Introduction
In eukaryotic cells, the process of proteolysis is
executed by a diverse group of enzymes known as proteases, and the
ubiquitin-proteasome pathway (UPP) is the most significant
intracellular proteolytic pathway. The degradation process mediated
by the UPP involves two steps: i) The target proteins are
ubiquitinated by multiple ubiquitin molecules; and ii) the tagged
proteins are recognized and degraded by the major ATP-dependent
protease, the 26S proteasome complex (1). The 26S proteasome is a biological
macromolecule containing a 20S catalytic core and two 19S
regulatory complexes (2).
Selective degradation of proteins by the UPP is a critical
determinant for maintaining cellular homeostasis. Numerous
proteasome target proteins are involved in the regulation of cancer
cell proliferation, differentiation and apoptosis (3,4).
Therefore, the aberrant degradation of oncoproteins or tumor
suppressor proteins can result in uncontrolled cell growth in
numerous cancer types.
In vitro and in vivo studies have
demonstrated that inhibition of proteasomes is an effective
anticancer therapeutic approach. Bortezomib, one of the first
proteasome inhibitors, which was designed to inhibit the activity
of the 26S proteasome by binding to the N-terminal threonine
residues at the active site of the catalytic region (5), was shown to be efficient against a
variety of malignancies, including myeloma, chronic lymphocytic
leukemia and certain solid tumors (6–8).
Osteosarcoma is the most common primary bone sarcoma and mostly
affects adolescent patients. Since osteosarcoma is metastatic and
highly aggressive, novel treatment strategies must be developed.
Bortezomib has been shown to suppress growth and induces apoptosis
in osteosarcoma cells and xenografts (8). The thiazole antibiotic thiostrepton,
which has been identified as a proteasome inhibitor in mammalian
tumor cells (9), induces apoptosis
in a wide variety of human cancer cell lines, including
osteosarcoma cells, on its own or in combination with bortezomib
(5,10). These data strongly suggest that
proteasome inhibition may also be effective as an adjuvant to
current treatment regimens for osteosarcoma.
NIN1/RPN12 binding protein 1 homolog (NOB1),
which was firstly identified in Saccharomyces cerevisiae,
encodes the essential protein Nin one binding protein (NOB1p)
(11). As a chaperone protein,
NOB1p joins the 20S proteasome with the 19S regulatory particle in
the nucleus and facilitates the maturation of the 20S proteasome,
thereby favoring the completion of 26S proteasome biogenesis
(12). Furthermore, NOB1,
along with five other genes, has been used as a diagnostic marker
discriminating chronic phase from blast crisis chronic myelogenous
leukemia (13). RNA interference
(RNAi)-mediated downregulation of NOB1 suppresses the growth
of human ovarian cancer cells and hepatocellular carcinoma cells
(14,15). In the present study, short hairpin
RNA (shRNA) was employed to knock down NOB1 in osteosarcoma
cells, and the effects of NOB1 silencing on cell growth and
migration were explored.
Materials and methods
Materials
SF-86, Saos-2, MG63, SW1353 and U2OS human
osteosarcoma cells and HEK-293T cells were obtained from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). TRIzol® reagent was purchased from Invitrogen
Life Technologies (Carlsbad, CA, USA). The SYBR® Green
Real-Time PCR assay kit was obtained from Applied Biosystems, Inc.
(Beijing, China). Rabbit anti-NOB1p antibody was purchased from
Sigma-Aldrich (St. Louis, MO, USA). Anti-GAPDH antibody and goat
anti-mouse immunoglobulin G conjugated with horseradish peroxidase
antibody were obtained from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA). Enhanced chemiluminescence reagents were purchased
from Amersham Life Science (Arlington Heights, IL, USA).
Cell culture
Cells were grown in Dulbecco’s Modified Eagle’s
medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine
serum (FBS; Invitrogen Life Technologies) at 37°C under 5%
CO2.
Lentivirus production and lentiviral
transduction
shRNA targeting the NOB1
(CCGGGCTGAACAATTTCAGTCATT TCTCGAGAAATGACTGAAATTGTTCAGCTTTTTG) and
negative control (TTCTCCGAACGTGTCACGT) sequences were cloned into
pFH-L (Shanghai Hollybio, Shanghai, China). The reconstructed
pFH-L-shNOB1 and pFH-L-shCon vectors were then
co-transfected into HEK-293T cells together with the helper
plasmids pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio) to generate
lentiviruses. After 96 h of incubation, the lentiviral particles
were harvested from the supernatant by ultracentrifugation
(16,17). The RNAi lentiviruses were referred
to as shNOB1 for the specific interference with the
NOB1 gene and shCon for the negative control. For lentiviral
transduction, 40% confluent Saos-2/U2OS osteosarcoma cells were
incubated with Lv-shNOB1 or Lv-shCon for 96 h, with a
replacement of the media 24 h after lentiviral treatment.
Quantitative polymerase chain reaction
(qPCR)
Saos-2 and U2OS cells were cultured in six-well
plates and then infected with the shNOB1 or shCon
lentiviruses. After 96 h of incubation, total RNA was isolated from
cultured cells using TRIzol® reagent and then cDNA was
synthesized from total RNA. Two sets of primers were used for PCR:
β-actin (ACTB) forward, 5′-GTGGACATCCGCAAAGAC-3′ and
reverse, 5′-AAAGGGTGTAACGCAACTA-3′; NOB1 forward,
5′-GAAAGAACAACGCCCTGGAG-3′ and reverse,
5′-CAGCCTTGAGATGACCTAAGC-3′. qPCR was performed according to the
manufacturer’s instructions (Applied Biosystems, Inc.). The
relative mRNA expression of NOB1 was calculated using the
2−ΔΔCt method (18).
Western blot analysis
Whole cell extracts were prepared with ice-cold cell
lysis buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.1% Triton X-100
and 0.1% SDS) and the protein concentration was determined using a
Bradford assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Protein extracts were separated using SDS-PAGE, transferred onto a
nitrocellulose membrane and incubated with anti-NOB1p and
anti-GAPDH antibodies. Immunodetection was performed using an
enhanced chemiluminescence western blotting kit (Amersham
Biosciences Inc., Piscataway, NJ, USA).
Cell proliferation assay
Cells collected from the three groups
(Lv-shNOB1, Lv-shCon and control) were trypsinized,
resuspended, seeded into 96-well plates at a density of 2,000 cells
per well and then incubated at 37°C for 96 h after lentiviral
treatment. The number of viable cells was assessed at indicated
time-points, when 20 μl MTT solution (5 mg/ml) was added into each
well. The plate was incubated for 4 h. The fixed plate was then
washed and 100 μl dimethyl sulfoxide was added. The absorbance of
each plate was measured at 595 nm using a spectrophotometer.
Colony formation assay
Following infection, cells in the three groups
(Lv-shNOB1, Lv-shCon and control) were seeded into a
six-well plate at a density of 300 cells per well and maintained at
37°C for 13 or 14 days (U2OS2 and Saos-2 cell lines, respectively).
The culture media were changed every 2–3 days. When the colonies
were formed, the plate was washed and fixed, stained with Giemsa
(Sigma Chemical Co.) for 10 min, and washed three times with
double-distilled H2O. The stained cells and colonies
(>50 cells/colony) were photographed and counted.
Cell cycle analysis
The cell cycle distribution (G0/G1, S or G2/M phase)
was characterized by differences in DNA content via flow cytometry.
Cells were collected by centrifugation at 404 × g for 5 min, washed
with phosphate-buffered saline (PBS) and fixed in ethanol. The
fixed cells were then resuspended in propidium iodide/RNase/PBS for
incubation in the dark (37°C, 30 min). The stained cells were
analyzed using the FACSCalibur™ II sorter and the CellQuest™
fluorescence-activated cell sorting system (BD Biosciences, San
Diego, CA, USA). The percentage of cells in each cell cycle phase
was analyzed. Each experiment was repeated three times and the
results are shown as the average.
Cell migration assay
The motility and migration of U2OS cells were
evaluated using the Transwell assay. Briefly, trypsinized U2OS
cells were transferred into the upper chambers of the Transwell
plates (8.0-μm pores, Corning Costar, Cambridge, MA, USA). The
lower chamber was filled with 500 ml DMEM supplemented with 10%
FBS. After 24 h at 37°C under 5% CO2/95% air, cell
migration was determined by counting cells in the bottom of the
membrane stained with crystal violet, and scored visually in five
random fields using light microscopy (magnification, ×100). In
addition, migrating cells were dissolved and quantified at 570 nm
using a spectrophotometer.
Statistical analysis
Data are expressed as the mean ± standard deviation
of at least three independent experiments. Statistical significance
was assessed using the Student’s t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Knockdown of NOB1 by the shRNA lentivirus
system in osteosarcoma cells
Expression levels of NOB1p in several osteosarcoma
cell lines were assessed using western blot analysis. NOB1p was
moderately expressed in Saos-2 and U2OS cells (Fig. 1A). A lentivirus-mediated RNAi
system was applied to specifically downregulate the expression of
NOB1. To ensure the lentiviral infection efficiency, the
expression of green fluorescent protein was detected using
fluorescence microscopy. Fig. 1B and
C show that, after 96 h of incubation, infection in Saos-2 and
U2OS cells was highly efficient (>80%). Knockdown efficiency was
determined using qPCR, which showed that the relative levels of
NOB1 mRNA transcripts were significantly decreased by almost
50% in the Lv-shNOB1 group as compared with those in the
Lv-shCon and Con groups in the two cell lines (Fig. 1D and E). These results demonstrated
that endogenous NOB1 expression was specifically inhibited
by the Lv-shNOB1 construct.
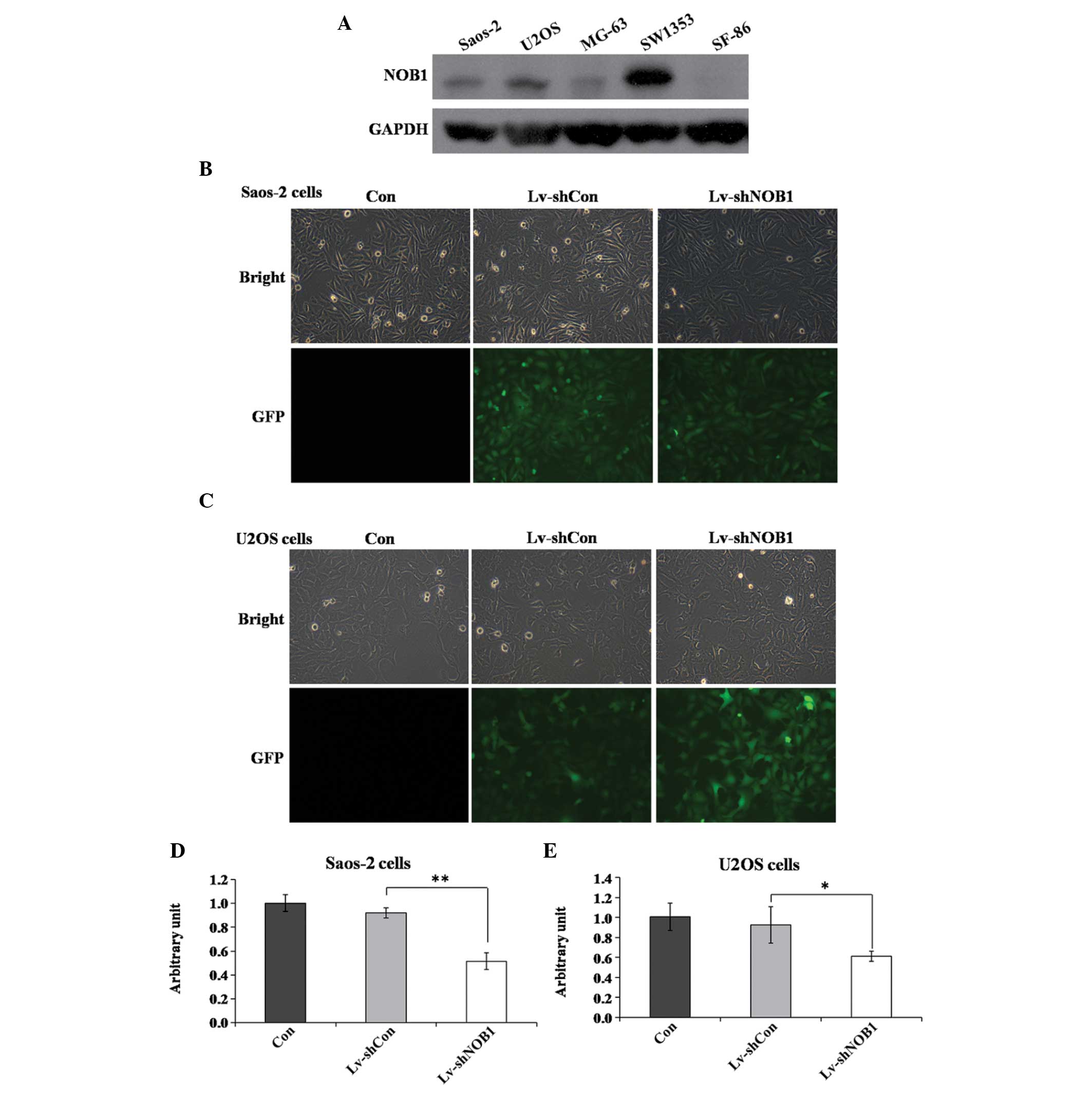 | Figure 1NOB1-knockdown by a
lentivirus-mediated RNA interference system. (A) Expression levels
of Nin one binding protein in five osteosarcoma cell lines (Saos-2,
U2OS, MG-63, SW1353 and SF-86) using western blot analysis. (B and
C) Fluorescence photomicrographs of (B) Saos-2 and (C) U2OS cells
infected by the lentivirus. Pictures were captured 96 h after
infection (magnification, ×100). (D and E) Identification of
NOB1-knockdown efficiency via quantitative polymerase chain
reaction in (D) Saos-2 and (E) U2OS cells. *P<0.05,
**P<0.01 compared with Lv-shCon. Con, no lentivirus
treatment; Lv-shCon, control lentivirus; Lv-shNOB1,
lentivirus containing short hairpin RNA targeting NOB1;
NOB1, NIN1/RPN12 binding protein 1 homolog (Saccharomyces
cerevisiae); GFP, green fluorescent protein. |
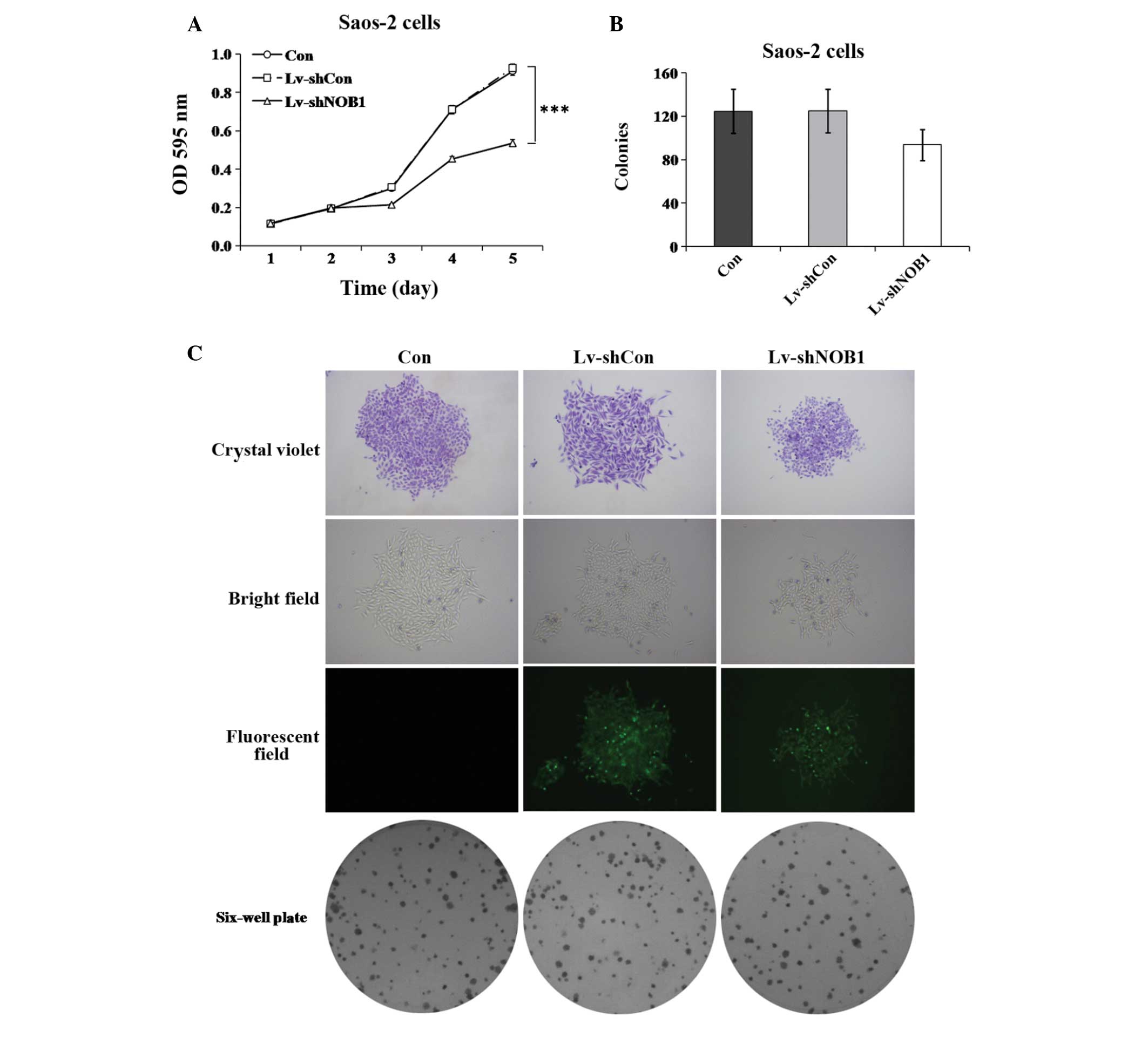
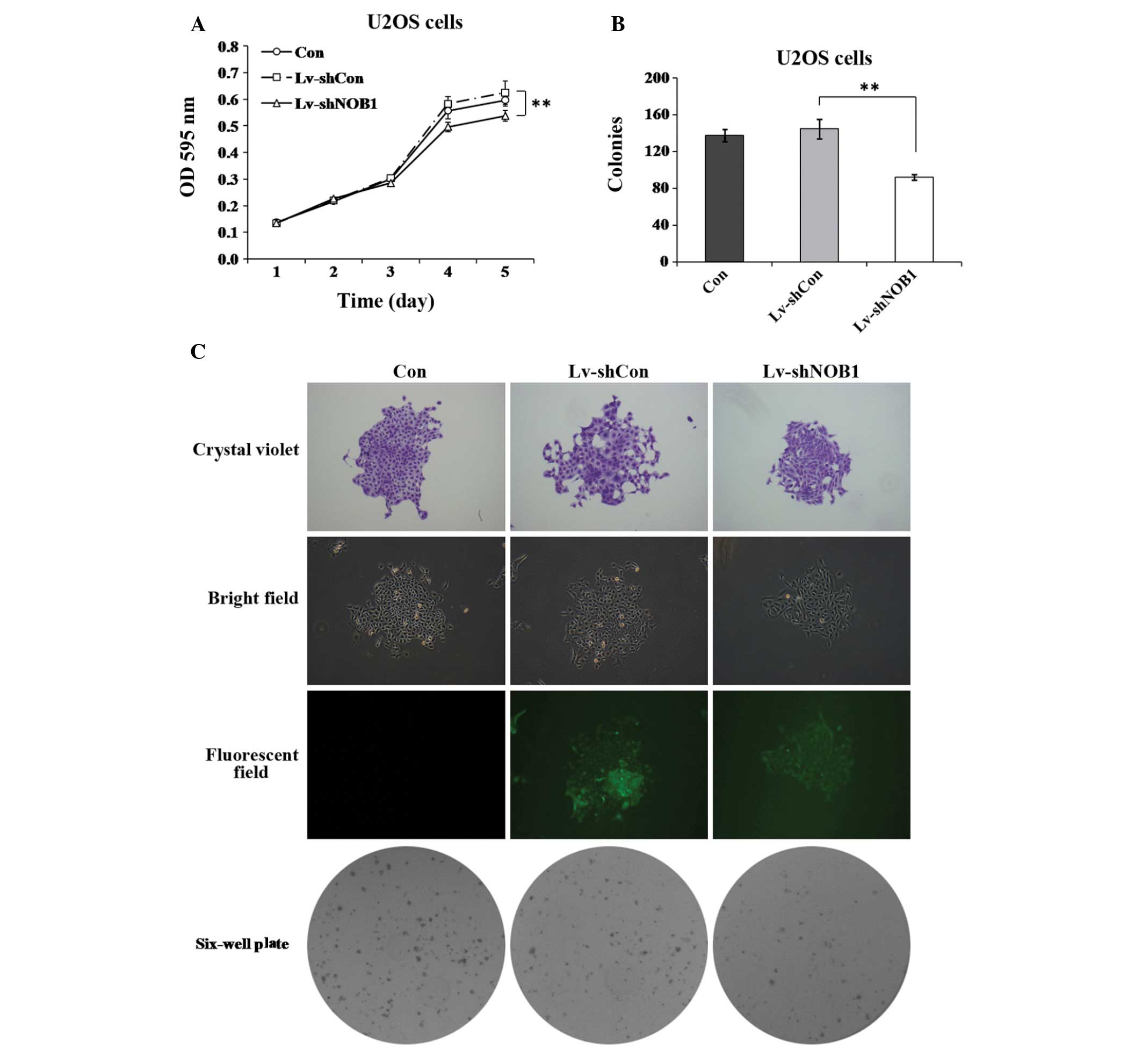
Effect of NOB1 silencing on proliferation
in human osteosarcoma cells
To further investigate the role of NOB1 in
regulating the proliferation of osteosarcoma cells, the MTT assay
and colony formation analysis were used. As shown in Fig. 2A, the proliferation of
Lv-shNOB1-treated Saos-2 cells at 96 h post-infection was
markedly inhibited as compared with that in the Lv-shCon and Con
groups (P<0.001). As shown in Fig.
2B and C, the colony formation ability of Saos-2 cells was also
slightly reduced by NOB1 inhibition (Lv-shNOB1, 94±14
colonies), as compared with that of the cells in the Lv-shCon
(125±20 colonies; P=0.1) or Con (125±20 colonies; P=0.1) groups. In
U2OS cells, similar results were obtained in the two assays
(Fig. 3), showing that
NOB1-knockdown can notably decrease the proliferation of
U2OS cells over a short or relatively long period of time. These
findings support the theory that NOB1 has an important role
in regulating the growth of osteosarcoma cells.
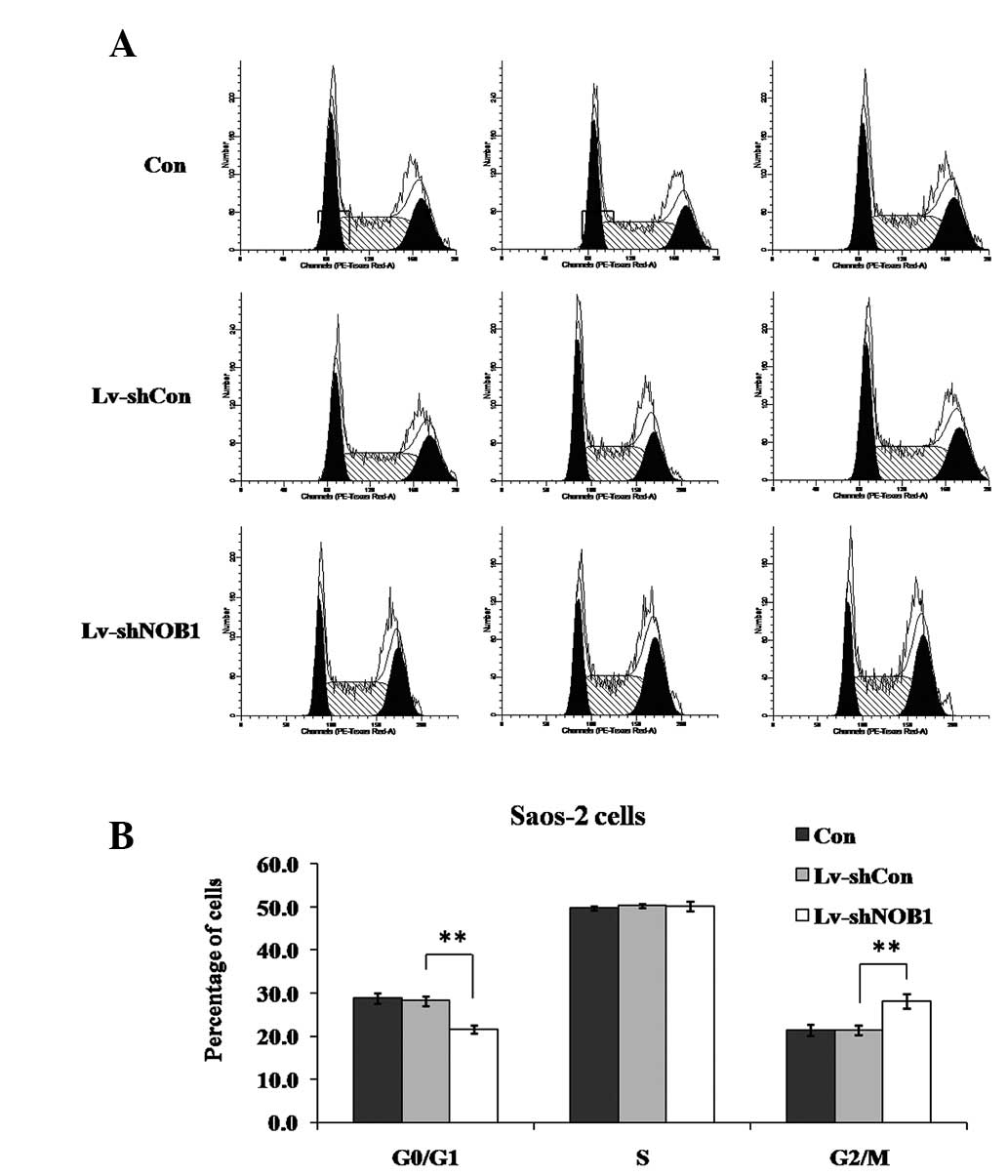
Effect of NOB1 silencing on the cell
cycle distribution in Saos-2 cells
To examine whether NOB1-knockdown suppressed
the growth of osteosarcoma cells through direct regulation of the
cell cycle, the cell cycle distribution following lentivirus
treatment was assessed. The cells were subjected to three different
treatments as described previously in the study (Lv-shNOB1,
Lv-shCon or Con), and the cell cycle distribution was analyzed
using flow cytometry (Fig. 4). In
Saos-2 cells, it was observed that, compared with Lv-shCon or Con,
Lv-shNOB1 significantly decreased the percentage of cells in
G0/G1 phase (P<0.01) and increased that in G2/M phase
(P<0.01), indicating that NOB1 suppression induced cell
cycle arrest at G2/M phase.
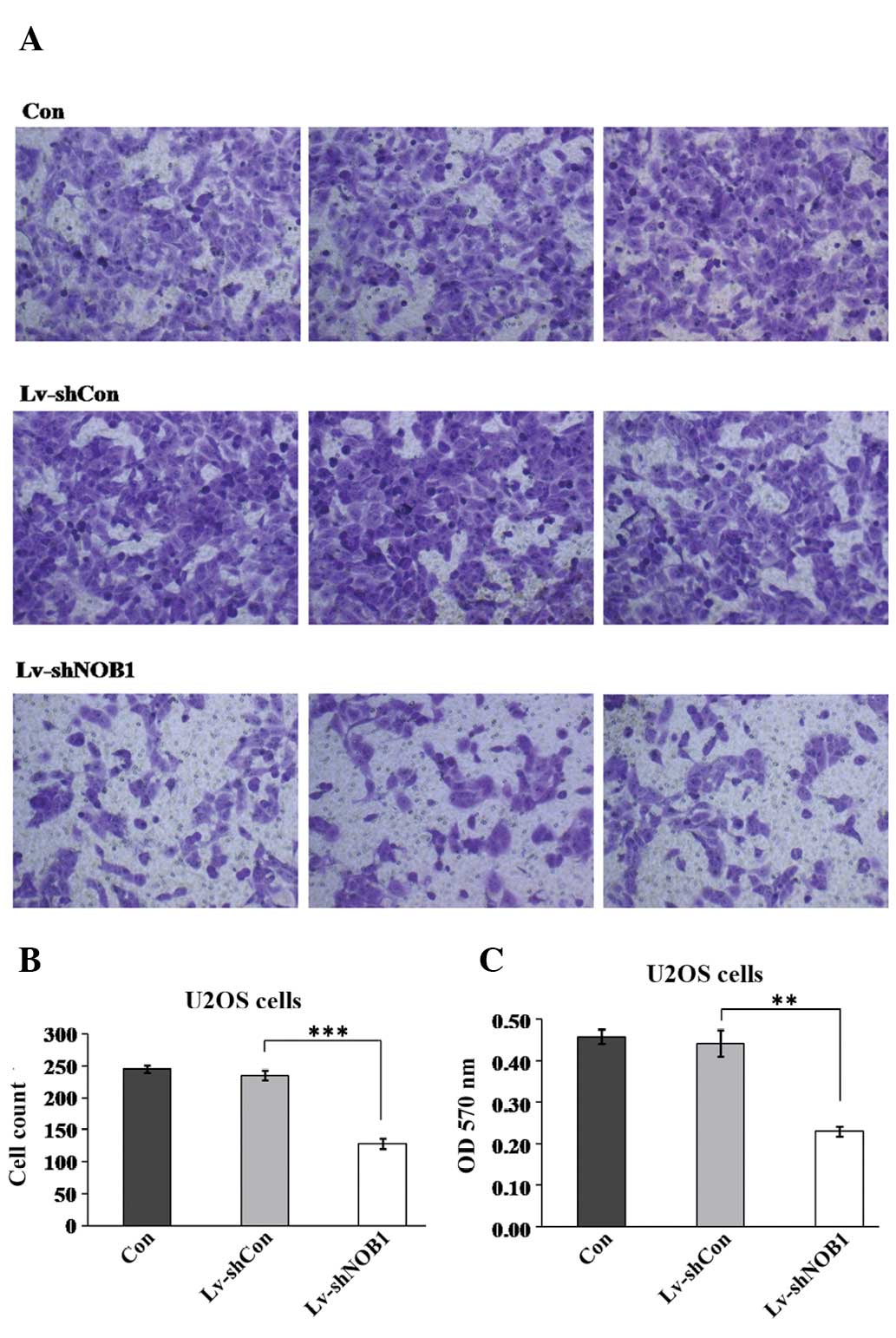
Effect of NOB1 silencing on cell
migration in U2OS cells
Cell migration is a critical step that occurs during
cancer progression. Therefore, the potential effect of NOB1
silencing in regulating U2OS cell migration was assessed using the
Transwell assay (Fig. 5). Fewer
cells in the Lv-shNOB1 group (128.2±8.2) migrated into the
lower filter as compared with the cells in the Lv-shCon (235.5±8.1)
and Con (245.3±5.8) groups. In addition, the crystal violet
staining intensity was significantly lower in the Lv-shNOB1
group (0.23±0.01) than that in the Lv-shCon (0.44±0.03) and Con
(0.46±0.02) groups. This assay indicated that NOB1-knockdown
strongly suppressed the migration of U2OS cells. Additionally, in
order to explore the underlying molecular mechanism of
Lv-shNOB1 suppressing the proliferation and migration of
osteosarcoma cells, the expression of several molecules, including
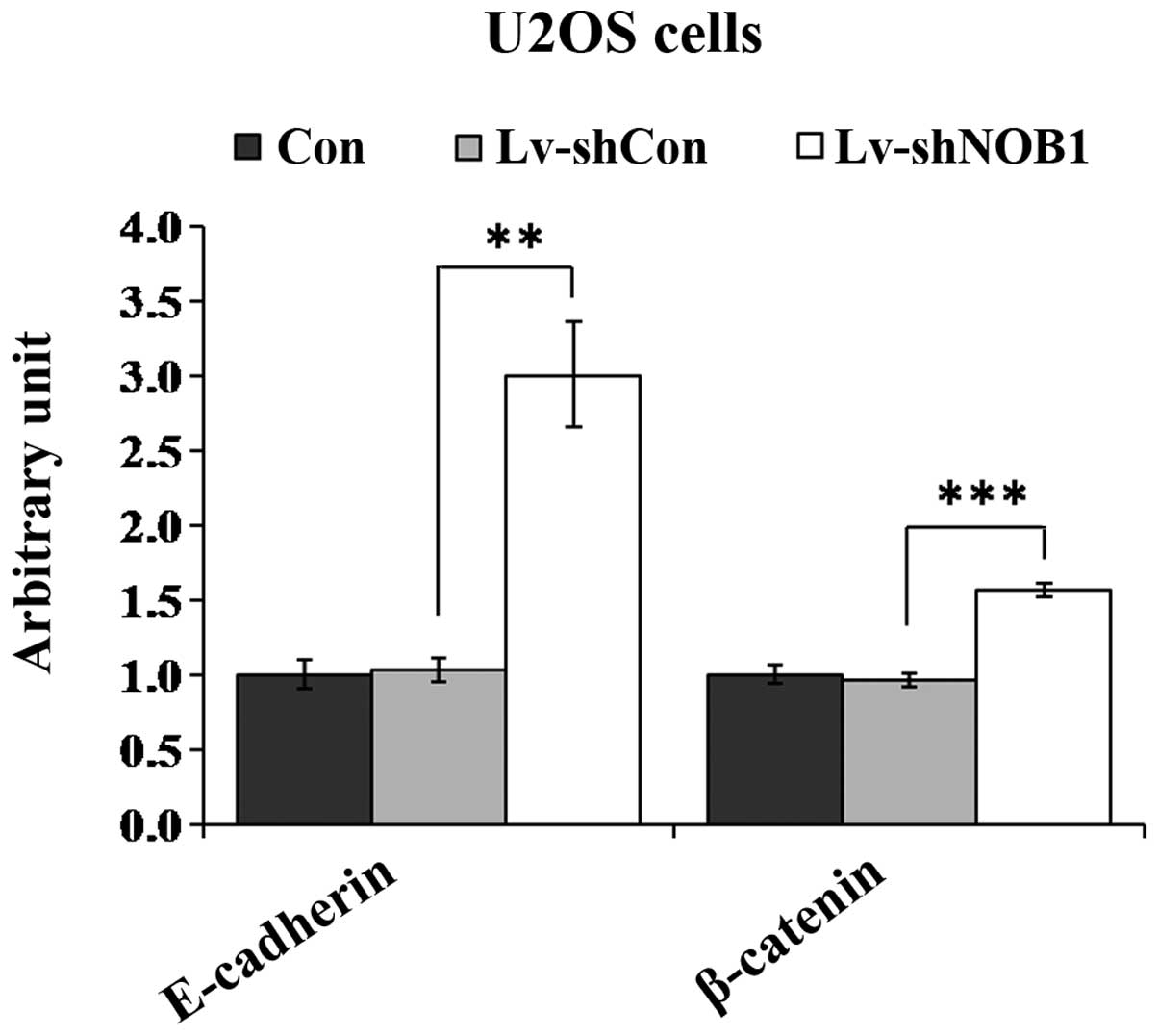
fibronectin, vimentin, N-cadherin, E-cadherin and β-catenin was
detected (data not shown). Downregulation of NOB1 was
associated with the increase expression of two regulators,
E-cadherin and β-catenin, as compared with expression in the Con or
Lv-con groups (Fig. 6). Thus, it
is likely that E-cadherin and β-catenin are involved in
Lv-shNOB1-mediated growth inhibition in human osteosarcoma
cells.
Discussion
NOB1 encodes a nuclear protein that regulates
the maturation of the 20S proteasome and favors 26S proteasome
biogenesis (12). Advances in the
investigation of the mechanisms underlying proteasome activity have
led to the exploration of proteasome inhibitors as effective drugs
against several human cancer and solid tumor types (6–8,10,19).
Several studies have demonstrated that bortezomib, alone or in
combination with other proteasome inhibitors, is by far the most
effective in the induction of apoptosis in osteosarcoma cells
(5), indicating that the
ubiquitin-proteasome complex (UPP) may have an important role in
osteosarcoma. However, the biological function and therapeutic
potential of NOB1, a key factor in the UPP and proteasome
complex, remain to be fully elucidated.
In the present study, a lenti-shRNA system was
applied, which effectively inhibited NOB1 expression at the
RNA and protein levels. NOB1-knockdown strongly suppressed
the growth of osteosarcoma cells and caused G2/M-phase arrest, as
confirmed by MTT, colony formation and cell cycle assays.
Furthermore, the absence of NOB1 inhibited osteosarcoma cell
motility and migration. It is of note that the expression levels of
E-cadherin and β-catenin were significantly increased when
NOB1 was downregulated. These two molecules have been
reported to be associated with the metastatic progression of
several types of cancer (20–24).
Loss of the tumor suppressor genes E-cadherin and β-catenin has
been suggested to enable metastasis by disrupting intercellular
contacts, which is an early step in metastatic dissemination
(21). E-cadherin is also known to
associate with a number of proteins, including three catenins (α, β
and p120), via its cytoplasmic domain, which links E-cadherin to
the actin cytoskeleton (21). Cai
et al (25) demonstrated
that the wingless-type MMTV integration site family (Wnt)/β-catenin
pathway is inactivated in osteosarcoma. Moreover, activation of the
Wnt/β-catenin pathway inhibits cell proliferation and promotes
osteogenic differentiation in osteosarcoma cells. The present
results indicate that NOB1 depletion may inhibit
osteosarcoma development by increasing E-cadherin and β-catenin
expression.
In conclusion, the present study reported the novel
finding that NOB1 inhibition is able to strongly suppress
cell growth and migration of human osteosarcoma cells. Therefore,
it is suggested that NOB1 may be a potential target in
developing specific UPP inhibitors for the treatment of
osteosarcoma.
References
|
1
|
Glickman MH and Ciechanover A: The
ubiquitin-proteasome proteolytic pathway: destruction for the sake
of construction. Physiol Rev. 82:373–428. 2002.PubMed/NCBI
|
|
2
|
Ferrell K, Wilkinson CR, Dubiel W and
Gordon C: Regulatory subunit interactions of the 26S proteasome, a
complex problem. Trends Biochem Sci. 25:83–88. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Frezza M, Schmit S and Dou QP: Targeting
the ubiquitin-proteasome pathway: an emerging concept in cancer
therapy. Curr Top Med Chem. 11:2888–2905. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yerlikaya A and Yöntem M: The significance
of ubiquitin proteasome pathway in cancer development. Recent Pat
Anticancer Drug Discov. 8:298–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pandit B and Gartel AL: Thiazole
antibiotic thiostrepton synergize with bortezomib to induce
apoptosis in cancer cells. PLoS One. 6:e171102011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Piperdi B, Ling YH, Liebes L, Muggia F and
Perez-Soler R: Bortezomib: understanding the mechanism of action.
Mol Cancer Ther. 10:2029–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dick LR and Fleming PE: Building on
bortezomib: second-generation proteasome inhibitors as anti-cancer
therapy. Drug Discov Today. 15:243–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shapovalov Y, Benavidez D, Zuch D and
Eliseev RA: Proteasome inhibition with bortezomib suppresses growth
and induces apoptosis in osteosarcoma. Int J Cancer. 127:67–76.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pandit B, Bhat UG and Gartel AL:
Proteasome inhibitory activity of thiazole antibiotics. Cancer Biol
Ther. 11:43–47. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhat UG, Zipfel PA, Tyler DS and Gartel
AL: Novel anticancer compounds induce apoptosis in melanoma cells.
Cell Cycle. 7:1851–1855. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tone Y, Tanahashi N, Tanaka K, Fujimuro M,
Yokosawa H and Toh-e A: Nob1p, a new essential protein, associates
with the 26S proteasome of growing Saccharomyces cerevisiae
cells. Gene. 243:37–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tone Y and Toh-E A: Nob1p is required for
biogenesis of the 26S proteasome and degraded upon its maturation
in Saccharomyces cerevisiae. Genes Dev. 16:3142–3157. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oehler VG, Yeung KY, Choi YE, Bumgarner
RE, Raftery AE and Radich JP: The derivation of diagnostic markers
of chronic myeloid leukemia progression from microarray data.
Blood. 114:3292–3298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu Z, Guo Q, Shi A, Xie F and Lu Q:
Downregulation of NIN/RPN12 binding protein inhibit the growth of
human hepatocellular carcinoma cells. Mol Biol Rep. 39:501–507.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin Y, Peng S, Yu H, Teng H and Cui M:
RNAi-mediated downregulation of Nob1 suppresses the growth and
colony-formation ability of human ovarian cancer cells. Med Oncol.
29:311–317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakoda T, Kasahara N, Hamamori Y and Kedes
L: A high-titer lentiviral production system mediates efficient
transduction of differentiated cells including beating cardiac
myocytes. J Mol Cell Cardiol. 31:2037–2047. 1999. View Article : Google Scholar
|
|
17
|
Soneoka Y, Cannon PM, Ramsdale EE, et al:
A transient three-plasmid expression system for the production of
high titer retroviral vectors. Nucleic Acids Res. 23:628–633. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30:e362002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adams BK, Ferstl EM, Davis MC, et al:
Synthesis and biological evaluation of novel curcumin analogs as
anti-cancer and anti-angiogenesis agents. Bioorg Med Chem.
12:3871–3883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SJ, Choi SY, Kim WJ, et al: Combined
aberrant expression of E-cadherin and S100A4, but not β-catenin is
associated with disease-free survival and overall survival in
colorectal cancer patients. Diagn Pathol. 8:992013.
|
|
21
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Derksen PW, Liu X, Saridin F, et al:
Somatic inactivation of E-cadherin and p53 in mice leads to
metastatic lobular mammary carcinoma through induction of anoikis
resistance and angiogenesis. Cancer Cell. 10:437–449. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Debald M, Kaiser C, Abramian A, et al:
Evaluation of E-cadherin, Ki-67 and lymphatic vessel invasion in
abdominal metastases of human breast cancer. Anticancer Res.
33:1971–1975. 2013.PubMed/NCBI
|
|
25
|
Cai Y, Mohseny AB, Karperien M, Hogendoorn
PC, Zhou G and Cleton-Jansen AM: Inactive Wnt/beta-catenin pathway
in conventional high-grade osteosarcoma. J Pathol. 220:24–33. 2010.
View Article : Google Scholar : PubMed/NCBI
|