Introduction
Systemic lupus erythematosus (SLE) is a typical
systemic autoimmune disease (1).
The pathogenesis and severity of SLE are associated with the
hereditary factor. Abnormalities of the immune system have a role
in the occurrence of this disease, which affects almost all
monocytes and a number of cytokines. Previous studies have shown
that interleukin (IL)-10 has an important role in the pathogenesis
of SLE (2–4). The mechanism may be associated with
the hypothesis that IL-10 induces immune dysfunction and loss of
immunotolerance, and increases the production of antibodies
implicated in organ damage (5).
As an inhibitive cytokine, IL-10 is produced by a
number of cells. Previous studies have indicated that the
regulatory T lymphocyte (Treg) and regulatory B lymphocyte (Breg)
are the primary origin of IL-10 in the body (6–8). As
with other cytokines, IL-10 is required to bind to the IL-10
receptor (IL-10R) prior to activation. Human and mouse IL-10R,
which are members of the interferon receptor family, are composed
of two subunits, IL-10R1 and IL-10R2 (9,10).
IL-10R2 is considered to be a member of the interferon (IFN)
receptor, does not bind to IL-10 and initiates intracellular
transduction signals. IL-10R1 has a significant role in IL-10R
binding to IL-10. Following binding to IL-10, IL-10R1 transfers the
excitability and inhibitory signal through the Janus kinase
(Jak)-signal transducer and activator of transcription (Stat)
signal transduction system (11).
Previous studies suggested that the distribution of the functional
regions of IL-10R is located in the cytoplasm (12).
In our previous study, the results of DNA sequencing
analysis of the IL-10R1 gene coding region indicated that, compared
with the C57BL/6 mouse, the New Zealand W (NZW) mouse has 18 base
displacements, among which, 7 regions are hypothesized to cause an
amino acid change. Notably, in the no. 1146 base of IL-10R1 of the
NZW mouse, a G was replaced with A (G1146A), which induces the
change of a coding amino acid from G to E (G356E) and the loss of
the Jak1 and tyrosine kinase 2 (Tyk2) phosphorylation binding
sequence of IL-10R1 (13). G356E
is hypothesized to be located at the 298–405 region of the IL-10R1
cytoplasm region, which is responsible for the negative signal
transduction of IL-10. Notably, the IL-10R1 gene type of the Murphy
Roths large (MRL) mouse, another type of typical SLE model mouse,
is the same as the NZE mouse, thus indicating that the defect of
IL-10R1 may cause SLE (14). The
purpose of the present study was to investigate the signal
transduction mechanism of the NZW type IL-10R1 and the effect on
the function of B lymphocytes.
Materials and methods
Cell culture
BA/F3 cells were purchased from the cell center of
Peking Union Medical College (Beijing, China). The cells were
cultured in Dulbecco’s modified Eagle medium (DMEM; Invitrogen,
Carlsbad, CA, USA) with 10 ng/ml rmIL-3 (R&D Systems, San
Diego, CA, USA) and 10% fetal calf serum (FCS; Invitrogen), and
were incubated.
Vector construction
The study was approved by the Ethics Committee of
China Medical University (Shenyang, China). Total RNA was extracted
from the spleen of C57BL/6 and NZW mice (purchased from Shanghai
Laboratory Animal Center of the Chinese Academy of Sciences and
maintained under specific pathogen-free conditions). The following
primers were designed by Takara Biotechnology (Dalian, China;
forward, 5′-GGATCCACC ATGTTGTCGCGTTTGCTCCC-3′ and reverse, 5′-CTC
GAGTCATTCTTCTACCTGCAG GCTGGAG-3′) were used to obtain the coding
sequence region of IL-10. DNA was cloned into the pMD19-T simple
vector and the DNA sequences were checked using agarose gel
electrophoresis. Using GeneBank to compare the results, it was
observed that the genes analyzed were not the target genes. Thus,
the genes were cloned into the pcDNA3.1(+) multiple clonal sites to
produce pcDNA3.1(+)-wild-type (WILD)-IL-10R1 and
pcDNA3.1(+)-NZW-IL-10R1. The designed primers (forward,
5′-GAGTCTCCAGAGCTACAGGCCACCTG-3′ and reverse,
5′-GCCTCTAGCTCTGGAGACTCTTCTTTTCC-3′) were used to perform the
G1146A point mutation in pcDNA3.1(+)-WILD-IL-10R1.
pcDNA3.1(+)-G1146A-IL-10R1 was confirmed by sequencing using
agarose gel electrophoresis.
Transfection
BA/F3 cells (1×105/well) were incubated
in 24-well plates for one day prior to transfection. According to
the manufacturer’s instruction for Lipofectamine 2000 (Invitrogen),
pcDNA3.1(+)-WILD-IL-10R1, pcDNA3.1(+)-NZW-IL-10R1 and
pcDNA3.1(+)-G1146A-IL-10R1 were transfected into the cells. On the
second day following transfection, 500 μg/ml G418 (Gibco, Grand
Island, NY, USA) was added to the culture medium. On the fourth day
following transfection, culture medium containing 200 μg/ml G418
was used to culture the cells for subsequent use.
Flow cytometric analysis
Phosphate-buffered saline (PBS) (pH 7.2–7.4) was
used with 1% bovine serum albumin to wash the cells three times and
cells were left in 0.5 ml PBS following washing. Next,
phycoerythrin anti-mouse IL-10R, fluorescein isothiocyanate
anti-mouse CD16/32 and allophycocyanin anti-mouse CD62L (BioLegend
Inc., San Diego, CA, USA) was added and the cells were incubated at
room temperature for 30 min in the dark. Next, the flow cytometer
FACScan (BD Biosciences, San Jose, CA, USA) was used to measure the
expression levels of IL-10R1, CD16/32 and CD62L. The results were
analyzed by WinMDI 2.9 (The Scripps Research Institute, San Diego,
CA, USA).
MTT assay
Cells (1,000 cells/well) were incubated in a 96-well
plate with 100 μl culture medium. Different concentrations of IL-10
(0, 0.01, 0.1, 1 and 10 ng/ml) were used to activate the cells for
12 h. A total of 1 ng/ml IL-10 was used to activate the cells for
different times (0, 4, 8, 12, 18 and 22 h). Next, 5 mg/ml MTT (10
μl per well) was added and cells were allowed to incubate at 37°C
under 5% CO2 for 3 h. Following the addition of
dimethylsulfoxide (100 μl/well), the plate was agitated for 10 min
and a CLARIOstar microplate reader (BMG Labtech, Ortenberg,
Germany) was used to read the optical density value (490 nm).
Western blot analysis
The cells were starved in DMEM without serum for 12
h, incubated with 10 ng/ml IL-10 for 15 min and lysed. Protein
solutes (70 μg) were purified using 7% SDS-PAGE. Next, the protein
was transferred to a Hybond-enhanced chemiluminescence membrane.
The membrane was then blocked for at least 1 h in 5% skimmed milk.
The membrane was incubated with anti-Jak1, p-Jak1, STAT3, p-STAT3,
Tyk2, p-Tyk2 or β-actin primary antibodies (Invitrogen) at 4°C
overnight. Following washing, the membrane was incubated with the
secondary antibodies for 1 h in the dark. Images were scanned using
a Typhoon 9400 scanner (GE Healthcare, Fairfield, CT, USA ).
Statistical analysis
Data are presented as the mean ± standard error of
the mean of at least three independent experiments. For statistical
analysis, a one-way analysis of variance was used for comparison of
one variance (ANOVA) among groups and two-way ANOVA was used for
comparison of two independent variances among groups followed by
Tukey’s post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
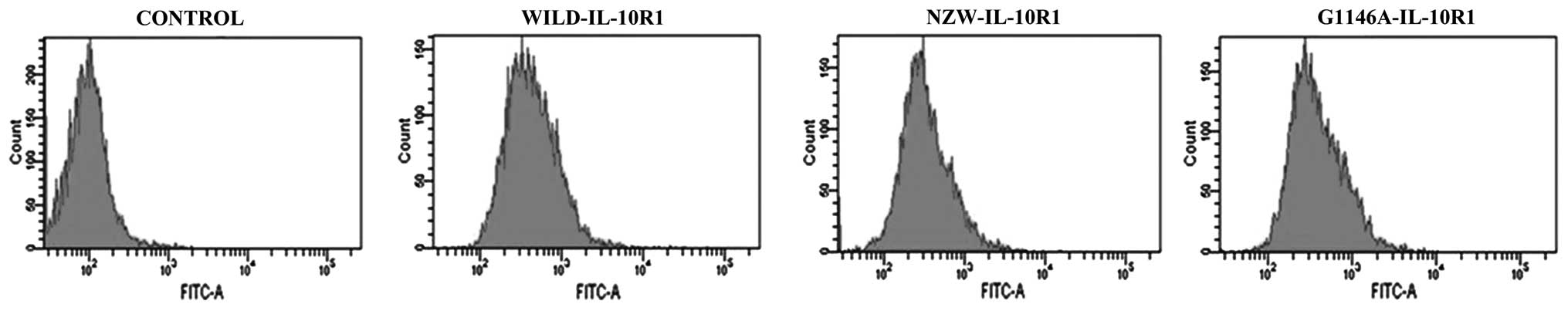
Transfected cell lines express
IL-10R1
The expression levels of IL-10R1 on the surface of
BA/F3 cells stably transfected with NZW, G1146 and WILD IL-10R1
expression vectors, respectively, were assessed by flow cytometry
and are presented in Fig. 1. The
moderately expressing cells were selected for subsequent usage.
IL-10 affects the proliferation of BA/F3
cells expressing different types of IL-10R1
IL-10 was capable of inducing as well as regulating
B-cell proliferation. The same dose of IL-10 (1 ng/ml) was used to
stimulate the BA/F3 cells expressing G1146A-IL-10R1, NZW-IL-10R1
and WILD-IL-10R1. The BA/F3 cells expressing G1146A-IL-10R1 and
NZW-IL-10R1 did not exhibit the ability to inhibit proliferation
with progressing stimulation time. However, IL-10 exhibited an
ability to induce proliferation. The proliferation curve was
steeper compared with the groups without (control cells) and with
IL-3. Notably, the proliferation of BA/F3 cells expressing
G1146A-IL-10R1 was significantly higher compared with the other
groups. IL-10 inhibited the proliferation of the BA/F3 cells
expressing WILD-IL-10R1 as the stimulation time was extended. The
proliferation curve was markedly lower compared with the group
without (control cells) and with IL-3 (Fig. 2A). Different doses of IL-10 (0.01,
0.1, 1.0 and 10 ng/ml) were used to stimulate the BA/F3 cells
expressing G1146A-IL-10R1, NZW-IL-10R1 and WILD-IL-10R1 for 12 h.
The proliferation rates of the BA/F3 cells expressing NZW-IL-10R1
and G1146A-IL-10R1 were positively correlated with the IL-10 levels
and were higher than those of the control group. The proliferation
rate of the BA/F3 cells expressing G1146A-IL-10R1 was markedly
higher than the proliferation rate of those expressing NZW-IL-10R1.
However, the proliferation rate of the BA/F3 cells expressing
WILD-IL-10R1 was negatively correlated with the concentration of
IL-10 and lower compared with the control group (Fig. 2B).
IL-10 affects the expression of CD16/32
and LECAM-1 in BA/F3 cells expressing different types of
IL-10R1
Fig. 3A shows
CD16/32 and LECAM-1 expression in BA/F3 cells expressing
G1146A-IL-10R1, NZW-IL-10R1 and WILD-IL-10R1 following stimulation
with IL-10. Compared with the BA/F3-expressing WILD-IL-10R1,
CD16/32 expression on the BA/F3 cells expressing G1146A-IL-10R1 and
NZW-IL-10R1 was decreased following stimulation with IL-10. The
G1146A-IL-10R1 cell expression was decreased compared with cells
expressing NZW-IL-10R1. Among the three cell types, the expression
of CD16/32 was lower compared with that of the control (Fig. 3B). The expression of LECAM-1 in the
BA/F3 expressing WILD-IL-10R1 was markedly decreased compared with
the control and the other two groups. The expression of LECAM-1 in
BA/F3 cells expressing G1146A-IL-10R1 was decreased compared with
the control. However, the expression of LECAM-1 in BA/F3 cells
expressing NZW-IL-10R1 was higher compared with the control
(Fig. 3C).
Intracellular signal transduction of
different types IL-10R1 on BA/F3 cells
Levels of the intracellular signal transduction
proteins Jak1, Stat3 and Tyk2, which are associated with IL-10R1,
in BA/F3 cells expressing G1146A-IL-10R1, NZW-IL-10R1 and
WILD-IL-10R1 without IL-10 stimulation were identical (Fig. 4). Following stimulation with 10
ng/ml IL-10 for 15 min, the levels of the intracellular signal
transduction proteins Jak1, Stat3 and Tyk2 were also identical in
BA/F3 cells expressing G1146A-IL-10R1, NZW-IL-10R1 and WILD-IL-10R1
(Fig. 5A). However, phosphorylated
(p)-Jak1 and p-Stat3 levels in BA/F3 cells expressing
G1146A-IL-10R1 and NZW-IL-10R1 were markedly lower compared with
BA/F3 cells expressing WILD-IL-10R1 following IL-10 stimulation
(Fig. 5B and C), while the levels
of p-Tyk2 in the three types of cells did not differ significantly
(Fig. 5D).
Discussion
As a cytokine with an inhibitory function in the
immune reaction, IL-10 has an important role in immunoregulation
(15). Treg and Breg are
negatively regulated by IL-10 secretion. IL-10 is now recognized as
a cytokine associated with SLE morbidity (16). IL-10 has the ability to inhibit
B-cell proliferation and differentiation. These effects are closely
associated with IL-10R1 and the distribution of the functional
regions is located in the cytoplasm (12). A comparison of IL-10 in CD57BL/6,
NZW and MRL mice indicated 18 base replacements, seven of which
induce amino acid changes. The most notable change was in G1146A,
which is located in the immune inhibition region and blocks the
linkage with the signal transduction protein to interrupt the
ability of immune inhibition. The proliferation rate of BA/F3
expressing WILD-IL-10R1 was observed to be negatively correlated
with the time and dosage of stimulation with IL-10, and was
consistently lower compared with BA/F3 cells without stimulation.
These results indicated that the intact immune inhibition region
may regulate the ability of the proliferation region effectively
and inhibit proliferation to downregulate the number of B cells and
decrease the output of antibodies. By contrast, the proliferation
rate of BA/F3 expressing G1146A-IL-10R1 was positively correlated
with the time and dosage of stimulation. The results indicated that
base replacement caused a loss in the regulation of proliferation.
The general effect of IL-10 on the proliferation rate of BA/F3
cells expressing NZW-IL-10R1 was similar to that of BA/F3 cells
expressing G1146A-IL-10R1. However, the proliferation rate of BA/F3
cells expressing NZW-IL-10R1 markedly lower, which may be
associated with intracellular signal transduction disorders induced
by base displacements located elsewhere. It is known that, due to
the base replacement in the immune inhibition region, IL-10R1 loses
the intracellular signal transduction ability necessary for
proliferation inhibition, which may impair the inhibition of B-cell
proliferation (17). In SLE,
B-cell functions cannot be inhibited and the output of antibodies
cannot be downregulated, which increases with the onset and
progression of SLE (18).
In the present study, which investigated cellular
signal transduction, the levels of p-Jak1 and p-Stat3 were markedly
downregulated in BA/F3 cells expressing NZW-IL-10R1 and
G1146A-IL-10R1 following IL-10 stimulation. Furthermore, the levels
of p-Tyk2 were not different from those in the BA/F3 expressing
WILD-IL-10R1. This indicated that the base replacement in the
inhibition region of NZW-IL-10R1 induced the loss of the Jak1
phosphorylation site and Jak1 and Stat3 could not be
phosphorylated. The results also indicated that the inhibition
ability of IL-10 proceeded via the Jak1-Stat3 pathway (19), but not the Ttk2 pathway (9). Activation of this pathway may restore
the inhibition ability of IL-10R1 for the treatment of SLE.
As described above, the base replacement in the
inhibition region of NZW-IL-10R1 induces a loss in Jak1-Stat3
signaling. NZW-IL-10R1 does not inhibit B-cell proliferation
induced by the proliferation region of IL-10R1 effectively and
downregulates the expression of CD32 to reduce inhibition of B-cell
activation. NZW-IL-10R1 does not readily inhibit the LECAM-1
expression to induce the B-cell migration to the inflammation
sites, thus potentially triggering SLE. Previous studies have
indicated that MRL mice exhibit a specific defect regarding B cell
stimulation by IL-10 (20). In
Fasplr mice, the antibody levels of B- and T-cell subgroups and the
progression of lupus were similar to the non-defect mice (20). A previous study showed that in
MRL/lpr mice into which T2-MZP activated by anti-CD40 in
vitro was transferred, the symptoms of SLE were ameliorated
(21). At present, the role of T
cells in the pathogenesis and progression of SLE is being
increasingly investigated. A previous study indicated that mouse
CD8+ T cells expressed IL-10R1 immediately following
activation (22). Mouse
CD4+ T cells were also observed to express IL-10R1
immediately following activation. These findings indicated that
IL-10 may regulate the function of mouse T cells immediately
following activation. This hypothesis, combined with the
observations of the present study, require further investigation
and indicate that NZW-IL-10R1 may be implicated in the pathogenesis
and development of SLE and may present a novel therapeutic
target.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (nos. 30571701 and 30600541).
References
|
1
|
Heinlen LD, McClain MT, Merrill J, et al:
Clinical criteria for systemic lupus erythematosus precede
diagnosis, and associated autoantibodies are present before
clinical symptoms. Arthritis Rheum. 56:2344–2351. 2007. View Article : Google Scholar
|
|
2
|
Beebe AM, Cua DJ and de Waal Malefyt R:
The role of interleukin-10 in autoimmune disease: systemic lupus
erythematosus (SLE) and multiple sclerosis (MS). Cytokine Growth
Factor Rev. 13:403–412. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Houssiau FA, Lefebvre C, Vanden Berghe M,
et al: Serum interleukin 10 titers in systemic lupus erythematosus
reflect disease activity. Lupus. 4:393–395. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mongan AE, Ramdahin S and Warrington RJ:
Interleukin-10 response abnormalities in systemic lupus
erythematosus. Scand J Immunol. 46:406–412. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mok CC and Lau CS: Pathogenesis of
systemic lupus erythematosus. J Clin Pathol. 56:481–490. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sprengers D, Stoop JN, Binda RS, et al:
Induction of regulatory T-cells and interleukin-10-producing cells
in non-responders to pegylated interferon-alpha therapy for chronic
hepatitis B. Antivir Ther. 12:1087–1096. 2007.PubMed/NCBI
|
|
7
|
Ray A, Basu S, Williams CB, et al: A novel
IL-10-independent regulatory role for B cells in suppressing
autoimmunity by maintenance of regulatory T cells via GITR ligand.
J Immunol. 188:3188–3198. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jankovic D, Kullberg MC, Feng CG, et al:
Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of
host-protective regulatory IL-10 during intracellular protozoan
infection. J Exp Med. 204:273–283. 2007.
|
|
9
|
Liu Y, Wei SH, Ho AS, et al: Expression
cloning and characterization of a human IL-10 receptor. J Immunol.
152:1821–1829. 1994.PubMed/NCBI
|
|
10
|
Ho AS, Wei SH, Mui AL, et al: Functional
regions of the mouse interleukin-10 receptor cytoplasmic domain.
Mol Cell Biol. 15:5043–5053. 1995.PubMed/NCBI
|
|
11
|
Riley JK, Takeda K, Akira S, et al:
Interleukin-10 receptor signaling through the JAK-STAT pathway.
Requirement for two distinct receptor-derived signals for
anti-inflammatory action. J Biol Chem. 274:16513–16521. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moore KW, de Waal Malefyt R, Coffman RL,
et al: Interleukin-10 and the interleukin-10 receptor. Annu Rev
Immunol. 19:683–765. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi ZM, Wang J, Sun ZR, et al: Polymorphism
of the mouse gene for the interleukin 10 receptor alpha chain
(Il10ra) and its association with the autoimmune phenotype.
Immunogenetics. 57:697–702. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin Z, Bahtiyar G, Zhang N, Liu L, Zhu P,
Robert ME, McNiff J, Madaio MP and Craft J: IL-10 regulates murine
lupus. J Immunol. 169:2148–2155. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akdis CA and Blaser K: Mechanisms of
interleukin-10-mediated immune suppression. Immunology.
103:131–136. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou M, Ding L, Peng H, et al: Association
of the interleukin-10 gene polymorphism (−1082A/G) with systemic
lupus erythematosus: a meta-analysis. Lupus. 22:128–135. 2013.
|
|
17
|
Donnelly RP, Dickensheets H and Finbloom
DS: The interleukin-10 signal transduction pathway and regulation
of gene expression in mononuclear phagocytes. J Interferon Cytokine
Res. 19:563–573. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peeva E, Venkatesh J, Michael D and
Diamond B: Prolactin as a modulator of B cell function:
implications for SLE. Biomed Pharmacother. 58:310–319. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Valencia-Pacheco G, Layseca-Espinosa E,
Niño-Moreno P, et al: Expression and function of IL-10R in
mononuclear cells from patients with systemic lupus erythematosus.
Scand J Rheumatol. 35:368–378. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Teichmann LL, Kashgarian M, Weaver CT, et
al: B cell-derived IL-10 does not regulate spontaneous systemic
autoimmunity in MRL.Fas(lpr) mice. J Immunol. 188:678–685. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blair PA, Chavez-Rueda KA, Evans JG, et
al: Selective targeting of B cells with agonistic anti-CD40 is an
efficacious strategy for the generation of induced regulatory
T2-like B cells and for the suppression of lupus in MRL/lpr mice. J
Immunol. 182:3492–3502. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Biswas PS, Pedicord V, Ploss A, et al:
Pathogen-specific CD8 T Cell responses are directly inhibited by
IL-10. J Immunol. 179:4520–4528. 2007. View Article : Google Scholar : PubMed/NCBI
|