Introduction
General anesthetics act on the central nervous
system, mainly the brain and the spinal cord, to induce general
anesthesia. General anesthesia has commonly been used in the
surgical arena for ~150 years, and its efficacy and safety have
been assured by clinical outcomes (1–3).
However, little is known regarding its comprehensive influence at
the genomic and molecular levels, which is not reflected by
mortality and morbidity. Sevoflurane is an inhalational anesthetic
frequently used in Japan (4). It
has low solubility in blood and tissues, thus there is usually a
rapid recovery from anesthesia. Propofol is the most commonly used
intravenous anesthetic and is administered as an alkylphenol
formulated in a lipid emulsion (5,6).
Propofol provides rapid and smooth induction of anesthesia and
exhibits rapid clearance from the body.
Due to rapid progress in the field of genomics,
microarray technology allows the analysis of genome-wide changes in
gene expression in cells and tissues. Several studies regarding
general anesthetics, including volatile and intravenous
anesthetics, have been performed and they have found that general
anesthetics affect gene expression in the rat central nervous
system. To elucidate cellular function, exhaustive analyses of the
expression of genes (genomics), proteins (proteomics) and
metabolites (metabolomics) have been performed in the brain of
rats, which is the main target of general anesthetics. In a
previous genomic study, the inhalation anesthetic sevoflurane
affected the expression of 1.5% of genes in various rat organs, as
detected using a microarray analysis (7). General anesthesia altered the
expression levels of genes involved in circadian rhythms;
persistent suppression of several circadian genes was identified
following treatment of rats with the inhalation anesthetic
sevoflurane (8) and the
intravenous anesthetics propofol and dexmedetomidine (9). In a proteomics study, the protein
expression in the rat brain was regulated differentially according
to the anesthetic agent used: The inhalation anesthetic sevoflurane
or the intravenous anesthetic propofol (10). In a metabolomics study, the
inhalation anesthetic isoflurane and the intravenous anesthetic
propofol exerted differential effects on the brain metabolism of
rats (11). Based on these
previous studies, it was hypothesized that gene expression, protein
expression and metabolism are different in the rat brain under
inhalation anesthesia and intravenous anesthesia.
MicroRNAs (miRNAs) are a class of highly conserved,
short, single-stranded, non-coding RNA molecules. miRNAs are 20–24
nucleotides in length and are encoded by the genome. miRNAs bind to
imperfectly complementary sequences in the 3′-untranslated region
(3′-UTR) of target messenger RNAs (mRNAs) and regulate gene
expression at the post-transcriptional level; that is, inhibiting
mRNA translation and/or inducing mRNA degeneration (12). A study has shown that miRNAs
negatively regulate >30% of mammalian gene expression (13). Furthermore, single miRNAs may bind
with hundreds of unique mRNAs, thus altering the expression of
multiple cellular pathways (14).
miRNAs are important in the central nervous system as they affect
various post-transcriptional regulation mechanisms, including
cytoskeletal/neuronal growth, cellular metabolism, biosynthesis,
stress/death responses, signal transduction and synaptic plasticity
(15–18). There is evidence that miRNAs have
specific temporal and spatial expression patterns in the mammalian
brain (19). However, the gene
regulatory networks of the majority of miRNAs in the brain have not
yet been fully elucidated.
General anesthetics were developed empirically in
clinical settings. This type of anesthetic is essential in
producing analgesia, immobility, unconsciousness and memory
blockade (amnesia). As certain patients experience unexpected
recall of events during surgery, intraoperative amnesia has been a
desired endpoint of general anesthesia from the perspectives of the
patient and the anesthesiologist. Understanding the mechanisms
underlying amnesia is a matter of particular importance. The
hippocampus is considered to be an important organ in cognitive
function and episodic memory processes and, as reported by Pan
et al (20), administration
of the inhalation anesthetic sevoflurane resulted in altered gene
expression levels of mRNAs that affect memory function in the rat
hippocampus.
To the best of our knowledge, no published studies
have addressed the association between sevoflurane anesthesia and
the expression of miRNAs in the rat hippocampus. It was
hypothesized that the miRNAs that influence mRNA expression also
alter gene expression levels in the hippocampus under sevoflurane
anesthesia. In addition, the inhalation anesthetic sevoflurane and
the intravenous anesthetic propofol were predicted to induce
different effects on the expression levels of miRNAs in the rat
brain. Thus, the aim of the present study was to compare the miRNA
expression profiles in the rat hippocampus in response to
anesthesia with representative volatile (sevoflurane) and
intravenous (propofol) anesthetics. To the best of our knowledge,
this is the first attempt to perform an exhaustive profiling
analysis of miRNA expression in the hippocampus under general
anesthesia.
Materials and methods
Experimental protocol
A previous study model (21–23)
on miRNA data acquisition, processing and statistical analysis was
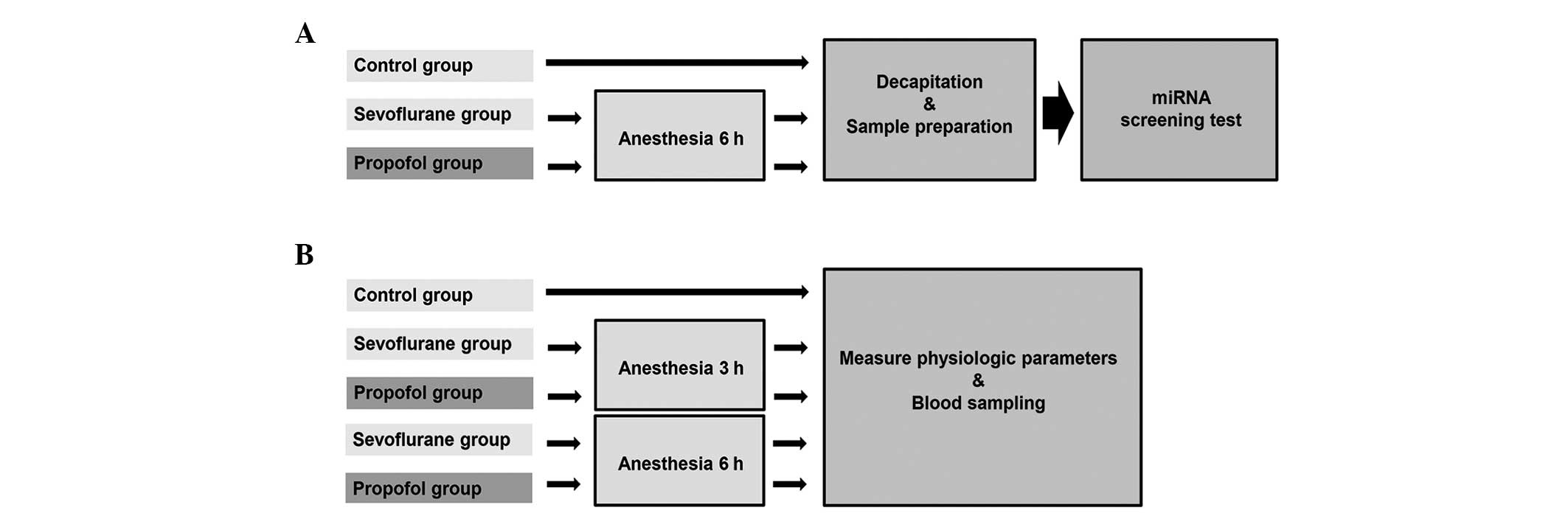
used. A flow chart of the experimental protocol is shown in
Fig. 1.
Animals
Animal protocols, including the method for
euthanasia, were approved by the Animal Experimental Ethics Review
Committee of Nippon Medical School (Tokyo, Japan; approval no.:
24-005). All procedures followed the guidelines of the
International Association for the Study of Pain (24). Specific pathogen-free male Wistar
rats aged six weeks old, (Saitama Experimental Animal Supply Co.,
Saitama, Japan) were purchased and acclimated under
temperature-controlled conditions on a 12-h light/dark cycle,
starting at 06:00 and 18:00, respectively, for a week prior to the
commencement of experiments. All rats received food and water ad
libitum, and the average starting weight of the rats was 250±10
g.
Procedures for administration of
anesthetic and termination
During the experiment, each rat was housed in a
plastic box (45×32×23 cm), which was large enough for the rat to
move and breathe fresh air. The rats were minimally handled in
order to reduce stress. The tails of the rats were placed through a
small hole in the wall of the box, a catheter was inserted into the
tail vein without anesthesia and normal saline was infused
continuously at a rate of 1.0 ml/h. The rats were allowed to
breathe spontaneously and were supplied with an air-oxygen mixture
[fraction of inspired oxygen (FiO2) = 0.40]. Body
temperature was maintained at 37.0°C with a heat lamp.
The rats were randomized into three groups (n=7 in
each group) and each group received either sevoflurane, propofol or
no anesthetic (control group). In the anesthesia groups, the rats
were anesthetized with sevoflurane or propofol for 6 h (between
9:00 and 15:00). Rats undergoing inhalational anesthesia
(sevoflurane) were placed in plastic boxes supplied with
sevoflurane (Sevofrane®; Maruishi Pharmaceutical Co.,
Ltd., Osaka, Japan; 2.4% gas-air mixture) (25) and 40% oxygen (6 l/min). Normal
saline was administered via a venous catheter at a rate of 1.0
ml/h. Rats undergoing intravenous anesthesia (propofol) were housed
in plastic boxes supplied with 40% oxygen at a rate of 6 l/min and
were administered 1% propofol (Diprivan®; AstraZeneca,
Osaka, Japan; 600 μg/kg/min) (26)
via a venous catheter (rate of 9.0 ml/h for rats weighing 250 g).
In the control group, the rats were not administered anesthetics;
however, they were similarly housed in plastic boxes supplied with
40% oxygen at a rate of 6 l/min and normal saline was administered
via a venous catheter at a rate of 1.0 ml/h. Rats in the control
group were not allowed access to food and water. All rats were
allowed to breathe spontaneously. Animals were sacrificed
immediately following completion of anesthesia at 15:00. Within 3
mins after decapitation, the whole brain was rapidly removed and
the bilateral hippocampus was directly removed from the brain, as
described previously (27,28).
Physiological parameters
Physiological variables were measured in three
separate groups of rats not used for hippocampal miRNA evaluation
(n=7 each), which were treated with sevoflurane, propofol or no
anesthetic (control group). For all rats, a catheter was inserted
into the tail vein without anesthesia and normal saline was infused
continuously at a rate of 1.0 ml/h. During the experiment, each rat
was placed in a plastic cage (45×32×23 cm) supplied with an
air-oxygen mixture [(FiO2) = 0.40] and allowed to
breathe spontaneously. The body temperatures of the rats were
maintained at 37.0°C with a heat lamp and their rectal temperatures
were measured. As a surgical procedure, the left femoral artery was
cannulated to obtain blood samples for measuring the arterial
PO2, arterial PCO2, arterial blood pH, heart
rate and arterial blood pressure (Table I). Anesthetics were administered as
described for the miRNA analysis groups. In the sevoflurane and
propofol physiological groups, anesthesia was maintained for an
additional 3 or 6 h after surgery. In the control group, the rats
were anesthetized by intraperitoneal injection of sodium
pentobarbital (50 mg/kg) for the surgery and it was maintained for
3 h. The rats were placed in a rat tunnel in the plastic cages and
allowed to recover from the anesthesia. Blood samples were obtained
from all five groups.
 | Table IPhysiological data for the control,
sevoflurane and propofol anesthesia groups. |
Table I
Physiological data for the control,
sevoflurane and propofol anesthesia groups.
| Control | Sevoflurane | Propofol |
|---|
|
|
|
|
|---|
| 3 h | 3 h | 6 h | 3 h | 6 h |
|---|
| pH | 7.44±0.02 | 7.43±0.03 | 7.42±0.02 | 7.41±0.02 | 7.41±0.03 |
| PaO2
(mmHg) | 137.5±10.5 | 135.6±12.5 | 131.2±14.5 | 129.3±10.6 | 130.5±8.6 |
| PaCO2
(mmHg) | 36.5±2.3 | 39.8±2.9 | 40.5±3.3 | 41.5±3.2 | 41.7±2.5 |
| Heart rate
(beats/min) | 331±17 | 324±19 | 331±21 | 315±12 | 320±22 |
| Mean arterial
pressure (mmHg) | 120±9 | 111±12 | 117±13 | 109±15 | 112±13 |
| Body temperature
(°C) | 37.1±0.2 | 37.0±0.1 | 37.0±0.1 | 36.9±0.2 | 37.0±0.2 |
RNA extraction and miRNA screening test;
Taqman Low-Density Arrays (TLDA)
Hippocampal samples were washed twice with cold
phosphate-buffered saline, immediately placed in RNAlater solution
(Applied Biosystems, Inc., Foster City, CA, USA) and stored at
−80°C for one day. Following storage, the samples were defrosted
and the RNAlater solution was rapidly separated from the samples by
centrifugation (10,000 × g, 4°C, 5 min). Total RNA was extracted
from each sample using a mirVana™ miRNA Isolation kit (Applied
Biosystems, Inc.) according to the manufacturer’s instructions. RNA
quantity and quality were assessed by measuring absorbance using a
NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). Any
RNA samples with a 260/280 nm absorbance ratio >1.8 were
subjected to quantitative analysis. Total RNA samples containing
miRNA were used for quantitative reverse transcription-polymerase
chain reaction (qRT-PCR).
The miRNA expression profiles in the hippocampal
samples were analyzed using TLDA Rodent miRNA cards v.3.0 A and B
(Applied Biosystems, Inc.), a set of miRNA low-density arrays which
enables simultaneous qRT-PCR for 373 preloaded rat miRNA targets
and three endogenous controls (Mamm U6, U87 and Y1). All miRNA
targets are cataloged in the miRBase database (http://www.mirbase.org/). All procedures were based on
those of a previous study (29).
Briefly, TLDA was performed as a two-step process. During the first
step, 1,200 ng of total RNA per sample was reverse-transcribed
using Megaplex RT Primer Pools A and B (Applied Biosystems, Inc.),
up to 381 stem-looped primers per pool and a TaqMan miRNA Reverse
Transcription kit (Applied Biosystems, Inc.). In the second step,
each of the resulting complementary DNA pools was diluted, mixed
with TaqMan Universal PCR Master mix (Applied Biosystems, Inc.) and
deionized distilled water (Wako Pure Chemical Industries, Ltd.,
Tokyo, Japan), and loaded into one of the eight fill ports on the
TLDA microfluidic card. The cards were briefly centrifuged for 1
min at 315 × g to distribute samples to the multiple wells
connected to the fill ports and were then sealed to prevent
well-to-well contamination. The cards were processed and analyzed
using a 7900HT Fast Real-Time PCR system (Applied Biosystems,
Inc.).
Data analysis was performed using DataAssist
software v2.0 (Applied Biosystems, Inc.). The resulting data were
expressed as threshold cycle (Ct) values, where Ct represents a
unitless value defined as the fractional cycle number at which the
sample fluorescence signal passes a fixed threshold above baseline.
The relative quantities of all the miRNAs were calculated by the
comparative Ct method. ΔCt is the difference in Ct values derived
from the experimental samples and the controls, and ΔΔCt represents
the difference between paired samples, as calculated by the
following formula: ΔΔCt = ΔCt of sample following anesthesia − ΔCt
of the control group. The expression ratio shows the relative
quantity of the target gene (Xtarget) to the control gene
(Xcontrol). The fold change was computed by the following formula:
Xtarget/Xcontrol = 2−ΔΔCt. The 2−ΔΔCt method
is a convenient method for analyzing the relative changes in gene
expression from qRT-PCR experiments. Relative quantification (RQ)
values (relative levels of miRNA expression) were calculated by
this comparative Ct method using RQ Manager version 1.2.1 (Applied
Biosystems, Inc.) (30). Graphic
displays were visualized as heat map results of hierarchical
clustering. Distances between samples and assays were calculated
for hierarchical clustering as determined by ΔCt values, using
Euclidean distance.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Analysis of variance (ANOVA) followed by Tukey-Kramer
test was used to compare the physiological data and miRNA
expression levels in the sevoflurane anesthesia, propofol
anesthesia and control groups. P<0.05 was considered to indicate
a statistically significant difference in all tests. Tukey-Kramer
test was applied to identify genes and miRNAs that were
significantly differentially expressed on exposure to anesthetic
agents. ANOVA followed by Tukey-Kramer test was performed using
Kyplot software, version 5.0 (KyensLab Incorporated, Tokyo,
Japan).
Results
Anesthesia of rats
Following induction of sevoflurane or propofol
anesthesia, all rats lost consciousness within 5 min, with loss of
corneal reflexes and the withdrawal response to pain. No rats moved
spontaneously or died until the end of anesthesia. Data for all
animals were used.
Physiological data
The possible effects of anesthesia on a number of
physiological parameters were investigated in three separate groups
of rats. Fig. 1 shows a flow chart
of the experimental protocol. The physiological data for the
treatment groups during anesthesia were compared with those of the
control group, in which no anesthesia was administered following
the termination of anesthesia for the cannulation of the femoral
artery. Low mean blood pressure and high PCO2 were
observed following the administration of anesthetic drugs. However,
no significant differences in the physiological parameters among
the administration groups and control group were identified
(Table I). Hypoxia,
hyper/hypocapnia, hypotension or hypothermia were not observed in
any of the three groups.
miRNA screening test: TLDA
In order to identify suitable endogenous controls,
three different candidate miRNAs were analyzed for variance in gene
expression with DataAssist software v2.0. The statistical method
ranked the candidate endogenous control genes with an excellent
correlation of raw stability values. The most stably expressed Y1
was selected as the endogenous control and the relative miRNA
expression levels were normalized against Y1. Ct values >35
indicated that their expression levels were too low for accurate
analysis. The miRNA expression Ct value was therefore standardized
at ≤35.
The results of the TLDA analysis of the hippocampi
are shown in Tables II–IV (n=7 for each group; significance
level, P<0.05). There were 750 miRNAs on the TLDA plate; 373
miRNAs were associated with rats and the rest with mice. TLDA
analysis revealed 279 expressed miRNAs (74.8%), which were
expressed in the sevoflurane, propofol and control groups.
Significant differences were observed in the levels of 33 of the
279 expressed miRNAs (11.8%) among the three groups in response to
the anesthetic agents, and when compared with the control group,
significant differences were found in 26 of the 279 expressed
miRNAs (9.3%). Following sevoflurane anesthesia, the levels of four
miRNAs were significantly increased and those of 12 were
significantly reduced compared with those of the control group. By
contrast, following propofol anesthesia, the levels of 11 miRNAs
were significantly reduced but no miRNAs exhibited significantly
elevated levels. Of the total 33 significantly differentially
expressed miRNAs, one miRNA was common to both anesthetics, whereas
14 miRNAs were significantly differentially expressed (Tables II–IV).
 | Table IIDifferential expression of miRNAs
between the sevoflurane anesthesia and control groups using
TLDA. |
Table II
Differential expression of miRNAs
between the sevoflurane anesthesia and control groups using
TLDA.
| Assay | Sevoflurane
(FC) | Raw P-value |
|---|
| hsa-miR-423-3P | 1.87±0.62 | 0.021 |
| mmu-miR-24-2# | 1.42±0.37 | 0.032 |
| rno-miR-664 | 1.50±0.50 | 0.033 |
| hsa-miR-9# | 1.25±0.24 | 0.048 |
| mmu-miR-103 | 0.71±0.15 | 0.007 |
|
mmu-miR-125b-5p | 0.71±0.15 | 0.010 |
| mmu-miR-142-3p | 0.63±0.14 | 0.010 |
| mmu-miR-101b | 0.72±0.48 | 0.010 |
| mmu-miR-27b | 0.77±0.14 | 0.019 |
| mmu-miR-499 | 0.37±0.12 | 0.019 |
| mmu-miR-30b | 0.64±0.18 | 0.029 |
| mmu-miR-19b | 0.70±0.23 | 0.030 |
| rno-miR-351 | 0.42±0.17 | 0.038 |
| mmu-miR-129-3p | 0.74±0.18 | 0.040 |
| mmu-miR-369-5p | 0.70±0.22 | 0.040 |
|
rno-miR-219-2-3p | 0.70±0.22 | 0.049 |
 | Table IVDifferential expression of miRNAs
between the sevoflurane and propofol anesthesia groups using
TLDA. |
Table IV
Differential expression of miRNAs
between the sevoflurane and propofol anesthesia groups using
TLDA.
| Assay | Sevoflurane
(FC) | Propofol (FC) | Raw P-value |
|---|
| hsa-miR-9# | 1.25±0.24 | 0.88±0.11 | 0.002 |
| rno-miR-125b# | 1.14±0.12 | 0.60±0.37 | 0.002 |
| hsa-miR-324-3p | 1.38±0.57 | 0.75±0.08 | 0.004 |
| mmu-miR-674# | 1.27±0.27 | 0.78±0.21 | 0.006 |
| mmu-miR-101b | 1.49±0.47 | 0.98±0.12 | 0.007 |
| hsa-miR-423-3P | 1.87±0.62 | 1.05±0.59 | 0.011 |
| hsa-miR-140-3p | 1.60±0.80 | 0.82±0.17 | 0.015 |
| hsa-miR-206 | 1.76±1.07 | 0.70±0.45 | 0.019 |
| mmu-miR-24-2# | 1.42±0.37 | 0.99±0.24 | 0.028 |
| rno-miR-664 | 1.50±0.50 | 0.99±0.21 | 0.033 |
| rno-miR-409-3P | 1.52±0.59 | 0.91±0.22 | 0.037 |
| mmu-miR-770-5p | 1.64±0.95 | 0.84±0.25 | 0.041 |
| mmu-miR-29c | 0.99±0.27 | 0.61±0.44 | 0.042 |
| mmu-miR-374-5p | 1.26±0.51 | 0.77±0.18 | 0.042 |
Cluster analysis from TLDA data is shown in Fig. 2. Significantly differentially
expressed miRNAs were converted into a heat map in order to improve
the understanding of the miRNA expression patterns in each group.
Heat maps for the calculated clusters are commonly used
gene-expression analyses and are constructed for visualization of
gene expression levels. In the present study, heat maps illustrated
the gene expression levels from the hippocampal samples following
treatment with anesthetics using ΔCT values. The color intensity on
the heat map corresponds to the absolute intensity of the miRNA
expression. Each column represents a sample and each row shows
miRNA in the heat map. By individually clustering columns and rows,
the heat map simultaneously presents the separate samples and miRNA
clustering in one graphic. The dendrograms of clustering analysis
for the samples and miRNAs are shown to the top and left of the
heat map. This indicated the relatedness of the samples as
determined by the overall miRNA expression values. The dendrogram
separates into three main branches: The sevoflurane anesthesia, the
propofol anesthesia and the control groups.
Discussion
The present study demonstrated that sevoflurane and
propofol anesthesia induced numerous changes in the miRNA
expression levels of the rat hippocampus. Using TLDA data, the
hierarchical clustering with heat map presented the semantic
similarities and expression patterns of miRNA. Hierarchical
clustering separated the samples into three distinct branches in
the dendrogram: The sevoflurane anesthesia group, the propofol
anesthesia group and the control group (no anesthetic). These
findings indicate that the miRNA expression patterns with general
anesthetic treatment were distinct from those of the control group.
In addition, the inhalation anesthetic sevoflurane and the
intravenous anesthetic propofol exerted different effects on the
miRNA expression in the rat hippocampus.
In the present study, experiments were conducted
using the inhaled anesthetic sevoflurane and the intravenous
anesthetic propofol. Sevoflurane was selected as it is one of the
most commonly used inhaled clinical anesthetic agents in Japan
(4). Propofol was selected as it
is a widely used intravenous anesthetic in everyday clinical
practice in a number of countries. The two drugs have been used
extensively in previous studies concerning the effects of
anesthetic agents on gene expression (7–9),
protein expression (10) and
metabolites (11) in the rat
brain. Changes in the miRNA expression levels in the hippocampi of
rats anesthetized with one minimum alveolar concentration of
inhaled anesthetics (sevoflurane at 2.4%; 22) or with continuous
infusion of intravenous anesthetics (propofol at 600 μg/kg/min; 23)
were investigated in the present study. Infusion of propofol at 600
μg/kg/min is the median effective dose for rats (26). The two doses were selected since
lower doses may not induce adequate anesthesia and higher doses may
produce hemodynamic changes (10,22)
that may independently alter hippocampal miRNA expression
levels.
A number of miRNAs determined to be differentially
expressed in the present study are known to be involved in stem
cell self-renewal, synaptic plasticity and memory consolidation in
the rat hippocampus. For example, miR-125b, a neuronal miRNA, is
important in proliferation and differentiation of neural stem cells
(31). miRNA-181a is rich in
mature nerve cells and bioinformatic analysis has revealed that the
cAMP response element-binding protein 1 (CREB1) mRNA 3′-UTR
contains a complementary sequence to the miR-181a seed region.
CREB1 is a key nuclear factor highly expressed in hippocampal
neurons on which numerous signaling pathways converge (32–34).
In the present study, the expression levels of these miRNAs were
found to be significantly affected by sevoflurane and/or propofol
anesthesia; therefore, general anesthesia may affect these cell
signaling pathways, including synaptic plasticity and memory
consolidation. In addition, the changes in the levels of miRNA may
be attenuated following general anesthesia.
It was hypothesized that sevoflurane and propofol
anesthesia induce different expression patterns of miRNA in the rat
hippocampus. Sevoflurane and propofol induce general anesthesia but
have different mechanisms of anesthetic action. Single miRNAs may
bind with hundreds of unique mRNAs, thus regulating the expression
of multiple cellular pathways, and individual miRNAs reciprocally
act on mRNA. The TLDA data of the present study were not
satisfactory to determine the validity of this hypothesis since
they provide only a snapshot of the changes in miRNA expression
patterns. Pan et al (20)
showed that sevoflurane anesthesia induces changes in the mRNA
expression levels in the rat hippocampus, which may be associated
with memory impairment or other neural disorders. Although changes
in the levels of mRNA and miRNA expression are likely to affect
subsequent protein expression, there is a lack of correlation
between them. The final functional entities in cells are not mRNAs
or miRNAs, but proteins that undergo numerous post-translational
modifications, thereby controlling cellular mechanisms (35). Investigation of the action
mechanisms of general anesthetics is a noteworthy field and further
studies using molecular approaches are required for the elucidation
of these mechanisms.
Due to advances in high-throughput technologies and
bioinformatics, there are several methods for gene expression
profiling. The results of microarray analyses are commonly
validated using qRT-PCR, which is the most sensitive and
reproducible method for quantifying gene expression. Single qRT-PCR
assays and TLDA analyses rely on identical amplification
technologies. Single qRT-PCR is a reliable method for the analysis
of miRNA expression as the reactions are run in triplicate; by
contrast, TLDA analyses are run individually. Hui et al
(36) demonstrated that the
technical reproducibility of TLDAs was high (intrasample
correlations >0.9, accuracy 92.8%). Furthermore, Mees et
al (37) demonstrated that
TLDA and single qRT-PCR correlated well with regards to the
quantity and quality of the measured miRNAs. The TLDA analysis of
the present study has the same procedures as those of previous
studies (21,22,38).
In these studies, the single qRT-PCR results of individual miRNA
were equivalent to the TLDA analysis results. These results
confirmed that the TLDA analysis data of the present study were as
accurate as those of single qRT-PCR and that the TLDA data were
reliable.
The present study has certain limitations. The study
was conducted in rats and innate differences in gene expression in
different species of animals mean that extrapolation of results
from rats to other species is difficult. Additionally, miRNA
microarray analyses were conducted using whole hippocampal samples.
The hippocampus is composed of the dentate gyrus and the cornu
ammonis. The cornu ammonis is separated into three predominant
divisions: CA1, CA2 and CA3, but the results of the present study
only reflected the whole hippocampal miRNA expression pattern. For
further analyses, localization of gene-expression changes is
required. Furthermore, a large number of miRNAs were measured;
therefore, it is possible that certain results were false
positives.
In conclusion, the data from the present study
indicated changes in the miRNA expression levels in the rat
hippocampus under general anesthesia. When compared with that of
the control group, the inhalation anesthetic sevoflurane and the
intravenous anesthetic propofol exerted differential effects on
miRNA expression. In subsequent studies, the time-dependent changes
in the levels of miRNA expression under general anesthesia may be
further investigated, along with the post-anesthetic influence of
miRNA at various time points.
Acknowledgements
This study was supported by a Grant-in-Aid for
Science Research (C) (project no. 20591820) from the Japan Society
for the Promotion of Science (Tokyo, Japan).
References
|
1
|
Forrest JB, Rehder K, Goldsmith CH, et al:
Multicenter study of general anesthesia. I. Design and patient
demography. Anesthesiology. 72:252–261. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown BR Jr and Frink EJ: The safety of
sevoflurane in humans. Anesthesiology. 79:201–203. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marik PE: Propofol: therapeutic
indications and side-effects. Curr Pharm Des. 10:3639–3649. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stachnik J: Inhaled anesthetic agents. Am
J Health Syst Pharm. 63:623–634. 2006. View Article : Google Scholar
|
|
5
|
Eger EI 2nd: Characteristics of anesthetic
agents used for induction and maintenance of general anesthesia. Am
J Health Syst Pharm. 61(Suppl 4): S3–S10. 2004.PubMed/NCBI
|
|
6
|
Simons PJ, Cockshott ID, Douglas EJ,
Gordon EA, Hopkins K and Rowland M: Disposition in male volunteers
of a subanaesthetic intravenous dose of an oil in water emulsion of
14C-propofol. Xenobiotica. 18:429–440. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sakamoto A, Imai J, Nishikawa A, et al:
Influence of inhalation anesthesia assessed by comprehensive gene
expression profiling. Gene. 356:39–48. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kobayashi K, Takemori K and Sakamoto A:
Circadian gene expression is suppressed during sevoflurane
anesthesia and the suppression persists after awakening. Brain Res.
1185:1–7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yoshida Y, Nakazato K, Takemori K,
Kobayashi K and Sakamoto A: The influences of propofol and
dexmedetomidine on circadian gene expression in rat brain. Brain
Res Bull. 79:441–444. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsuboko Y and Sakamoto A: Propofol
anaesthesia alters the cerebral proteome differently from
sevoflurane anaesthesia. Biomed Res. 32:55–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawaguchi H, Hirakawa K, Miyauchi K, Koike
K, Ohno Y and Sakamoto A: Pattern recognition analysis of proton
nuclear magnetic resonance spectra of brain tissue extracts from
rats anesthetized with propofol or isoflurane. PLoS One.
5:e111722010. View Article : Google Scholar
|
|
12
|
Bak M, Silahtaroglu A, Møller M, et al:
MicroRNA expression in the adult mouse central nervous system. RNA.
14:432–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miranda KC, Huynh T, Tay Y, et al: A
pattern-based method for the identification of MicroRNA binding
sites and their corresponding heteroduplexes. Cell. 126:1203–1217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wayman GA, Davare M, Ando H, et al: An
activity-regulated microRNA controls dendritic plasticity by
down-regulating p250GAP. Proc Natl Acad Sci USA. 105:9093–9098.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Visvanathan J, Lee S, Lee B, Lee JW and
Lee SK: The microRNA miR-124 antagonizes the anti-neural REST/SCP1
pathway during embryonic CNS development. Genes Dev. 21:744–749.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bredy TW, Lin Q, Wei W, Baker-Andresen D
and Mattick JS: MicroRNA regulation of neural plasticity and
memory. Neurobiol Learn Mem. 96:89–94. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee K, Kim JH, Kwon OB, et al: An
activity-regulated microRNA, miR-188, controls dendritic plasticity
and synaptic transmission by downregulating neuropilin-2. J
Neurosci. 32:5678–5687. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosik KS: The neuronal microRNA system.
Nat Rev Neurosci. 7:911–920. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan Z, Lu XF, Shao C, et al: The effects
of sevoflurane anesthesia on rat hippocampus: a genomic expression
analysis. Brain Res. 1381:124–133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ishikawa M, Tanaka S, Arai M, Genda Y and
Sakamoto A: differences in microRNA changes of healthy rat liver
between sevoflurane and propofol anesthesia. Anesthesiology.
117:1245–1252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tanaka S, Ishikawa M, Arai M, Genda Y and
Sakamoto A: Changes in microRNA expression in rat lungs caused by
sevoflurane anesthesia: a TaqMan® low-density array
study. Biomed Res. 33:255–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Genda Y, Arai M, Ishikawa M, Tanaka S,
Okabe T and Sakamoto A: microRNA changes in the dorsal horn of the
spinal cord of rats with chronic constriction injury: A
TaqMan® Low Density Array study. Int J Mol Med.
31:129–137. 2013.PubMed/NCBI
|
|
24
|
Ethical standards for investigations of
experimental pain in animals. The Committee for Research and
Ethical Issues of the International Association for the Study of
Pain. Pain. 9:141–143. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Obal D, Preckel B, Scharbatke H, et al:
One MAC of sevoflurane provides protection against reperfusion
injury in the rat heart in vivo. Br J Anaesth. 87:905–911. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Orth M, Barter L, Dominguez C, Atherley R,
Carstens E and Antognini JF: Halothane and propofol differentially
affect electroencephalographic responses to noxious stimulation. Br
J Anaesth. 95:477–484. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiu K, Lau WM, Lau HT, So KF and Chang
RC: Micro-dissection of rat brain for RNA or protein extraction
from specific brain region. J Vis Exp. 7:2692007.PubMed/NCBI
|
|
28
|
Glowinski J and Iversen LL: Regional
studies of catecholamines in the rat brain. I. The disposition of
[3H]norepinephrine, [3H]dopamine and [3H]dopa in various regions of
the brain. J Neurochem. 13:655–669. 1966.
|
|
29
|
Wang B, Howel P, Bruheim S, et al:
Systematic evaluation of three microRNA profiling platforms:
microarray, beads array, and quantitative real-time PCR array. PLoS
One. 6:e171672011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
31
|
Skalnikova H, Vodicka P, Gadher SJ and
Kovarova H: Proteomics of neural stem cells. Expert Rev Proteomics.
5:175–186. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cui Y, Xiao Z, Han J, et al: MiR-125b
orchestrates cell proliferation, differentiation and migration in
neural stem/progenitor cells by targeting Nestin. BMC Neurosci.
13:1162012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saba R, Störchel PH, Aksoy-Aksel A, et al:
Dopamine-regulated microRNA MiR-181a controls GluA2 surface
expression in hippocampal neurons. Mol Cell Biol. 32:619–632. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Zhao Z, Yang F, Gao Y, Song J and
Wan Y: microRNA-181a is involved in insulin-like growth
factor-1-mediated regulation of the transcription factor CREB1. J
Neurochem. 126:771–780. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mozzachiodi R and Byrne JH: More than
synaptic plasticity: role of nonsynaptic plasticity in learning and
memory. Trends Neurosci. 33:17–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hui AB, Shi W, Boutros PC, et al: Robust
global micro-RNA profiling with formalin-fixed paraffin-embedded
breast cancer tissues. Lab Invest. 89:597–606. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mees ST, Schleicher C, Mardin WA,
Senninger N, Colombo-Benkmann M and Haier J: Analyzing miRNAs in
ductal adenocarcinomas of the pancreas. J Surg Res. 169:241–246.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Arai M, Genda Y, Ishikawa M, Shunsuke T,
Okabe T and Sakamoto A: The miRNA and mRNA changes in rat
hippocampi after chronic constriction injury. Pain Med. 14:720–729.
2013. View Article : Google Scholar : PubMed/NCBI
|