Introduction
Ovarian cancer is one of the most common types of
fatal tumors of the female reproductive tract and also a major
cause of mortality resulting from gynecological malignancies
(1). Epithelial ovarian cancer is
the predominant form among ovarian cancers and accounts for 90% of
incidences; the majority of mortalities are due to this malignancy
(2). There is a lack of effective
screening and early detection strategies; therefore, the majority
of females are diagnosed with advanced-stage metastatic cancer for
which surgical and pharmaceutical treatment options are
significantly less effective (3,4).
Standard treatment options include debulking followed by
chemotherapy with platinum agents. Although there is a good
response to primary surgery and chemotherapy treatments, the
recurrence rates are high (>60%) and salvage therapies available
are not curative (5–8). Therefore, it is important to
understand the molecular mechanisms underlying this disease in
order to develop novel treatment strategies to improve the clinical
outcomes for these patients.
Cyclooxygenase-2 (COX-2) is inducible by
inflammatory stimuli, including cytokines, growth factors and tumor
promoters, and is upregulated in a variety of malignancies. COX-2
upregulation favors the growth of malignant cells by stimulating
proliferation and angiogenesis (9,10). A
large number of previous studies demonstrated that COX-2 is
overexpressed in ovarian cancer (11–13).
Furthermore, Arico et al (14) found that COX-2 can induce
angiogenesis via vascular endothelial growth factor (VEGF) and
prostaglandin production and can also inhibit apoptosis by inducing
the anti-apoptotic factor B-cell lymphoma 2 as well as activating
anti-apoptotic signaling through Akt/protein kinase B (one of the
serine/threonine kinases). These results suggest that COX-2 has a
significant role in the generation and progression of solid tumors
and the inhibition of COX-2 may inhibit the growth of a variety of
solid malignancies. Therefore, downregulation of COX-2 in cancer
cells may prove useful in improving clinical outcomes in cancer
patients.
RNA interference (RNAi) is a powerful method for
gene inactivation (15–17) and cancer gene therapy (18). The basic mechanism of RNAi starts
with a long double-stranded RNA that is processed into small
interfering RNAs (siRNAs) of ~21 nt (19,20).
The advantage of RNAi technology is that it can be used to target a
large number of different genes, which are involved in a number of
distinct cellular pathways. The technology of RNA silencing is
poised to have a major impact on the treatment of human disease,
particularly cancer (19).
The aim of the present study was to investigate the
role of COX-2 in the growth of human ovarian cancer cells. The
effect of RNAi-induced COX-2 suppression on the proliferation,
invasion and migration of ovarian cancer cells was also
evaluated.
Materials and methods
Cell culture
SKOV3 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and cultured in Modified
McCoy’s 5A Medium (Sigma, St. Louis, MO, USA) supplemented with 10%
fetal bovine serum (FBS) (Gibco-BRL, Carlsbad, CA, USA). The cells
were maintained in a humidified 37°C incubator with 5%
CO2.
siRNA design and conduct
In total, three different siRNAs were designed
against COX-2 mRNA as suggested by the method used in a study by
Elbashir et al (21). The
targeting sequences corresponding to the siRNAs against COX-2
(GeneBank accession no. NM_000963.2) were as follows: Bases 290–310
(siRNA-1, 5′-AAACTGCTCAACACCGGAATT-3′), bases 456–477 (siRNA-2,
5′-TCACATTTGATTGACAGTCCA-3′) and 517–538 (siRNA-3,
5′-CCTTCTCTAACCTCTCCTATT-3′). The interference vector
pGenesil-COX-2 was constructed using the pGenesil-1 vector. In
brief, three pairs of oligonucleotide fragments were designed,
synthesized and annealed. pGenesil-1, which consisted of human U6
short hairpin RNA (shRNA) promoter, was used to generate a series
of siRNA expression vectors by inserting three pairs of annealed
oligonucleotides. The recombinant plasmid vectors
pGenesil-1-COX-2(1),
pGenesil-1-COX-2(2) and
pGenesil-1-COX-2(3) were
repeatedly excised and ligated successively. Thus, the tandem
recombinant vector pGenesil-1-COX-2(1+2+3) was constructed and
called pGenesil-1-COX-2. All the sequences inserted were verified
by DNA sequencing. Plasmid pGenesil-1-KB was used to serve as a
control for the empty vector. All the siRNA sequences were checked
in terms of their specificity using the Basic Local Alignment
Search Tool (National Library of Medicine, Bethesda, MD, USA)
database and did not exhibit any homology to other human genes.
The SKOV3 cells were randomly divided into three
groups: Untransfected control group, empty vector transfected
vector group that served as a blank control, and the
pGenesil-1-COX-2 siRNA expression plasmid transfected siRNA group.
In total, 5×105 cells were seeded in six-well plates and
grown overnight prior to transfection. All the plasmids were
transiently transfected into SKOV3 cells by Lipofectamine™ 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions and incubated in serum-starved media at
37°C in 5% CO2. At 8 h post-transfection, the medium was
replaced with fresh complete medium containing 10% FBS. The cells
were harvested 48 h after transfection and used for the evaluation
of COX-2 expression. The transfection efficiency was monitored by
measuring the percentage of fluorescent cells among a total of
1,000 cells using fluorescence microscopy.
RNA isolation and quantitative polymerase
chain reaction (qPCR)
To evaluate COX-2 mRNA expression, the cells were
harvested following transfection. The total RNA was extracted from
the cells using TRIzol reagent (Invitrogen Life Technologies) for
reverse transcription. The RNA was transcribed to cDNA using the
Superscript First-Strand Synthesis kit (Takara, Dalian, China)
following the manufacturer’s instructions. qPCR assays were
performed using SYBR-Green Real-Time PCR Master Mix (Toyobo, Osaka,
Japan) and RT-PCR amplification equipment (7300 Real-Time PCR
System; Applied Biosystems, Foster City, CA, USA) using specific
primers: COX-2 sense, 5′-CCCTTGGGTGTCAAAGGTAAA-3′ and antisense,
5′-AAACTGATGCGTGAAGTGCTG-3′; and β-actin sense,
5′-GCGAGCACAGAGCCTCGCCTTTG-3′ and antisense,
5′-GATGCCGTGCTCGATGGGGTAC-3′. The PCR conditions were as follows:
Pre-denaturation at 95°C for 2 min, followed by 40 cycles of
denaturation at 95°C for 10 sec and annealing/extension at 58°C for
20 sec. The amplification specificity was checked by melting curve
analysis. The expression of the genes of interest was determined by
normalization of the threshold cycle (Ct) of these genes to that of
the β-actin control.
Western blot analysis
The cells were harvested and lysed in Triton X-100
in Hepes buffer [150 mM NaCl, 50 mM Hepes, 1.5 mM MgCl2,
1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS) and a protease
inhibitor cocktail (Sigma)]. Western blot analysis was performed
using conventional protocols. Briefly, the protein concentration of
the extracts was determined using a bicinchoninic acid kit (Sigma)
with bovine serum albumin used as the standard. The total protein
samples (40 μg) were separated by 8% SDS-PAGE, proteins were
transferred to nitrocellulose membranes (Millipore, Bedford, MA,
USA), immunoblotted with specific primary antibodies and incubated
with corresponding horseradish peroxidase (HRP)-conjugated
secondary antibody. The other primary antibodies used in the
western blots were as follows: Antibodies against COX-2, β-actin,
VEGF (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); MMP-9
and MMP-2 (Sigma Aldrich, St. Louis, MO, USA); secondary antibodies
used for immunodetection were as follows: HRP-conjugated goat
anti-mouse immunoglobulin (Ig) G and goat anti-rabbit IgG (Amersham
Biosciences, Uppsala, Sweden). All the immunoblots were visualized
by enhanced chemiluminescence (Pierce Biotechnology, Inc.,
Rockford, IL, USA). All the assays using COX-2 knockdown SKVO3
cells were performed after the third day of siRNA transfection.
Cell viability assay
At 24, 48 and 72 h post-transfection with COX-2
siRNAs, the cells were seeded in quadruplicate into 96-well plates
(5,000 cells/well in 100 ml of medium). At the indicated times, the
cells were incubated with 1 mg/ml MTT in normal culture medium for
6 h at 37°C. The medium was then aspirated and the formazan was
dissolved in 200 ml dimethyl sulfoxide. The absorbance was measured
at 570 nm by a STAKMAX™ microplate reader (Molecular Devices,
Sunnyvale, CA, USA) and the experiments were repeated five
times.
Cell cycle analysis
The SKVO3 cells were treated with siRNA for 24 h in
Dulbecco’s modified Eagle’s medium (DMEM) containing 5% FBS. All
the cells were collected, and 1×106 cells were
centrifuged, resuspended in ice-cold 70% ethanol and stored at
−20°C until further analysis. Washed cells were stained by 0.1%
Triton X-100 in 0.01 M phosphate-buffered saline (pH 7.2) with 50
μg/ml propidium iodide (Sigma-Aldrich) and 1 mg/ml RNase A
(Invitrogen), and incubated at 37°C for 30 min in the dark. Samples
of the cells were then analyzed for their DNA content using FACScan
flow cytometry (Beckman, Miami, FL, USA), and cell cycle phase
distributions were analyzed by the Cell Quest acquisition software
(BD Biosciences, Franklin Lanes, NJ, USA). All experiments were
performed in duplicate and repeated twice.
Cell migration assay
The migration assay was performed using a 12-well
Boyden Chamber (Neuro Probe, Inc., Gaithersburg, MD, USA) with an 8
μm pore size. In total, ~1×105 cells were seeded into
upper wells of the Boyden Chamber and incubated for 6 h at 37°C in
medium containing 1% FBS. DMEM with 10% FBS was used as a
chemoattractant in the bottom wells. The cells that did not migrate
through the pores of the Boyden Chamber were manually removed with
a rubber swab. The cells that migrated to the lower side of the
membrane were stained with hematoxylin and eosin and photographed
using an inverted microscope.
Transwell invasion assay
The invasiveness of SKVO3 cancer cells was assessed
using 24-well Transwell plates (Corning, Lowell, MA, USA). In
brief, 2×105 cells in DMEM with 0.5% FBS were added to
the upper chamber containing a 8-mm pore polycarbonate coated with
1 mg/ml Matrigel; the lower chamber was filled with media
containing 5% FBS. Subsequent to 16 h incubation, the upper surface
of the membrane was scrubbed with a cotton-tipped swab. The
invading cells on the lower surface of the membrane were fixed and
stained with 0.5% crystal violet dye. A total of five random fields
per membrane were photographed at magnification, ×40 for
calculating the cell number using an inverted phase-contrast
microscope (Leica, Solms, Germany). In addition, cells were
quantified by measuring the absorbance of dye extracts at 570 nm in
100 ml of Sorenson’s solution (9 mg trisodium citrate, 305 ml
distilled water, 195 ml 0.1 N HCl and 500 ml 90% ethanol). All
experiments were performed in triplicate and repeated four
times.
Determination of prostaglandin E2 (PGE2)
synthesis by ELISA
PGE2 synthesis was determined according to methods
that were previously described (22). In brief, the SKVO3 cells were grown
in 12-well plates overnight. The culture media of the cells were
changed to new DMEM for 30 min prior to harvesting of culture media
and then these culture media were centrifuged to remove cell
debris. Cell-free culture media were collected at indicated time
periods, then PGE2 levels were determined using a Human PGE2 ELISA
kit according to the manufacturer’s instructions (Cayman Chemical,
Ann Arbor, MI, USA) and an ELISA reader (μQuant; Biotek
Instruments, Inc., Winooski, VT, USA).
Statistical analysis
The values were expressed as the mean ± standard
deviation. The data were analyzed by one-way analysis of variance
and P<0.05 was considered to be statistically significant.
Results
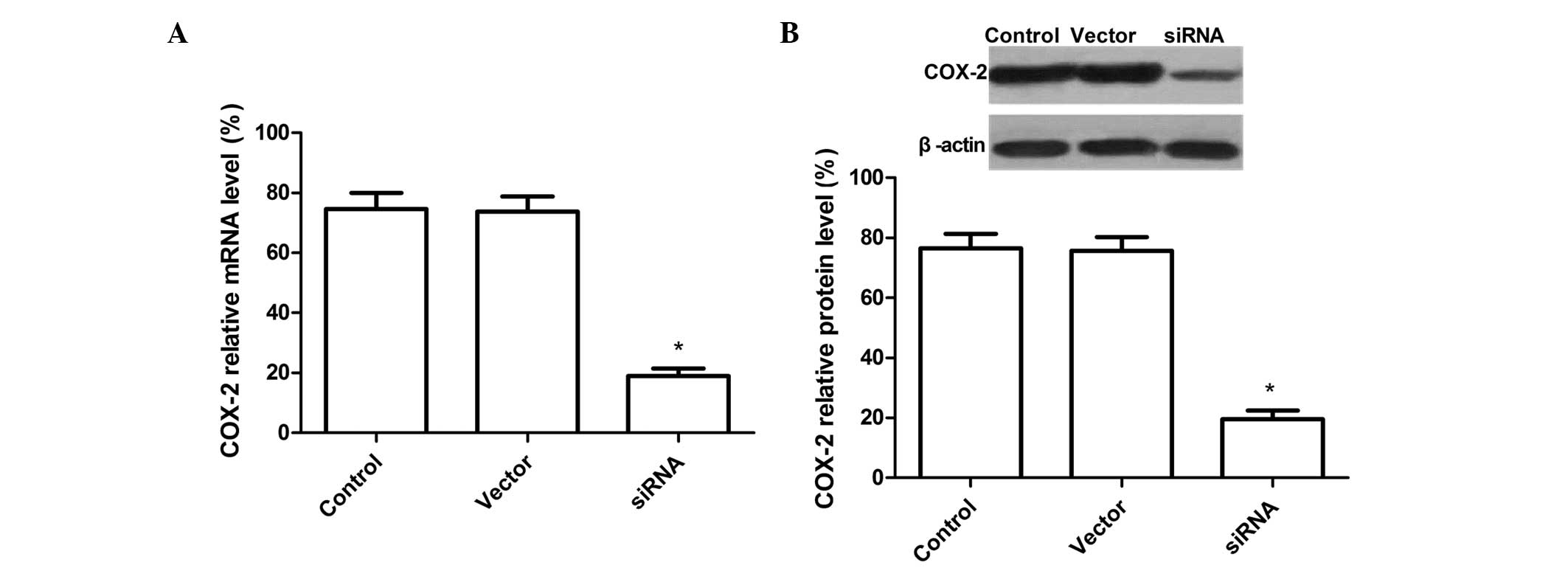
Downregulation of COX-2 mRNA and protein
expression levels by COX-2-siRNA
A recombinant vector was designed and constructed,
which expresses three siRNAs targeting the COX-2 gene in tandem,
and transfected it into SKOV3 cells. Subsequent to 72 h
transfection, the cells were harvested and the COX-2 mRNA levels
were analyzed by qPCR. COX-2 expression was significantly
downregulated in SKOV3 cells transfected with siRNA compared with
the cells transfected with the control vector and untransfected
cells (P<0.01). There was no significant difference between the
untransfected and empty vector groups (P>0.05) (Fig. 1A). The results indicated that most
of the COX-2 mRNA was degraded by COX-2 siRNA in the SKOV3
cells.
The effect of COX-2 siRNA treatment on protein
expression was assessed by western blot analysis. As shown in
Fig. 1B, there was no difference
between the untransfected and empty vector group (P>0.05), while
the band density clearly decreased in the COX-2 siRNA group as
compared with the untransfected and empty vector group. These
results demonstrated that siRNA targeting COX-2 significantly
silenced COX-2 protein expression in SKOV3 ovarian cancer cells
(P<0.01).
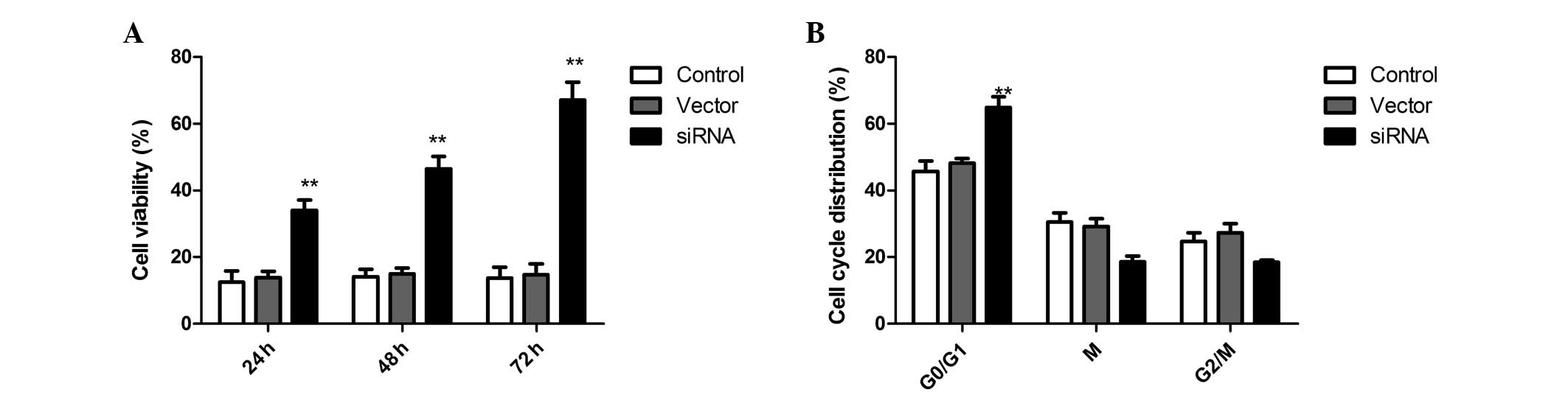
COX-2 silencing affects SKVO3 cell
proliferation and the cell cycle
To elucidate the effects of the siRNA on SKOV3
ovarian cancer cell proliferation, the MTT assay was used and cell
proliferation was determined by counting the number of viable
cells. SKOV3 ovarian cancer cells transfected with COX-2-siRNA
proliferated at a much lower rate compared with the control cells
at 24, 48 and 72 h post-transfection. Compared with the
untransfected cells, the proliferation properties of SKOV3 cells
were significantly inhibited (Fig.
2A). These data demonstrate that the inhibition of COX-2 by
RNAi can inhibit the proliferation of SKOV3 cells.
In addition, to determine the effects of SKVO3 cell
cycle progression following COX-2 silencing, flow cytometry was
performed in the present study. In the siRNA therapy group, the
percentage of cells in G0/G1 phase was significantly increased as
compared with the scrambled-treated and control cells. These
results indicated that COX-2 silencing can induce cell cycle arrest
in G0/G1 phase in SKVO3 cells (Fig.
2B).
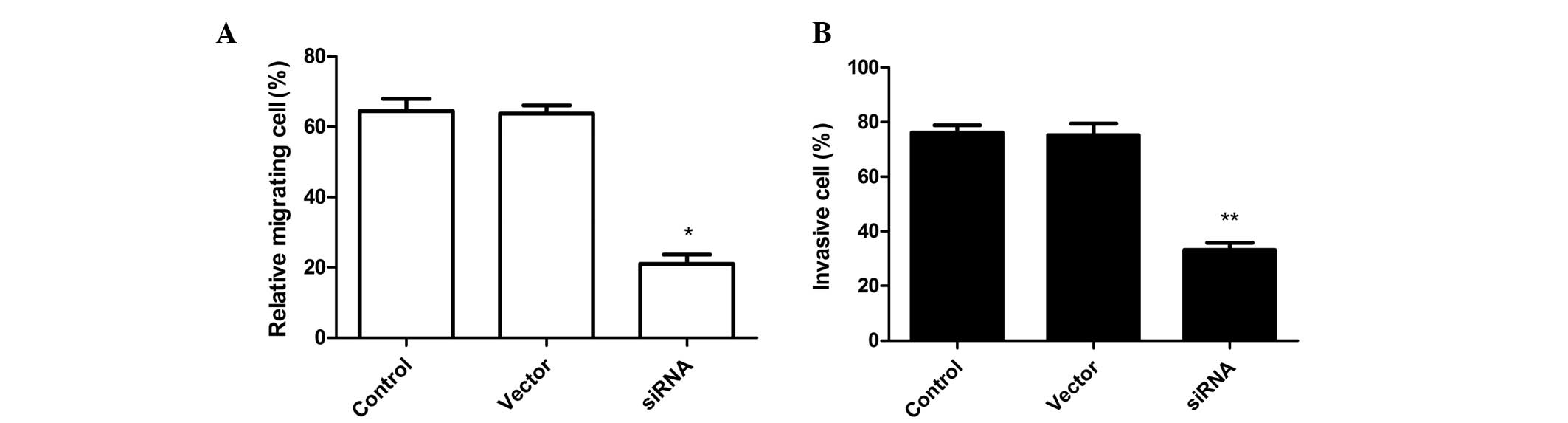
COX-2 silencing inhibits SKVO3 cell
invasion and cell migration
To analyze whether siRNA affects SKVO3 cell
migration, migration assays were performed using Boyden chambers.
RNAi-mediated COX-2 silencing significantly inhibited SKVO3 cell
migration as compared with the control (untransfected) and negative
control-transfected (Vector) groups (Fig. 3A). On the other hand, COX-2
knockdown in SKVO3 cells markedly inhibited invasion in
vitro as compared with the vector-treated and control groups
(Fig. 3B). The control cells and
vector cells remained invasive and no statistically significant
differences were observed.
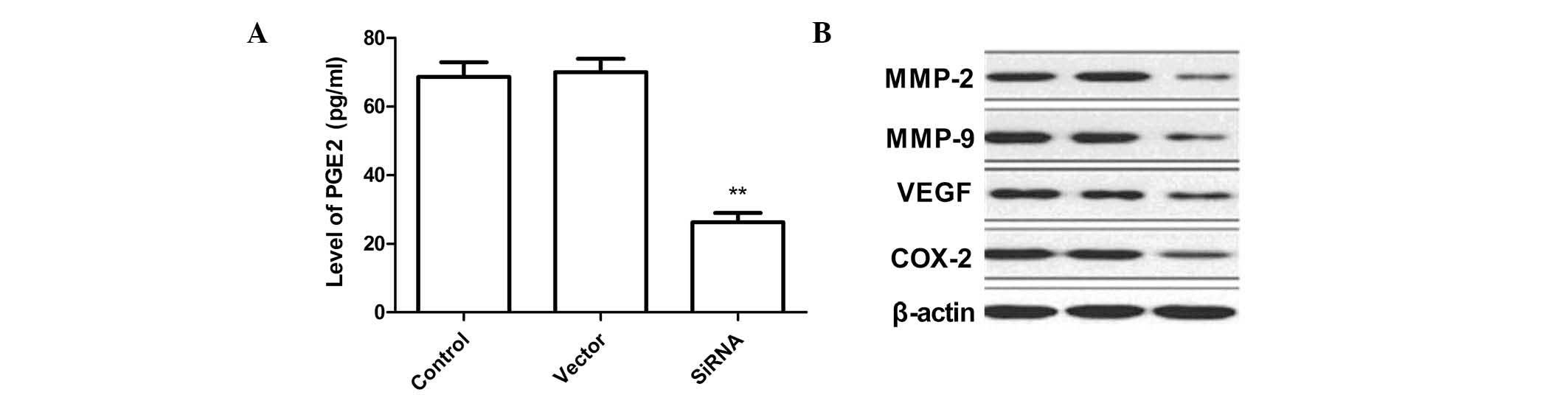
Silencing of COX-2 decreases levels of
PGE2 and other proteins in SKVO3 cells
PGE2 levels in SKVO3 cells were determined by ELISA
analysis. As shown in Fig. 4A, the
PGE2 levels in the siRNA therapy groups were significantly lower
compared with those in the untransfected and empty vector groups.
In addition, VEGF protein expression levels were determined
following COX-2 silencing. It was found that COX-2 silencing was
found to significantly inhibit VEGF expression in the SKVO3 tumor
cells compared with the control and empty vector cells (Fig. 4B).
In order to investigate the mechanisms involved in
the inhibition of the invasion and migration ability by
downregulation of COX-2 silencing of SKVO3 cells, western blot
analysis was performed to evaluate the activity of MMP-2 and MMP-9.
COX-2 silencing caused a significant decrease in MMP-9 and MMP-2
levels compared with the control cells as determined by western
blot analysis (Fig. 4B).
Discussion
RNAi is a fundamental cellular mechanism used for
silencing gene expression (16,23–25).
RNAi has been widely used in cancer therapy to silence the
expression of oncogenes and growth factors or their receptors,
which may result in inhibition of the cell cycle, cell
proliferation and tumor angiogenesis as well as induction of cell
apoptosis (26–28). To date, RNAi has been regarded as
an effective and useful approach for therapeutic applications of
cancer (26,39). In the present
study, RNAi strategies were used to reduce the expression of COX-2
in the ovarian cancer cell line SKOV3 and to evaluate the role of
COX-2 in SKOV3 cells. A total of three siRNA sequences were
designed to target the COX-2 coding region and inserted into the
pGenesil-1 vector in tandem to efficiently suppress the expression
of COX-2. The present study demonstrated that this vector was
efficiently silenced COX-2. Downregulation of COX-2 reduced
proliferation, cell cycle, cell migration and invasion in SKOV3
cells. The results were consistent with those of previous studies
revealing that the number of viable cells was significantly
decreased following transfection with COX-2 siRNA (28,30,31).
COX-2 is known to be involved in multiple
pathophysiological processes, including inflammation and
tumorigenesis (32,33). COX-2 is undetectable in numerous
normal tissues, yet it is commonly overexpressed in various human
cancers, including ovarian cancer (14), with its downstream product being
PGE2, which is linked to more aggressive behavior of tumors and
thus contributing to ovarian cancer progression (34,35).
PGE2 is a significant mediator in tumor-promoting inflammation
(36). Additionally, PGE2 promotes
tumor cell proliferation, induces VEGF upregulation and inhibits
tumor cell apoptosis as well as immune function (37). The present study revealed that
downregulation of the COX-2 gene by silencing decreased the levels
of PGE2 and VEGF (Fig. 4B) and
inhibit tumor cell proliferation and the cell cycle. These results
implied that the major mechanism of COX-2 in stimulating
tumorigenesis is via its product PGE2.
The poor prognosis of ovarian cancer is mainly due
to dissemination caused by the aggressive migration activity of the
cancer cells (38). In the present
study, siRNA-mediated downregulation of COX-2 expression in human
ovarian cancer cells lead to a significant decrease in SKOV3 cell
invasion and migration. These results were consistent with previous
studies (28,30,31),
demonstrating that COX-2 mediates the invasive and metastatic
potential of ovarian cancer cells.
In conclusion, the vector expressing three siRNAs
targeting the COX-2 gene in tandem was able to silence the
expression of COX-2 in ovarian cancer cells. The COX-2 knockdown
not only resulted in a decrease of cell proliferation and
inhibition of the cell cycle in ovarian tumor cells, but it also
suppressed cell migration and invasion of ovarian cancer cells.
These results indicated that COX-2 is a potential therapeutic
target for the prevention or treatment of ovarian cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of Jilin (no. 83657432).
References
|
1
|
Lin Y, Peng S, Yu H, Teng H and Cui M:
RNAi-mediated downregulation of NOB1 suppresses the growth and
colony-formation ability of human ovarian cancer cells. Med Oncol.
29:311–317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Auersperg N, Wong AS, Choi KC, Kang SK and
Leung PC: Ovarian surface epithelium: biology, endocrinology, and
pathology. Endocr Rev. 22:255–258. 2001.PubMed/NCBI
|
|
3
|
Ziebarth AJ, Landen CN Jr and Alvarez RD:
Molecular/genetic therapies in ovarian cancer: future opportunities
and challenges. Clin Obstet Gynecol. 55:156–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Linghu H, Wang J, Che YL, Xiang
TX, Tang WX and Yao ZW: The role of Crk/Dock180/Rac1 pathway in the
malignant behavior of human ovarian cancer cell SKOV3. Tumour Biol.
31:59–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fader AN and Rose PG: Role of surgery in
ovarian carcinoma. J Clin Oncol. 25:2873–2883. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salzberg M, Thurlimann B, Bonnefois H,
Fink D, Rochlitz C, Von Moos R and Senn H: Current concepts of
treatment strategies in advanced or recurrent ovarian cancer.
Oncology. 68:293–298. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McGuire WP and Ozols RF: Chemotherapy of
advanced ovarian cancer. Semin Oncol. 25:340–348. 1998.PubMed/NCBI
|
|
8
|
Fung-Kee-Fung M, Oliver T, Elit L, Oza A,
Hirte HW and Bryson P: Optimal chemotherapy treatment for women
with recurrent ovarian cancer. Curr Oncol. 14:195–208. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dempke W, Rie C, Grothey A and Schmoll HJ:
Cyclooxygenase-2: a novel target for cancer chemotherapy? J Cancer
Res Clin Oncol. 127:411–417. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Williams CS, Mann M and DuBois RN: The
role of cyclooxygenases in inflammation, cancer, and development.
Oncogene. 18:7908–7916. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Miner K, Fannin R, Carl Barrett J
and Davis BJ: Cyclooxygenase-1 and 2 in normal and malignant human
ovarian epithelium. Gynecol Oncol. 92:622–627. 2004. View Article : Google Scholar
|
|
12
|
Denkert C, Köbel M, Pest S, Koch I, Berger
S, Schwabe M, Siegert A, Reles A, Klosterhalfen B and Hauptmann S:
Expression of cyclooxygenase-2 is an independent prognostic factor
in human ovarian carcinoma. Am J Pathol. 160:893–903. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Erkinheimo TL, Lassus H, Finne P, van Rees
BP, Leminen A, Ylikorkala O, Haglund C, Butzow R and Ristimäki A:
Elevated cyclooxygenase-2 expression is associated with altered
expression of p53 and SMAD4, amplification of HER-2/neu, and poor
outcome in serous ovarian carcinoma. Clin Cancer Res. 10:538–545.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arico S, Pattingre S, Bauvy C, Gane P,
Barbat A, Codogno P and Ogier-Denis E: Celecoxib induces apoptosis
by inhibiting 3-phosphoinositide-dependent protein kinase-1
activity in the human colon cancer HT-29 cell line. J Biol Chem.
277:27613–27621. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cerutti H: RNA interference: traveling in
the cell and gaining functions? Trends Genet. 19:39–46. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature.
391:806–811. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hannon GL and Rossi JJ: Unlocking the
potential of the human genome with RNA interference. Nature.
431:371–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Izquierdo M: Short interfering RNAs as a
tool for cancer gene therapy. Cancer Gene Ther. 12:217–227. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caplen NJ: Gene therapy progress and
prospects. Downregulating gene expression: the impact of RNA
interference. Gene Ther. 11:1241–1248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elbashir SM, Harborth J, Weber K and
Tuschl T: Analysis of gene function in somatic mammalian cells
using small interfering RNAs. Methods. 26:199–213. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tai MH, Weng CH, Mon DP, Hu CY and Wu MH:
Ultraviolet C irradiation induces different expression of
cyclooxygenase 2 in NIH 3T3 cells and A431 cells: the roles of
COX-2 are different in various cell lines. Int J Mol Sci.
13:4351–4366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim DH, Longo M, Han Y, Lundberg P, Cantin
E and Rossi JJ: Interferon induction by siRNAs and ssRNAs
synthesized by phage polymerase. Nat Biotechnol. 22:321–325. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meister G and Tuschi T: Mechanisms of gene
silencing by double-stranded RNA. Nature. 431:343–349. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mello CC and Conte D Jr: Revealing the
world of RNA interference. Nature. 431:338–342. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang R, Wang X, Lin F, Gao P, Dong K and
Zhang HZ: shRNA-targeted cyclooxygenase (COX)-2 inhibits
proliferation, reduces invasion and enhances chemosensitivity in
laryngeal carcinoma cells. Mol Cell Biochem. 317:179–188. 2008.
View Article : Google Scholar
|
|
27
|
Chen Q, Pan Q, Cai R and Qian C: Prospects
of RNA interference induced by RNA Pol II promoter in cancer
therapy. Prog Biochem Biophys. 34:806–815. 2007.
|
|
28
|
Strillacci A, Griffoni C, Spisni E, Manara
MC and Tomasi V: RNA interference as a key to knockdown
overexpressed cyclooxygenase-2 gene in tumour cells. Br J Cancer.
94:1300–1310. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vogler M, Walczak H, Stadel D, Haas TL,
Genze F, Jovanovic M, Gschwend JE, Simmet T, Debatin KM and Fulda
S: Targeting XIAP bypasses Bcl-2-mediated resistance to TRAIL and
cooperates with TRAIL to suppress pancreatic cancer growth in vitro
and in vivo. Cancer Res. 68:7956–7965. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Wu YD, Li P, Tu J, Niu YL, Xu CM
and Zhang ST: Effects of cyclooxygenase-2 on human esophageal
squamous cell carcinoma. World J Gastroenterol. 17:4572–4580. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Denkert C, Fürstenberg A, Daniel PT, Koch
I, Köbel M, Weichert W, Siegert A and Hauptmann S: Induction of
G0/G1 cell cycle arrest in ovarian carcinoma cells by the
anti-inflammatory drug NS-398, but not by COX-2-specific RNA
interference. Oncogene. 22:8653–8661. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park W, Oh YT, Han JH and Pyo H: Antitumor
enhancement of celecoxib, a selective cyclooxygenase-2 inhibitor,
in a Lewis lung carcinoma expressing cyclooxygenase-2. J Exp Clin
Cancer Res. 27:662008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Müller-Decker K and Fürstenberger G: The
cyclooxygenase-2-mediated prostaglandin signaling is causally
related to epithelial carcinogenesis. Mol Carcinog. 46:705–710.
2007.PubMed/NCBI
|
|
34
|
Leahy KM, Ornberg RL, Wang Y, Zweifel BS,
Koki AT and Masferrer JL: Cyclooxygenase-2 inhibition by celecoxib
reduces proliferation and induces apoptosis in angiogenic
endothelial cells in vivo. Cancer Res. 62:625–631. 2002.PubMed/NCBI
|
|
35
|
Mamdani M, Juurlink DN, Lee DS, Rochon PA,
Koop A, Naglie G, Austin PC, Laupacis A and Stukel TA:
Cyclo-oxygenase-2 inhibitors versus non-selective non-steroidal
anti-inflammatory drugs and congestive heart failure outcomes in
elderly patients: a population-based cohort study. Lancet.
363:1751–1756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rasmuson A, Kock A, Fuskevåg OM, Kruspig
B, Simón-Santamaría J, Gogvadze V, Johnsen JI, Kogner P and
Sveinbjörnsson B: Autocrine prostaglandin E2 signaling promotes
tumor cell survival and proliferation in childhood neuroblastoma.
PLoS One. 7:e293312012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pockaj BA, Basu GD, Pathangey LB, Gray RJ,
Hernandez JL, Gendler SJ and Mukherjee P: Reduced T-cell and
dendritic cell function is related to cyclooxygenase-2
overexpression and prostaglandin E2 secretion in patients with
breast cancer. Ann Surg Oncol. 11:328–339. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Iñiguez MA, Rodríguez A, Volpert OV,
Fresno M and Redondo JM: Cyclooxygenase-2: a therapeutic target in
angiogenesis. Trends Mol Med. 9:73–78. 2003.
|