Introduction
Brain arteriovenous malformations (bAVMs) were first
described ~200 years ago and are defined as the direct
communication of arteries to abnormally tortuous and dilated veins
without interposing capillaries (1,2).
bAVMs may present as hemorrhage, epilepsy, headache,
non-hemorrhagic neurological deficits or are asymptomatic. Although
it has been associated with certain well-defined genetic disorders,
including ataxia telangiectasia, Osler-Weber-Rendu syndrome,
Sturge-Weber syndrome and Wyburn-Mason syndrome (3–6), the
molecular etiology of bAVM has not been fully elucidated. A
previous study has suggested that bAVM may be congenital but
dynamic with the possibility to grow, regress and even reappear
(7). Several studies have found
that there is an association between the upregulation of the Notch
signaling pathway and bAVM (8–11).
However, whether the pathway is involved in the hemorrhage of
bAVMs, has yet to be fully elucidated.
The Notch signaling pathway is a transmembrane,
evolutionarily conserved intercellular signaling pathway in
vertebrates and non-vertebrates. It affects a wide variety of
developmental processes and cell-fate determination during the
embryonic period and postnatally (12–14).
The Notch signaling pathway consists of four receptors (Notch1–4)
and five ligands [Jagged 1 and 2, Delta-like (DLL) 1, 3 and 4] in
mammals. It controls lymphatic endothelial quiescence, endothelial
cell (EC) selection and the morphogenesis of vascular branches, as
well as the interaction between the developing endothelium and
endothelial basement membrane and arterial maturation (15–19).
It has also been reported to coordinate with vascular endothelial
growth factor (VEGF), transforming growth factor-β (TGF-β), the
anaplastic lymphoma kinase signaling pathway, the Wnt/β-catenin
pathway and nuclear factor-κB (16,17,20–23).
The pathway is initiated with the binding of adjacent signaling
cell ligands and receptors on receiving cells, followed by two
proteolytic cleavage events. The second cleavage event releases the
Notch intracellular domain (NICD), which is then translocated to
the nucleus and forms a complex with DNA-binding proteins,
including CBF1, Su(H) and LAG-1(CSL), displaces previously bound
co-repressors and recruits co-activators. In addition, the Notch
signaling pathway functions as a transcriptional activator of the
downstream targets, hairy and enhancer of split (Hes) and
Hes-related protein gene families (24–26).
Furthermore, the Notch pathway has been found to be associated with
syndromes presenting with abnormal vascular phenotypes, including
Alagille (27) and Cerebral
Autosomal Dominant Arteriopathy with Subcortical Infarcts and
Leukoencephalopathy syndromes (28,29).
The present study aimed to assess the expression
levels of NOTCH1, NOTCH4, DLL4 and JAGGED1 in the
nidus of patients with bAVM by quantitative polymerase chain
reaction (qPCR) and immunohistochemical staining to determine
whether there is an association between the Notch signaling pathway
receptor or ligand expression levels and the clinical hemorrhage of
bAVM.
Materials and methods
Subjects
A total of 35 nidus tissues from patients with bAVM
were obtained from the Capital Medical University Affiliated
Beijing Tiantan Hospital (Beijing, China) from June 2011 to June
2012. All patients were diagnosed with bAVM according to digital
subtraction angiography and magnetic resonance imaging results
combined with postoperative pathology diagnosis. The patients
ranged in age from 7 to 57 years with a mean age of 29.29±14.64
years (median ± standard deviation). The majority of patients
presented with the clinical symptoms of bAVM, including
intracranial hemorrhage, epilepsy and headaches. Only one patient
was recruited via regular medical examination. Among the 35
patients, five underwent endovascular treatment prior to surgery.
The patients with bAVM were further divided into two groups: The
hemorrhage group and the non-hemorrhage group, based on their first
clinical presentation. The mean age of the hemorrhage and
non-hemorrhage groups were 25.68±13.38 and 33.56±15.12 years,
respectively. The clinical features of patients with bAVM are shown
in Table I. Ten normal human
superficial temporal arteries (STAs) were obtained from head trauma
patients with a mean age of 45.7±16.3 years (range, 23–60 years).
Informed consent was obtained from all patients and controls,
either directly from the individual or their legal guardian. The
protocol of the present study was approved by the ethics committee
of the Beijing Tiantan Hospital (Beijing, China).
 | Table IClinical features of patients with
brain arteriovenous malformation. |
Table I
Clinical features of patients with
brain arteriovenous malformation.
| Clinical
variables | Values |
|---|
| Age, years | 30.10±14.97 (range,
8–57) |
| Gender, cases
(%) |
| Male | 19 (54.29) |
| Female | 16 (45.71) |
| Clinical
presentation, cases (%) |
| Epilepsy | 17 (48.57) |
| Hemorrhage | 16 (45.71) |
| Headache | 5 (14.29) |
| Asymptomatic | 1 (2.86) |
| Other cerebarl
vascular diseases, cases |
| Aneurysm | 3 |
| Ateriovenous
fistula | 1 |
| Achnoid cyst | 1 |
| Prior embolism,
cases (%) | 5 (14.29) |
| S-M score | 2.46±0.65 |
qPCR
Expression levels of mRNA were normalized against
hypoxanthine phosphoribosyl-transferase-1 (HPRT-1), using the
following specific PCR primers designed by the NCBI Basic Local
Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi, 2013);
primers are shown in Table II.
Total RNA was isolated using TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA), and cDNA was acquired according
to the Takara procedure (Takara Bio, Inc., Shiga, Japan) with 2 μg
of total RNA. qPCR reactions for the four genes, NOTCH1, NOTCH4,
DLL4 and JAGGED1, were performed using the Takara
Thermal Cycler Dice Real-Time System (TP800; Takara Bio, Inc.) and
the conditions were as follows: 95°C for 30 sec; 40 cycles
comprising 95°C for 5 sec, 55°C for 30 sec, 72°C for 30 sec; and a
final hold at 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec.
All reactions were performed in triplicate. After completion of
qPCR, the amplification products underwent electrophoresis
immediately on agarose gel. The target gene was confirmed according
to the position in the DNA marker. Absorbance data were collected
at the end of every extension, and cycle threshold (Ct) values were
analyzed by the ΔCt method (30).
The efficiency of amplification of the target genes and the
internal control (HPRT-1) were examined.
 | Table IIPrimers designed for quantitative
polymerase chain reaction. |
Table II
Primers designed for quantitative
polymerase chain reaction.
| Genes | Forward
primers | Reverse
primers |
|---|
| HPRT1 |
GCCCTGGCGTCGTGATTAGT |
GGGCTACAATGTGATGGCCTCC |
| NOTCH1 |
GCCGCAGTTGTGCTCCTGAA |
TGTCGTGATGCATGCGCTCC |
| NOTCH4 |
TGGAGAAGGGGCTGTGGAATG |
CACACTGGCAGGTCCCTTGT |
| DLL4 |
CGCAATGACCACTTCGGCCA |
ATTCGTTACACAGCCGGCCC |
| JAGGED1 |
TCGAGTCTGAGGCCGTTGCT |
GAGACCGTGTCGGCTGCAAG |
Immunohistochemistry
Brain AVM specimens and STAs were embedded in
paraffin and cut into 5-μm sections. Specimens were deparaffinized
with xylene and rehydrated with ethanol following antigen retrieval
with antigen unmasking solution according to the manufacturer’s
instructions (Vector Laboratories, Burlingame, CA, USA). Endogenous
peroxidase activity was blocked by incubation with 3%
H2O2 at room temperature for 10 min.
Specimens were then washed with phosphate-buffered saline (PBS) and
incubated in blocking solution for 3 min at a high pressure
(pressure cooker; Biocare Medical, Inc., Concord, CA, USA). The
primary antibodies used were as follows: Mouse monoclonal
anti-Notch1 intracellular domain antibody (MAB5352, 1:300;
Millipore, Billerica, MA, USA), rabbit polyclonal anti-Notch4
intracellular domain antibody (sc-5594, 1:100; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), rabbit polyclonal
anti-CD31 (ab28364, 1:200; Abcam, Cambridge, MA, USA), rabbit
polyclonal anti-Jagged-1 (ab7771, 1:200; Abcam), rabbit polyclonal
anti-Hes-1 (ab71559, 1:100; Abcam) and rabbit polyclonal anti-DSL
domain of DLL4 (ab7280, 1:150; Abcam). Primary antibodies were
added to the blocking buffer and incubated with the tissue sections
at 4°C overnight. Sections were then washed with PBS and incubated
with horseradish peroxidase-labeled goat anti-rabbit or anti-mouse
antibody for 1 h at room temperature. 3,3′-diaminobenzidine
solution (Vector Laboratories) was used to obtain a visible
reaction product. The staining process was performed blind from the
source. A Leica microscope and Leica digital color camera (DM4000;
Leica, Mannheim, Germany) were used to examine and photograph the
slides, respectively.
Statistical analyses
Statistical analysis was performed using a two sided
t-test and one-way analysis of variance (ANOVA). All data are
presented as the mean ± standard deviation. Data were analyzed
using SPSS software, version 19.0 (SPSS Inc., Chicago, IL, USA).
P≤0.05 was considered to indicate a statistically significant
difference.
Results
Expression levels of the four genes in
bAVM and STA specimens detected by immunohistochemistry
Elevated expression levels of NOTCH1 and
NOTCH4 were detected in the hemorrhage bAVM group compared
with that of the STA group. HPRT-1 was selected as the internal
control for normalization. The expression levels of NOTCH1
and NOTCH4 were elevated by 2.26-fold (P=0.033) and
2.80-fold (P=0.002), respectively; however, the expression of
JAGGED1 and DLL4 were slightly different but without
significance (P=0.106 and P=0.492, respectively). In order to
further examine the ligands involved in the Notch signaling
pathway, immunohistochemical analysis was perfomed using a positive
control, the middle cerebral artery (MCA). To determine the
phenotype of expressing cells, anti-CD31 was selected as a
representative. Immunohistological analysis demonstrated that
Notch1 and Notch4 were weakly expressed in ECs in MCA and STA
(Fig. 2), but overexpressed in the
hemorrhage and non-hemorrhage groups. Jagged1 and DLL4 demonstrated
similar expression levels in the two control and bAVM groups. The
downstream target gene of the Notch signaling pathway, HES1,
was predominantly expressed in nuclei of ECs and smooth muscle
cells in bAVM, but was negatively expressed in the STAs and
MCAs.
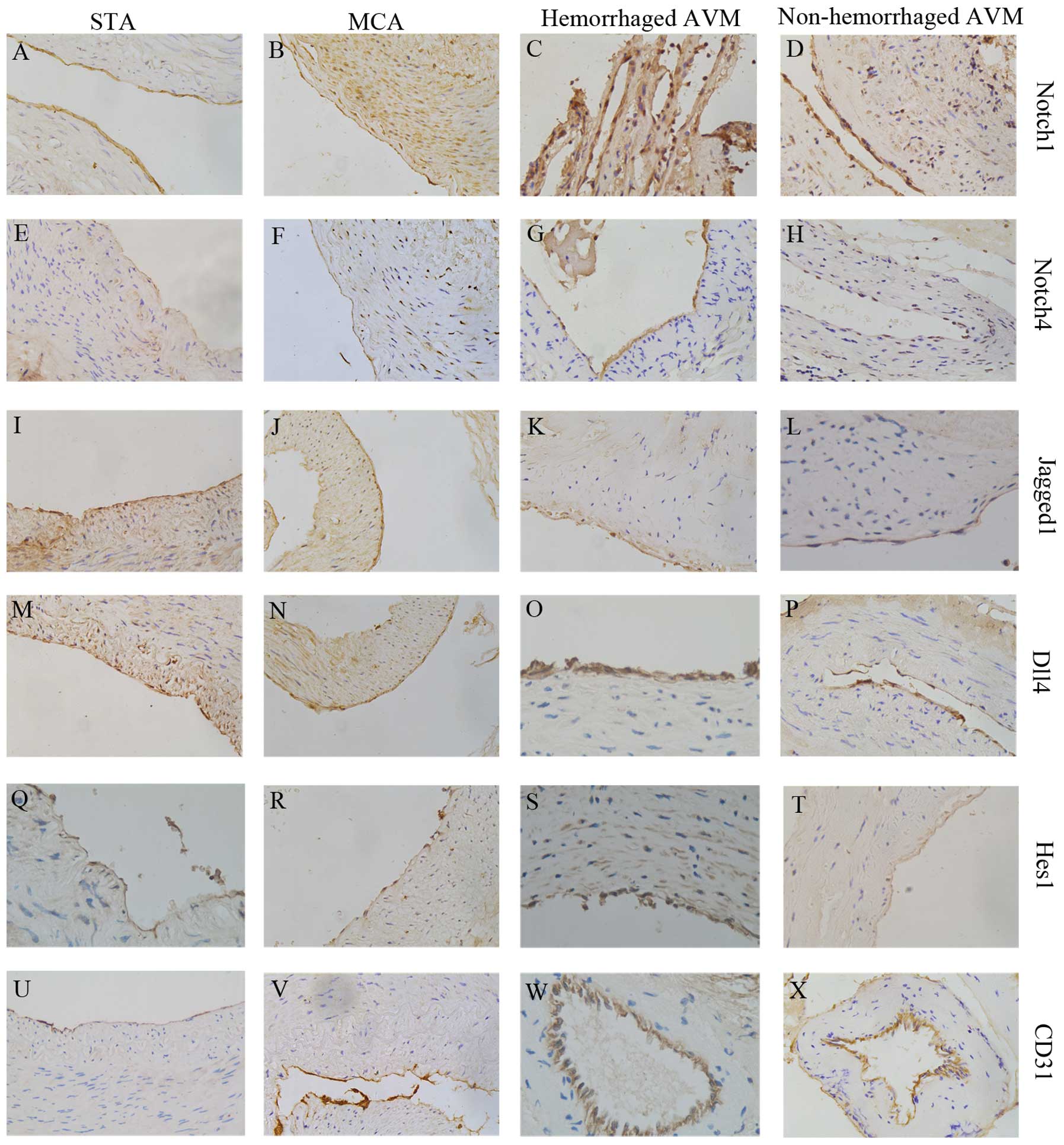 | Figure 2IH staining of STA, MCA, hemorrhaged
AVM and non-hemorrhaged AVM samples. (A–D) Notch1, (E–H) in STA,
MCA, hemorrhaged AVM and non-hemorrhaged AVM, respectively. (E–H)
Notch4 in STA, MCA hemorrhaged AVM and non-hemorrhaged AVM. (I–L)
Jagged1 in STA, MCA hemorrhaged AVM and non-hemorrhaged AVM. (M–P)
IH stain for DLL4. (Q–T) IH stain for Hes1. There were no
significant differences between the STA and MCA groups in (A,B)
Notch1, (E,F) Notch4, (I,J) Jagged1, (M,N) DLL4, (Q,R) Hes1 and
(U,V) CD31. In concordance with the quantitative polymerase chain
reaction results, Notch1 and Notch4 expression were significantly
increased in the AVM groups, whereas DLL4 and Jagged1 expression
were similar in the STA control group. Compared with the hemorrhage
bAVM and non-hemorrhage bAVM groups, only Notch1 was upregulated in
hemorrhage brain AVM, other components remained unchanged in bAVM
groups. (U–X) CD31 was stained to demonstrate endothelial cells.
IH, immunohistochemical; STA, superficial temporal artery; MCA,
middle cerebral artery; bAVM, brain arteriovenous malformation;
DLL, delta-like; Hes-1, hairy and enhancer of split-1. |
Expression levels of the four genes
detected by qPCR
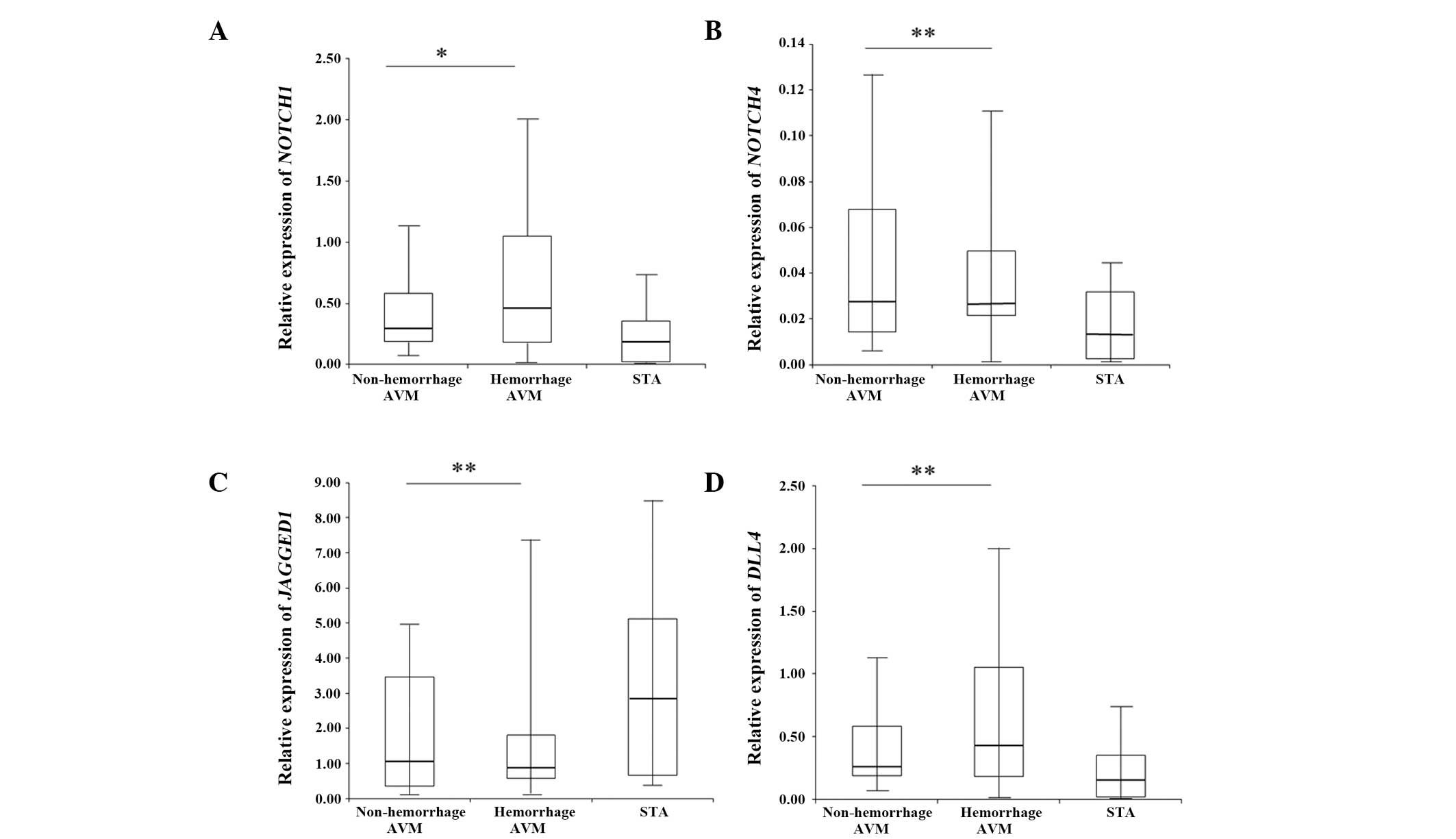
qPCR detected a significant increase (2.82-fold;
0.629±0.596) in NOTCH1 expression in the hemorrhage group
compared with that of the non-hemorrhage group (P=0.023). The
expression levels of NOTCH4 and DLL4 remained
unchanged between the two groups, while JAGGED1 was
overexpressed in the non-hemorrhage group (1.24-fold) (Fig. 1). However, the overexpression of
JAGGED1 lacked significance when analyzed by ANOVA. The
expression levels detected by qPCR of the key components of the
Notch signaling pathway in the bAVM groups were in concordance with
the immunohistochemical results. The lack of significant
differences in the expression of JAGGED1 between the
hemorrhage and non-hemorrhage groups indicated that the abnormal
expression levels may be caused by the overall expression levels of
JAGGED1 in neural cells.
Discussion
bAVMs are commonly detected in patients aged ~30
years and there have been a number of cases reporting the regrowth
of bAVM following complete surgery or embolism, reflecting the
enigmatic etiology of bAVM (2,31).
Due to the scarce data available on these cases, it is not possible
to arbitrarily conclude that bAVM is a complete congenital
lesion.
Previous studies have suggested that NOTCH1
is an oncogene as its upregulation is associated with the presence
of solid tumors in the digestive system, breast and hematological
system (32–34). Uyttendaele et al (10) found that the introduction of the
active form of NOTCH4 and knockout of either NOTCH1
or NOTCH1/NOTCH4 in ECs produced similar phenotypes of
vascular deficiency with the loss of fine branches or excessive
branches. In addition, Murphy et al (9) observed the increased activation of
endothelial Notch signaling with the presence of bAVM-like
structures in mice, which suggested that the activation of the
Notch signaling pathway is potentially a crucial molecular
candidate in bAVM pathogenesis. In addition, Zhuge et al
(11) identified the upregulation
of Notch1 in human samples, confirming that abnormal Notch
signaling activation has a vital role in the pathogenesis of bAVM.
In a further study to elucidate the mechanisms of the Notch
signaling pathway and determine potential treatment for bAVM,
Murphy et al (9) focused on
the normalization of the receptor Notch4. In his study, the
high-flow AV shunts, caused by tetracycline-responsive system,
decreased in blood flow as well as vessel diameter following
repression of Notch4. Based on these findings, therapy that targets
the Notch signaling pathway may be a promising approach for bAVM
treatment (8,11,35).
In addition, a previous study using mice demonstrated that a
complete and safe normalization of Notch4 in animals reduced blood
vessel size in bAVM, suggesting a treatment option of bAVM
alternative to surgery, embolism or radiation therapy (8,35).
In the present study, elevated expression levels of
NOTCH1 and NOTCH4 were found in the bAVM nidus at the
RNA and protein levels compared with those of normal STA and normal
MCA, which was in accordance with previous research (10,11,35).
To the best of our knowledge, the present study was the first to
determine whether the Notch signaling pathway is associated with
the clinical presentation of hemorrhage in bAVM. Patients with bAVM
were stratified into the hemorrhage and non-hemorrhage groups and
the expression levels of key factors in the Notch signaling pathway
were compared between the groups. It was found that the expression
levels of NOTCH1 significantly increased in the hemorrhage
group, suggesting that NOTCH4, DLL4 and JAGGGED1 may
not be involved in the pathogenesis of hemorrhage in bAVM nidus.
Recently, Liebler et al (36) reported that while Notch1 NICD
markedly repressed endothelial migration and sprouting
angiogenesis, the intracellular domain of Jagged1 and DLL4 did not
influence EC adhesion. It is therefore concluded that the relevance
of ligand intracellular domains in ECs remains unclear. However,
our findings also showed different results compared with those of
Zhuge et al (11). It was
demonstrated that the expression levels of DLL4 and JAGGED1
appeared unchanged between patients with bAVM and the controls. As
a large sample size was used to contribute to the power of
detection, the data are considered reliable.
In the present study, it was demonstrated that the
expression levels of NOTCH1 and NOTCH4 were increased
in patients with bAVM, and the correlation between hemorrhage and
Notch1/4 expression was analyzed. It was demonstrated that the
increased expression of key components of the Notch signaling
pathway are a potential cause of hemorrhage in human bAVM. The
findings also suggested that, although the continuous activation of
the Notch signaling pathway was reported to lead to hemorrhage in
the brain, liver or other organs in studies of mice and embryos
(8,9), there was no significant association
between hemorrhage of bAVM and Notch4, DLL4 and Jagged1; however,
there was an association between hemorrhage and Notch1. Further
studies targeting the remaining receptors and ligands of the Notch
signaling pathway are required to conclude whether Notch1 is the
key factor involved in hemorrhage in bAVM. This would elucidate the
importance of anti-Notch signaling drugs and their efficiency to
reduce the risk of hemorrhage in patients with bAVM.
Abbreviations:
|
AVMs
|
arteriovenous malformations
|
|
PCR
|
polymerase chain reaction
|
|
STA
|
superficial temporal artery
|
|
MCA
|
middle cerebral artery
|
|
NICD
|
Notch intracellular domain
|
|
EC
|
endothelial cell
|
References
|
1
|
Porter AJ and Bull J: Some aspects of the
natural history of cerebral arteriovenous malformation. Br J
Radiol. 42:667–675. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gross BA and Du R: Natural history of
cerebral arteriovenous malformations: a meta-analysis. J Neurosurg.
118:437–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nussbaum ES, Heros RC, Madison MT, Awasthi
D and Truwit CL: The pathogenesis of arteriovenous malformations:
insights provided by a case of multiple arteriovenous malformations
developing in relation to a developmental venous anomaly.
Neurosurgery. 43:347–351. 1998. View Article : Google Scholar
|
|
4
|
Ponce FA, Han PP, Spetzler RF, Canady A
and Feiz-Erfan I: Associated arteriovenous malformation of the
orbit and brain: a case of Wyburn-Mason syndrome without retinal
involvement. Case report J Neurosurg. 95:346–349. 2001.PubMed/NCBI
|
|
5
|
Stapf C, Mohr JP, Pile-Spellman J, Solomon
RA, Sacco RL and Connolly EJ Jr: Epidemiology and natural history
of arteriovenous malformations. Neurosurg Focus. 11:e12001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wohlwill FJ and Yakovlev PI:
Histopathology of meningo-facial angiomatosis (Sturge-Weber’s
disease); report of four cases. J Neuropathol Exp Neurol.
16:341–364. 1957.PubMed/NCBI
|
|
7
|
Sturiale CL, Puca A, Sebastiani P, et al:
Single nucleotide polymorphisms associated with sporadic brain
arteriovenous malformations: where do we stand? Brain. 136:665–681.
2013. View Article : Google Scholar
|
|
8
|
Murphy PA, Kim TN, Lu G, Bollen AW,
Schaffer CB and Wang RA: Notch4 normalization reduces blood vessel
size in arteriovenous malformations. Sci Transl Med. 4:117r–118r.
2012.PubMed/NCBI
|
|
9
|
Murphy PA, Lam MT, Wu X, et al:
Endothelial Notch4 signaling induces hallmarks of brain
arteriovenous malformations in mice. Proc Natl Acad Sci USA.
105:10901–10906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Uyttendaele H, Ho J, Rossant J and
Kitajewski J: Vascular patterning defects associated with
expression of activated Notch4 in embryonic endothelium. Proc Natl
Acad Sci U S A. 98:5643–5648. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
ZhuGe Q, Zhong M, Zheng W, et al: Notch-1
signalling is activated in brain arteriovenous malformations in
humans. Brain. 132:3231–3241. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Irvine KD: Fringe, Notch, and making
developmental boundaries. Curr Opin Genet Dev. 9:434–441. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li JL and Harris AL: Notch signaling from
tumor cells: a new mechanism of angiogenesis. Cancer Cell. 8:1–3.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Muskavitch MA: Delta-notch signaling and
Drosophila cell fate choice. Dev Biol. 166:415–430. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alberi L, Hoey SE, Brai E, Scotti AL and
Marathe S: Notch signaling in the brain: in good and bad times.
Ageing Res Rev. 12:801–814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Holderfield MT and Hughes CC: Crosstalk
between vascular endothelial growth factor, notch, and transforming
growth factor-beta in vascular morphogenesis. Circ Res.
102:637–652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lawson ND, Scheer N, Pham VN, et al: Notch
signaling is required for arterial-venous differentiation during
embryonic vascular development. Development. 128:3675–3683.
2001.PubMed/NCBI
|
|
18
|
Weng AP, Ferrando AA, Lee W, et al:
Activating mutations of NOTCH1 in human T cell acute lymphoblastic
leukemia. Science. 306:269–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng W, Tammela T, Yamamoto M, et al:
Notch restricts lymphatic vessel sprouting induced by vascular
endothelial growth factor. Blood. 118:1154–1162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clément N, Gueguen M, Glorian M, et al:
Notch3 and IL-1beta exert opposing effects on a vascular smooth
muscle cell inflammatory pathway in which NF-kappaB drives
crosstalk. J Cell Sci. 120:3352–3361. 2007.PubMed/NCBI
|
|
21
|
Corada M, Nyqvist D, Orsenigo F, et al:
The Wnt/beta-catenin pathway modulates vascular remodeling and
specification by upregulating Dll4/Notch signaling. Dev Cell.
18:938–949. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Domigan CK and Iruela-Arispe ML: Recent
advances in vascular development. Curr Opin Hematol. 19:176–183.
2012. View Article : Google Scholar
|
|
23
|
Larrivée B, Prahst C, Gordon E, et al:
ALK1 signaling inhibits angiogenesis by cooperating with the Notch
pathway. Dev Cell. 22:489–500. 2012.PubMed/NCBI
|
|
24
|
Lasky JL and Wu H: Notch signaling, brain
development, and human disease. Pediatr Res. 57:104R–109R. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bray SJ: Notch signalling: a simple
pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roca C and Adams RH: Regulation of
vascular morphogenesis by Notch signaling. Genes Dev. 21:2511–2524.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rocha R, Soro I, Leitão A, Silva ML and
Leão M: Moyamoya vascular pattern in Alagille syndrome. Pediatr
Neurol. 47:125–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Joutel A, Corpechot C, Ducros A, et al:
Notch3 mutations in CADASIL, a hereditary adult-onset condition
causing stroke and dementia. Nature. 383:707–710. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Louvi A and Artavanis-Tsakonas S: Notch
and disease: a growing field. Semin Cell Dev Biol. 23:473–480.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
31
|
Halim AX, Johnston SC, Singh V, et al:
Longitudinal risk of intracranial hemorrhage in patients with
arteriovenous malformation of the brain within a defined
population. Stroke. 35:1697–1702. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reedijk M, Odorcic S, Chang L, et al:
High-level coexpression of JAG1 and NOTCH1 is observed in human
breast cancer and is associated with poor overall survival. Cancer
Res. 65:8530–8537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weijzen S, Rizzo P, Braid M, et al:
Activation of Notch-1 signaling maintains the neoplastic phenotype
in human Ras-transformed cells. Nat Med. 8:979–986. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Gao X, Liu J, et al: Differential
Notch1 and Notch2 expression and frequent activation of Notch
signaling in gastric cancers. Arch Pathol Lab Med. 135:451–458.
2011.PubMed/NCBI
|
|
35
|
Murphy PA, Lu G, Shiah S, Bollen AW and
Wang RA: Endothelial Notch signaling is upregulated in human brain
arteriovenous malformations and a mouse model of the disease. Lab
Invest. 89:971–982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liebler SS, Feldner A, Adam MG, Korff T,
Augustin HG and Fischer A: No evidence for a functional role of
bi-directional Notch signaling during angiogenesis. PLoS One.
7:e530742012. View Article : Google Scholar : PubMed/NCBI
|