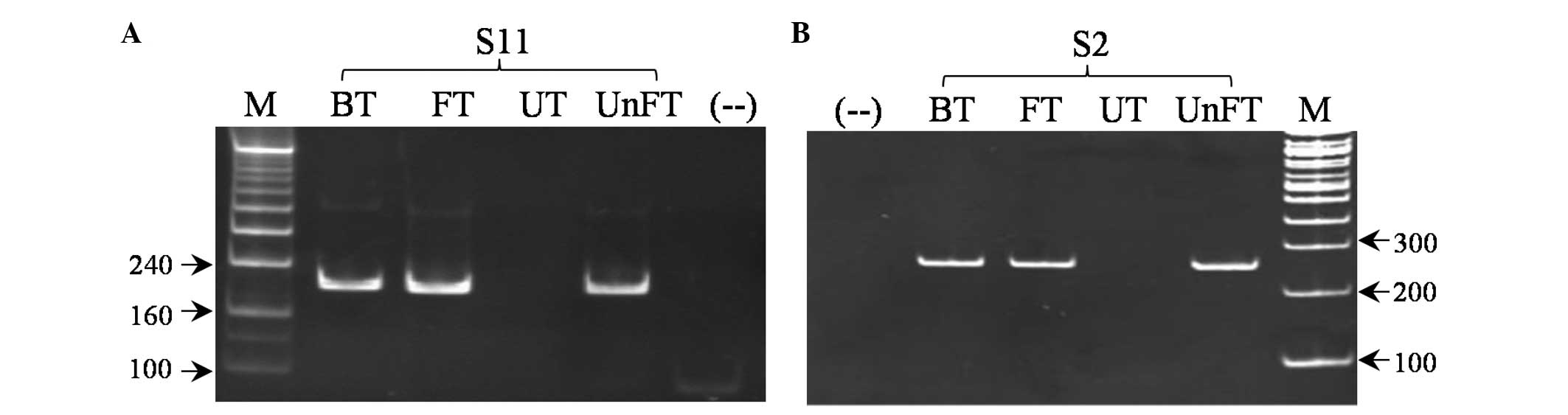
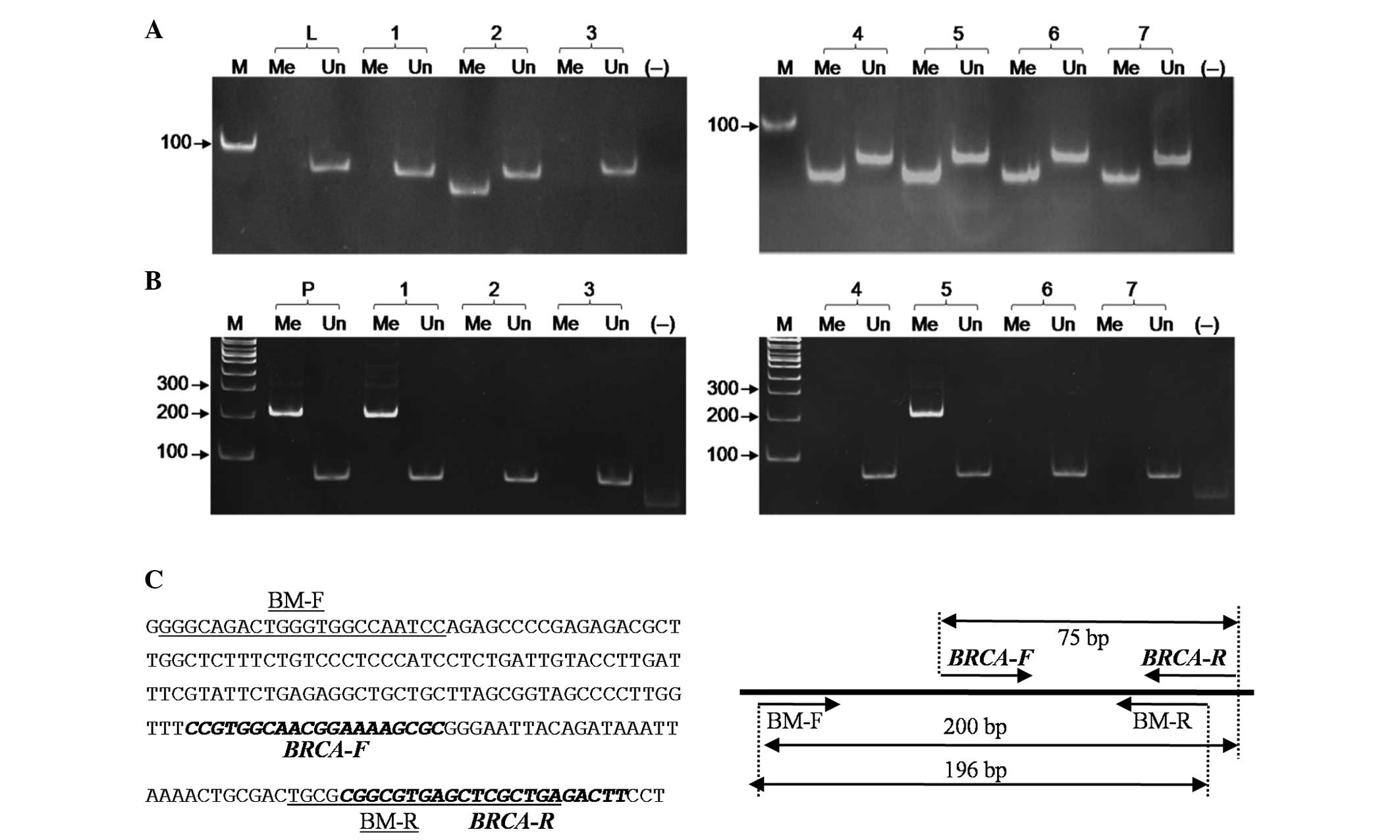
|
1
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dworkin AM, Huang TH and Toland AE:
Epigenetic alterations in the breast: Implications for breast
cancer detection, prognosis and treatment. Semin Cancer Biol.
19:165–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel A, Groopman JD and Umar A: DNA
methylation as a cancer-specific biomarker: from molecules to
populations. Ann NY Acad Sci. 983:286–297. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brooks J, Cairns P and Zeleniuch-Jacquotte
A: Promoter methylation and the detection of breast cancer. Cancer
Causes Control. 20:1539–1550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer - a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visvanathan K, Sukumar S and Davidson NE:
Epigenetic biomarkers and breast cancer: cause for optimism. Clin
Cancer Res. 12:6591–6593. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andrews J, Kennette W, Pilon J, Hodgson A,
Tuck AB, Chambers AF and Rodenhiser DI: Multi-platform whole-genome
microarray analyses refine the epigenetic signature of breast
cancer metastasis with gene expression and copy number. PLoS One.
5:e86652010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng S, Rubbi L, Jacobsen SE and
Pellegrini M: Determining DNA methylation profiles using
sequencing. Methods Mol Biol. 733:223–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kristensen LS and Hansen LL: PCR-based
methods for detecting single-locus DNA methylation biomarkers in
cancer diagnostics, prognostics, and response to treatment. Clin
Chem. 55:1471–1483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clark SJ, Harrison J, Paul CL and Frommer
M: High sensitivity mapping of methylated cytosines. Nucleic Acids
Res. 22:2990–2997. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hughes S and Jones JL: The use of multiple
displacement amplified DNA as a control for methylation specific
PCR, pyrosequencing, bisulfite sequencing and methylation-sensitive
restriction enzyme PCR. BMC Mol Biol. 8:912007. View Article : Google Scholar
|
|
14
|
Kagan J, Srivastava S, Barker PE, Belinsky
SA and Cairns P: Towards clinical application of methylated DNA
sequences as cancer biomarkers: A joint NCI’s EDRN and NIST
workshop on standards, methods, assays, reagents and tools. Cancer
Res. 67:4545–4549. 2007.PubMed/NCBI
|
|
15
|
Esteller M, Silva JM, Dominguez G, et al:
Promoter hypermethylation and BRCA1 inactivation in sporadic breast
and ovarian tumors. J Natl Cancer Inst. 92:564–569. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lapidus RG, Nass SJ, Butash KA, et al:
Mapping of ER gene CpG island methylation by methylation-specific
polymerase chain reaction. Cancer Res. 58:2515–2519.
1998.PubMed/NCBI
|
|
17
|
Cottrell SE and Laird PW: Sensitive
detection of DNA methylation. Ann NY Acad Sci. 983:120–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rabiau N, Thiam MO, Satih S, et al:
Methylation analysis of BRCA1, RASSF1, GSTP1 and EPHB2 promoters in
prostate biopsies according to different degrees of malignancy. In
Vivo. 23:387–391. 2009.PubMed/NCBI
|
|
19
|
Yamashita M, Toyota M, Suzuki H, et al:
DNA methylation of interferon regulatory factors in gastric cancer
and noncancerous gastric mucosae. Cancer Sci. 101:1708–1716. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou JL, Su HY, Chen LY, et al: Promoter
hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and
tumor suppressor, is associated with poor prognosis in human
ovarian cancer. Lab Invest. 90:414–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansmann T, Pliushch G, Leubner M, et al:
Constitutive promoter methylation of BRCA1 and RAD51C in patients
with familial ovarian cancer and early-onset sporadic breast
cancer. Hum Mol Genet. 21:4669–4679. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu NC, Huang YF, Yokoyama KK, Chu PY,
Chen FM and Hou MF: Methylation of BRCA1 promoter region is
associated with unfavorable prognosis in women with early-stage
breast cancer. PLoS One. 8:e562562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Archey WB, McEachern KA, Robson M, et al:
Increased CpG methylation of the estrogen receptor gene in
BRCA1-linked estrogen receptor-negative breast cancers. Oncogene.
21:7034–7041. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mann M, Cortez V and Vadlamudi RK:
Epigenetics of estrogen receptor signaling: Role in hormonal cancer
progression and therapy. Cancers (Basel). 3:1691–1707. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marquis ST, Rajan JV, Wynshaw-Boris A, et
al: The developmental pattern of BRCA1 expression implies a role in
differentiation of the breast and other tissues. Nat Genet.
11:17–26. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hayashi S, Niwa T and Yamaguchi Y:
Estrogen signaling pathway and its imaging in human breast cancer.
Cancer Sci. 100:1773–1778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tapia T, Smalley SV, Kohen P, et al:
Promoter hypermethylation of BRCA1 correlates with absence of
expression in hereditary breast cancer tumors. Epigenetics.
3:157–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YQ, Zhang JR, Li SD, He YY, Yang YX,
Liu XL and Wan XP: Aberrant methylation of breast and ovarian
cancer susceptibility gene 1 in chemosensitive human ovarian cancer
cells does not involve the phosphatidylinositol 3′-kinase-Akt
pathway. Cancer Sci. 101:1618–1623. 2010.PubMed/NCBI
|
|
29
|
Harder J, Engelstaedter V, Usadel H, et
al: CpG-island methylation of the ER promoter in colorectal cancer:
analysis of micrometastases in lymph nodes from UICC stage I and II
patients. Br J Cancer. 100:360–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei M, Xu J, Dignam J, et al: Estrogen
receptor alpha, BRCA1, and FANCF promoter methylation occur in
distinct subsets of sporadic breast cancers. Breast Cancer Res
Treat. 111:113–120. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Senturk E, Cohen S, Dottino PR and
Martignetti JA: A critical re-appraisal of BRCA1 methylation
studies in ovarian cancer. Gynecol Oncol. 119:376–383. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wilcox CB, Baysal BE, Gallion HH, Strange
MA and DeLoia JA: High-resolution methylation analysis of the BRCA1
promoter in ovarian tumors. Cancer Genet Cytogenet. 159:114–122.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Zhou J, Xu Y, et al: BRCA1
promoter methylation associated with poor survival in Chinese
patients with sporadic breast cancer. Cancer Sci. 100:1663–1667.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sasaki M, Anast J, Bassett W, Kawakami T,
Sakuragi N and Dahiya R: Bisulfite conversion-specific and
methylation-specific PCR: a sensitive technique for accurate
evaluation of CpG methylation. Biochem Biophys Res Commun.
309:305–309. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shaw RJ, Akufo-Tetteh EK, Risk JM, Field
JK and Liloglou T: Methylation enrichment pyrosequencing: combining
the specificity of MSP with validation by pyrosequencing. Nucleic
Acids Res. 34:e782006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rand K, Qu W, Ho T, Clark SJ and Molloy P:
Conversion-specific detection of DNA methylation using real-time
polymerase chain reaction (ConLight-MSP) to avoid false positives.
Methods. 27:114–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warnecke PM, Stirzaker C, Song J, Grunau
C, Melki JR and Clark SJ: Identification and resolution of
artifacts in bisulfite sequencing. Methods. 27:101–107. 2002.
View Article : Google Scholar : PubMed/NCBI
|