Introduction
Cholera has been widely studied over the past
several decades (1). Following the
development of gene recombination technology and awareness of the
importance of intestinal immunity, orally recombinant live cholera
vaccines have been investigated (2). A clinical episode of cholera induces
long lasting protection (at least three years) (3). Therefore, an orally recombinant live
vaccine that simulates natural cholera infection and, thus,
protection is warranted.
At present, numerous orally administered cholera
vaccines are being investigated in large-scale clinical studies
worldwide (4,5). However, the residual virulence of the
vaccine strains has affected the clinical application of these
vaccines. The three key virulence factors of Vibrio cholerae
are the cholera toxin (CTX) genetic element, the Vibrio
pathogenicity island (VPI) and the repeats in toxin (RTX) cluster
(6). CTX, RTX and the toxin-linked
cryptic element (TLC) are the chief virulence gene clusters and are
grouped together. Thus, they are able to be deleted together
through homologous recombination in order to construct a suitable
candidate vaccine strain.
The O1 or O139 groups of cholera vaccines do not
exhibit cross-protection. Thus, it is necessary to construct a
special or bivalent vaccine. In the present study, a V.
cholerae serogroup O139 vaccine was constructed and evaluated
using genetic engineering.
Materials and methods
Bacterial strains, plasmids and cultural
conditions
The live attenuated cholera vaccine strain utilized
in this study was based on the V. cholerae serogroup
O139-ZJ9693, a virulent strain. The bacterial strains and plasmids
used in this study are described in Table I (7,8).
Sucrose medium (SM; 1% tryptone, 0.5% yeast extract, 1.5% agar
powder and 10% sucrose) and AKI medium (1.5% peptone, 0.4% yeast
extract, 0.5% NaCl and 0.3% NaHCO3) were used in the
study. Antibiotics were utilized as follows: Ampicillin (Amp), 100
μg/ml and chloromycetin (Cm), 30 μg/ml for Escherichia. coli
and 15 μg/ml for V. cholerae. These reagents were purchased
from Oxoid (Oxoid, Ltd., Basingstoke, Hampshire, UK).
 | Table IBacterial strains and plasmids used in
the present study. |
Table I
Bacterial strains and plasmids used in
the present study.
| Strains and
plasmids | Characterization | Source |
|---|
| Vibrio
cholerae |
| N16961 | V. cholerae
O1El Tor, CTXΦ+ | Laboratory Medicine
Center |
| O139-ZJ9693-1 | ZJ199693
ΔTLC-CTX-RTX: :cat | Present study |
| NFYY101 | ZJ199693
ΔTLC-CTX-RTX: :ctxB+rstR | Present study |
| Escherichia
coli |
| JM109 | recA1supE44 endA1
hsdR17thi Δ(Lac-proAB) F′ (traD36
proAB+ LacIq LacZ ΔM15) | Laboratory Medicine
Center |
| SM10λpir | supE, recA:
:RP4-2-Tc: :Mu, Kmrλpir | Reference (7) |
| Plasmids |
| pUC18 | Clone vector,
oriMB1, lacZ+, Ampr | Laboratory Medicine
Center |
| pUC18-TLCup | pUC18 : :TLCup,
Ampr | Present study |
|
pUC18-TLCup-RTXdown | pUC18-TLCup:
:RTXdown, Ampr | Present study |
|
pUC18-TLCup-cat-RTXdown |
pUC18-TLCup-RTXdown: :cat,
Ampr, Cmr | Present study |
| pDS132 | Suicide plasmid,
mob, ori, sacB, Cmr | Reference (8) |
|
pDS132-TLCup-cat-RTXdown | pDS132:
:TLCup-cat-RTXdown, Cmr | Present study |
|
pUC18-TLCup-rstR-ctxB-RTXdown |
pUC18-TLCup-RTXdown: : rstR,
ctxB, Ampr | Present study |
| pCVD442 | Suicide plasmid,
mob, ori, bla, sacB, IS1,
Ampr | Reference (8) |
|
pCVD442-TLCup-rstR-ctxB-RTXdown | pCVD442:
:TLCup-rstR-ctxB-RTXdown, Amprr | Present study |
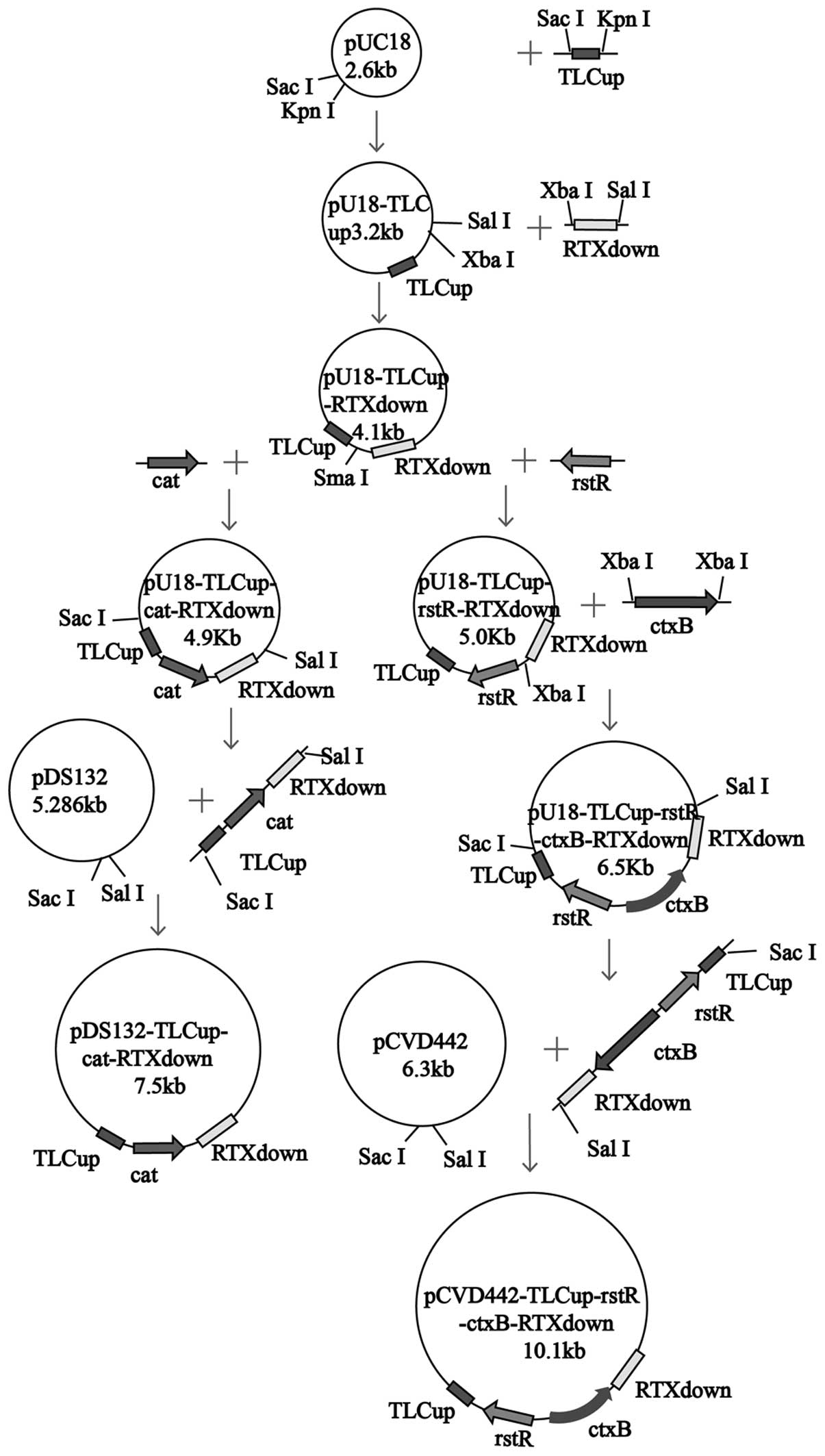
Construction of the O139 vaccine
candidate
The construction process for the O139 vaccine
candidate consisted of two steps. Initially, recombinant suicide
plasmids were constructed using genetic engineering (Fig. 1). Polymerase chain reaction (PCR)
was performed using the primer pairs stated in Table II to amplify the corresponding
gene fragments, and plasmids treated with specific enzymes were
utilized to construct recombinant plasmids.
pDS132-TLCup-cat-RTXdown and
pCVD442-TLCup-rstR-ctxB-RTXdown served as the
recombinant suicide plasmids.
 | Table IIPrimers used in the present
study. |
Table II
Primers used in the present
study.
| Primer | Primer sequence
(5′-3′) | RE site |
|---|
| TLCup-F | CAGGAGCTCATCCGCAACGTATTCCCACACC | SacI |
| TLCup-R | TGGGGTACCTGCTCCGAGTTATTTCGAAACC | KpnI |
| RTXdown-F | GCTCTAGATGACTCATGACCCAATG | XbaI |
| RTXdown-R | ACGCGTCGACATCACACGTCGTTTATC | SalI |
| cat-F |
CGTAGCACCAGGCGTTTAAG | - |
| cat-R |
GATCGGCACGTAAGAGGTTC | - |
| rstR-F |
CCGAATTCACTCACCTTGTATTCG | - |
| rstR-R |
CGGAATTCTCGACATCAAATGGCATG | - |
| ctxB-F |
AGTTCCATGGGGCAGATTCTAGACCTC | XbaI |
| ctxB-R | GATCTAGACGGTTGCTTCTCATCATCG | XbaI |
Following the construction of the recombinant
suicide plasmids, a vaccine candidate was constructed by homologous
recombination (Fig. 2).
SM10λpir carrying pDS132-TLC up-cat-RTXdown was
utilized as the donor strain to transfect O139-ZJ9693.
Transconjugants were propagated on lysogeny broth (LB) agar plates
overnight at 37°C, and then cultured on SM containing Cm. The
recombinant strain, O139-ZJ9693-1, was thereby obtained.
A process similar to the aforementioned method was
utilized to construct the NFYY101 candidate. O139-ZJ9693-1 was
utilized as the recipient bacterium to transfect with
SM10λpir carrying
pCVD442-TLCup-rstR-ctxB-RTXdown. The transconjugants
were cultured on SM and then streaked on general and Cm-resistant
medium, respectively. The colony that exhibited growth in general
medium only, as identified by PCR, was the O139-ZJ9693-2 candidate
and was then named NFYY101.
Monosialotetrahexosylganglioside
(GM1)-ELISA
Cholera toxin B subunit (CTB) protein exhibits the
ability to bind GM1. Thus, CTB expression of the recombinant strain
that re-acquired ctxB can be detected by ELISA (R&D
Systems Inc., Minneapolis, MN, USA). CTB extraction, preparation
and analysis were conducted as described previously (9). A 96-well plate was coated with 100 μl
GM1 (2 μg/ml; Sigma, St. Louis, MO, USA) per well, incubated
overnight at 4°C, and then washed three times with
phosphate-buffered saline (PBS)-Tween-20. Each well was blocked
with 3% bovine serum albumin (300 μl/well) for 1 h at 37°C and then
washed as described for the previous step. The supernatant of the
cell lysates of JM109, O139-ZJ9693, O139-ZJ9693-1 and NFYY101 was
added to triplicate wells (200 μl/well), and the plate was
incubated at 37°C for 1 h and then washed as described in the
previous step.
The plate was then incubated with a 200 μl 1:1,000
dilution of polyclonal mouse anti-CT antiserum (Sigma) per well for
1 h at 37°C and then washed as described for the previous step.
This step was repeated following addition of a 1:1,000 dilution of
horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G.
Tetramethyl benzidine was added (100 μl per well). After 15–30 min,
the reaction was blocked by the addition of 2 M
H2SO4. The absorption was assessed at 492 nm.
Results were considered positive when the ratio of the sample to
the control was ≥2.
Cytotoxic effect assay
Fresh mammalian Hep-2 cells [105 colony
forming units (CFU)/ml] were added to a 24-well cell culture plate,
and fresh RPMI-1640 (Gibco) containing 10% fetal bovine serum
(Gibco) without antibiotics was added. N16961, O139-ZJ9693 and
NFYY101 were cultured overnight at 37°C, washed twice with PBS and
diluted to achieve 109 CFU/ml. This was followed by
addition of 10 μl liquid containing bacteria to the wells of the
cell culture plate (three wells per strain) and the plate was
incubated at 37°C with 5% CO2 for 2 h. The cytotoxic
effects were then observed and documented.
Ligated ileal loop toxin test in
rabbits
N16961, O139-ZJ9693, O139-ZJ9693-1 and NFYY101 were
cultured overnight in LB. In total, 10 adult New Zealand White
rabbits (weight, 2 kg; Beijing Vital River Laboratory Animal
Technology Co., Ltd., Beijing, China) were fasted (water ad
libitum) for 24 h, and then their abdomens were opened. The
ilea of the rabbits were tied into 4–5-cm-long loops with 1-cm
intervals, and the four above strains (106 CFU) and
normal saline were injected into each loop, respectively. Saline
solution was used for the negative control and N16961 was used as
the positive control. The rabbits were sacrificed following closure
of the abdomen for 16–18 h. Congestion and effusion in the ileum
segments were then assessed. The present study was approved by the
Medical Ethics Committee of Nanfang Hospital, Southern Medical
University (Guangzhou, Guangdong, China) and conducted in
compliance with the Declaration of Helsinki.
Results
Construction and characterization of
O139-ZJ9693-1 and NFYY101
The O139-ZJ9693-1 and NFYY101 serum agglutination
tests were positive and the biochemical reactions of each were
identical to those of O139-ZJ9693. TLCup-cat-RTXdown was
detected by PCR and confirmed by sequencing in O139-ZJ9693-1;
rstR and ctxAB were not detected in the strain.
TLCup-rstR-ctxB-RTXdown of NFYY101 was
detected by PCR and sequenced. The sequencing results were used to
perform sequence alignment with rstR and ctxB of
N16961; the open reading frame consistency was verified for both.
These findings indicated that rstR and ctxB were
successfully cloned and integrated into the chromosome of the
vaccine candidate strain, NFYY101, as these genes are conserved in
V. cholerae.
Results of ligated ileal loop toxin
test
CT is encoded by the ctxAB gene and is the
most important factor to cause cholera. The presented study
demonstrated that the ileal loop containing N16961 and O139-ZJ9693
strains exhibited marked congestion and effusion, whereas no
similar effect appeared in any of the other ileal loop specimens
(Fig. 3). This phenomenon
indicated that the deletion mutant O139-ZJ9693-1 and vaccine
candidate NFYY101 were attenuated and that the chief toxin
ctxAB gene was deleted.
Cytotoxic effect assay
Cell rounding was observed following co-incubation
of Hep-2 cells with N16961 and O139-ZJ9693, whereas no similar
phenomena were observed following addition of NFYY101 and PBS
(Fig 4). This finding correlates
with the theoretical cytotoxicity of rtxA and toxin
activation of rtxC.
Determination of ctxB expression by
GM1-ELISA
GM1-ELISA analysis revealed that CTB was expressed
in the O139-ZJ9693, JM109 carrying
pUC18-TLCup-rstR-ctxB-RTXdown and NFYY101 strains,
and absent in O139-ZJ9693-1 and JM109 (Table III). Results of the GM1-ELISA
analysis indicated that ctxB was expressed in the NFYY101
vaccine candidate.
 | Table IIITest results by microplate
reader. |
Table III
Test results by microplate
reader.
| Samples |
OD492nm | P/N |
|---|
| O139-ZJ9693 | 0.396±0.090 | 5.74 |
| NFYY101 | 0.368±0.051 | 5.33 |
|
JM109-pUC18-TLCup-rstR-ctxB-RTXdown
(1:5) | 0.409±0.011 | 5.93 |
| O139-ZJ9693-1 | 0.072±0.010 | 1.04 |
| JM109 | 0.065±0.003 | 0.942 |
| PBS | 0.069±0.002 | - |
Discussion
Gene-knockout technology was first developed in the
late 1980s and has exhibited the potential for broad application in
the life sciences (10).
Homologous recombination is one method of gene-knockout. Factors
that affect the efficiency of homologous recombination include the
length of the two end fragments of the target gene, the presence of
the vector linearization-knockout and the position and structure of
the target genes (11). The length
of TLCup (562 bp) and RTXdown (888 bp) facilitates PCR and
successful fusions. Using the ‘upstream fragment-inserted
gene-downstream fragment’ model allows for the avoidance of the
polar effects of strain and inactivates the target genes completely
(12).
The suicide plasmid is an effective carrier that
builds a seamless mutant through homologous recombination,
precisely positioning the integration site using homologous
fragments at the two ends of the target gene. The pDS132 in the
present study was from pCVD442, with the insertion sequence 1
element removed and the bla gene replaced with the
cat gene. The recombination efficiency of pDS132 is higher
than that of pCVD442. The sacB gene contained in pDS132
encodes fructosan-sucrase, which induces sucrose to hydrolyze into
fructosan, a toxic compound fatal to a number of Gram-negative
bacteria. Thus, the deletion mutant was selected in the present
study using SM supplemented with chlormycetin. This strain is able
to grow on SM with chlormycetin as cat specifically replaces
the target gene by homologous recombination and the pDS132 vector
is then ablated.
The cat gene replaced the long segment (39
kb) between TLC and RTX; thus, the TLC, CTX and RTX clusters were
deleted from O139-ZJ9693 through homologous recombination.
O139-ZJ9693-1 was obtained using the aforementioned process.
According to previous studies of O1, the CTX element is derived
from the bacteriophage CTXΦ and is likely to be transferred among
diverse V. cholerae strains in the environment (13). CTXΦ is composed of two parts, a
core region of 4.6 kb and a repeat sequence region. The core region
harbors six genes, including the ctxAB gene, which encodes
the CT. TLC is located upstream of the CTX element, and its
function is associated with the acquisition and duplication of
CTXΦ. RTX is located downstream of the CTX element and is
associated with cytotoxicity; its activity is independent of CTX,
as it belongs to the RTX toxin family.
The construction of the suicide plasmid was
continued by replacing the cat gene with the rstR and
ctxB genes in O139-ZJ9693-1. O139-ZJ9693-1 carries Cm
resistance and pCVD442 exhibits ampicillin resistance, whereas
pDS132 exhibits Cm resistance, therefore the recombinant suicide
plasmid pCVD442-TLCup-rstR-ctxB-RTXdown was
constructed and then conjugated with O139-ZJ9693-1. The conjugant
was streaked onto SM, and then the strains were transferred to the
general medium and to the plate with Cm. The colony growing
exclusively in the general medium was the candidate strain,
NFYY101; it was identified and confirmed by PCR.
The rstR gene in the RS2 region of the CTX
element encodes RstR, a repressed protein that mediates CTXΦ
immunity via repression of rstA and rstB expression.
The immunity maintains the lysogenized state of the strain;
however, it represses the duplication of DNA in superinfection with
CTXΦ (14–16). This type of CTXΦ-immunity is
biotype-specific (14), therefore
the present study utilized the rstR gene of El Tor type to
prevent El Tor-derived CTXΦ (CTXETΦ) in the globally epidemic OEl
Tor strain from potentially infecting the novel vaccine strain that
was developed. This step enhanced the biological safety of the
candidate vaccine strain.
The virulent CTB subunit encoded by ctxB
conjugates ganglioside GM1 with close affinity in cellular
membranes of karyocytes, including epithelial cells,
lympholeukocytes and antigen-presenting cells (APCs). CTB is able
to adjust the APC reaction, the T-cell reaction, antibody
production and immune activation, and it also exhibits high
immunogenicity that can induce the production of antibodies against
CTB to efficiently prevent cholera. Therefore, it is necessary to
integrate ctxB into the cholera vaccine (17).
Virulent V. cholerae carrying CT can cause
congestion and effusion in the ileal loop of rabbits (18). The toxin test was used to
investigate the effect of homologous recombination. The reaction of
the ileal loop to CT is highly variable among individuals; thus, a
minimum of two rabbits was necessary. A control in the ligated
ileal loop of every rabbit was also used. The test results
indicated that O139-ZJ9693-1 and NFYY101 were markedly attenuated,
and that the chief toxin gene ctxAB was deleted from the two
strains.
In the TLC-CTX-RTX clusters, the
independent-activity RTX toxin encoded by the RTX gene, one of the
virulence factors, causes actin cross-linking and cell rounding
(19). In the cytotoxic effect
assay, N16961 and O139-ZJ9693 caused Hep-2 rounding, whereas
O139-ZJ9693-1 did not. This result indicated that O139-ZJ9693-1
without the RTX cluster loses cytotoxicity against Hep-2.
It has been previously reported that RTX in V.
cholerae is only excreted by the type I secretory system during
the growth phase, but not in the dormant phase (20). Thus, the cytotoxic effect assay in
the present study required fresh strains able to react with Hep-2.
Additionally, as N16961 grows faster than O139-ZJ9693, the cell
rounding is more obvious in N16961 during the same period of
culture.
In the present study, NFYY101, a novel V.
cholerae O139 live attenuated vaccine candidate strain was
constructed and characterized. Together, the results demonstrate
that NFYY101 is attenuated and ctxB is highly expressed in
the recombinant chromosomal DNA. No enterotoxigenic or cytotoxic
effects were observed in this strain. A recombinant strain with the
protective antigen genes that replaced virulence-associated genes
was successfully constructed; this candidate strain could
potentially be utilized to further evaluate immune response.
Further evaluation of the stability of the vaccine is warranted, as
well as further investigation regarding its ability to protect
against CTXΦ infection in vivo (21,22).
Acknowledgements
This study was supported by funds from the National
Natural Science Foundation of China (grant no. 30470099).
References
|
1
|
Harris JB, LaRocque RC, Qadri F, et al:
Cholera. Lancet. 379:2466–2476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leung DT, Chowdhury F, Calderwood SB, et
al: Immune responses to cholera in children. Expert Rev Anti Infect
Ther. 10:435–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harris AM, Bhuiyan MS, Chowdhury F, et al:
Antigen-specific memory B-cell responses to Vibrio cholerae
O1 infection in Bangladesh. Infect Immun. 77:3850–3856. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qadri F1, Chowdhury MI, Faruque SM, et al:
Peru-15, a live attenuated oral cholera vaccine, is safe and
immunogenic in Bangladeshi toddlers and infants. Vaccine.
25:231–238. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trach DD, Cam PD, Ke NT, et al:
Investigations into the safety and immunogenicity of a killed oral
cholera vaccine developed in Viet Nam. Bull World Health Organ.
80:2–8. 2002.PubMed/NCBI
|
|
6
|
Davis BM, Moyer KE, Boyd EF and Waldor MK:
CTX prophages in classical biotype Vibrio cholerae:
functional phage genes but dysfunctional phage genomes. J
Bacteriol. 182:6992–6998. 2000.PubMed/NCBI
|
|
7
|
Simon R, Priefer U and Pühler A: A broad
host range mobilization system for in vivo genetic engineering:
transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol.
1:784–791. 1983. View Article : Google Scholar
|
|
8
|
Philippe N, Alcaraz JP, Coursange E,
Geiselmann J and Schneider D: Improvement of pCVD442, a suicide
plasmid for gene allele exchange in bacteria. Plasmid. 51:246–255.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin W, Fullner KJ, Clayton R, et al:
Identification of a vibrio cholerae RTX toxin gene cluster
that is tightly linked to the cholera toxin prophage. Proc Natl
Acad Sci USA. 96:1071–1076. 1999.
|
|
10
|
Smithies O, Gregg RG, Boggs SS, et al:
Insertion of DNA sequences into the human chromosomal beta-globin
locus by homologous recombination. Nature. 317:230–234. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fullner KJ and Mekalanos JJ: In vivo
covalent cross-linking of cellular actin by the Vibrio
cholerae RTX toxin. EMBO J. 19:5315–5323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boardman BK, Meehan BM and Fullner
Satchell KJ: Growth phase regulation of Vibrio cholerae RTX
toxin export. J Bacteriol. 189:1827–1835. 2007. View Article : Google Scholar
|
|
13
|
Karaolis DK, Somara S, Manevel DR Jr, et
al: A bacteriophage encoding a pathogenicity island, a type-IV
pilus and a phage receptor in cholera bacteria. Nature.
399:375–379. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chattopadhyay K and Banerjee KK: Unfolding
of Vibrio cholerae hemolysin induces oligomerization of the
toxin monomer. J Biol Chem. 278:38470–38475. 2003.PubMed/NCBI
|
|
15
|
Karunasagar I, Rivera I, Joseph B, Kennedy
B, Shetty VR, Huq A, Karunasagar I and Colwell RR: ompU genes in
non-toxigenic Vibrio cholerae associated with aquaculture. J
Appl Microbiol. 95:338–343. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Olivier V, Haines GK III, Tan Y and
Satchell KJ: Hemolysin and the multifunctional autoprocessing RTX
toxin are virulence factors during intestinal infection of mice
with Vibrio cholerae El Tor O1 strains. Infect Immun.
75:5035–5042. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang W, Wang S, Yu F, Zhang L, Qi G, Liu
Y, Gao S and Kan B: Construction and evaluation of a safe, live,
oral Vibrio cholerae vaccine candidate, IEM108. Infect
Immun. 71:5498–5504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Spangler BD: Structure and function of
cholera toxin and the related Escherichia coli heat-labile
enterotoxin. Microbiol Rev. 56:622–647. 1992.PubMed/NCBI
|
|
19
|
Pearson GD, Woods A, Chiang SL and
Mekalanos JJ: CTX genetic element encodes a site-specific
recombination system and an intestinal colonization factor. Proc
Natl Acad Sci USA. 90:3750–3754. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kimsey HH and Waldor MK: CTXphi immunity:
application in the development of cholera vaccines. Proc Natl Acad
Sci USA. 95:7035–7039. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan M, Liu G, Diao B, Qiu H, Zhang L,
Liang W, Gao S and Kan B: A Vibrio cholerae serogroup O1
vaccine candidate against CTX ET Phi infection. Vaccine.
25:4046–4055. 2007.PubMed/NCBI
|
|
22
|
Ledón T, Ferrán B, Pérez C, Suzarte E,
Vichi J, Marrero K, Oliva R and Fando R: TLP01, an mshA mutant of
Vibrio cholerae O139 as vaccine candidate against cholera.
Microbes Infect. 14:968–978. 2012.PubMed/NCBI
|