Introduction
Infectious diseases caused by a variety of
Salmonella enterica serotypes are widespread worldwide,
representing a severe public health concern (1). Infection with Salmonella
enterica serotype paratyphi A (SPA) is an emerging global
public health problem due to the increase in enteric fever cases
caused by SPA and the lack of protective vaccines (2–4). In
Southeast and Southwest China, the infection rate of SPA has
increased in the past several decades with the development of
tourism, where >80% of the enteric fever outbreaks are caused by
SPA (5). In recent years, Yuxi
City of Yunnan Province has become one of the most severely endemic
areas of SPA in China (6).
Subtyping and tracking individual strains involved
in SPA outbreak or sporadic cases are important for the control and
prevention of SPA transmission in Yuxi. The technique of
pulsed-field gel electrophoresis (PFGE) is currently the standard
method for molecular typing and epidemic surveillance of
Salmonella spp., including SPA (7,8).
However, PFGE is not a routine method for SPA surveillance due to
the expense of the equipment and the requirement of highly trained
technicians (9). Multi-locus
variable number tandem repeat (VNTR) analysis (MLVA), a genotyping
method based on polymerase chain reaction (PCR) and sequencing,
which distinguishes tandem sequence repeats that vary in copy
numbers (10,11), may be practical for subtyping SPA
due to the simple operation, low cost, high-speed and weak
laboratory-dependence (12).
Furthermore, MLVA genotyping is becoming an important DNA-based
typing tool for investigating strains that are related or unrelated
to outbreaks (13).
Although one study has previously investigated the
use of MLVA for subtyping SPA, the information of VNTRs for MLVA of
SPA in this investigation is limited as the VNTRs were examined
from the genomes of one strain of SPA (ATCC9150) and two strains of
S. enterica serovar Typhi (S. Typhi; CT18 and Ty2) (14). Although the genomes of S. Typhi and
SPA are closely related (15),
their tandem repeats (TRs) are different. The present study
searched for TR loci from two SPA genomes, ATCC9150 (NC_006511) and
AKU_12601 (NC_011147), and determined nine VNTR loci for MLVA
typing of SPA. We aimed to identify the type of epidemic clone in
Yuxi and whether the Yuxi SPA isolates were phylogenetically
distant from the 20 strains of SPA isolates collected by the
Chinese Medical Culture Collection Center (CMCC).
Materials and method
Strains and extraction of bacterial
genomic DNA
A total of 215 strains of SPA, including 195 Yuxi
isolates and 20 CMCC strains were used in the present study. Among
the 20 CMCC strains, one strain was ATCC9150 while the other 19
were collected from various research organizations with limited
background information and stored by CMCC (Table I). Among the 195 Yuxi isolates, 48
were separated from the patients of the SPA outbreak in 2007 while
the others were isolated from sporadic cases between 2005 and
2009.
 | Table IInformation of 20 SPA strains
collected by CMCC. |
Table I
Information of 20 SPA strains
collected by CMCC.
| Strain | Source |
|---|
| ATCC9150 | ATCC |
| 50001 | Denmark |
| 50002 | Denmark |
| 50084 | USA |
| 50101 | Former Soviet
Union |
| 50154 | France |
| 50433 | Bulgaria |
| 50434 | Bulgaria |
| 50672 | Poland |
| 50674 | Poland |
| 50701 | Czech Republic |
| 50078 | Beijing, China |
| 50501 | Dalian, China |
| 50502 | Dalian, China |
| 50504 | Dalian, China |
| 50505 | Dalian, China |
| 50506 | Dalian, China |
| 50507 | Lanzhou, China |
| 50508 | Guangdong,
China |
| 50509 | Guangdong,
China |
Genomic DNA of SPA was extracted as previously
described (16,17). Briefly, the bacteria were streaked
on brain heart infusion agar (BHIA) plates and grown at 37°C
overnight in 5% CO2 incubator. A loop of typical
colonies was removed from the BHIA plates and boiled for 10 min in
200 μl Tris-EDTA buffer (10 mM Tris-Cl and 1 mM EDTA, pH 8.0). The
supernatant was obtained by centrifugation at 8,000 × g for 10 min
and used directly for PCR (18).
Identification of VNTRs
Potential TRs were first exploited from the genomes
of ATCC9150 and AKU_12601 using the Tandem Repeats Finder (TRF)
program (19,20) and the http://tandem.bu.edu/trf/trf.htlm website (21). The candidates were scored as
match(+2), mismatch(−3) and indel(−5) for pattern alignment
(22). The potential TRs were
selected by alignment scores ≥80, or homology of repeat locus ≥85%.
A total of 51 TRs (TR1-51) were screened from the genomes of
ATCC9150 and AKU_12601 (data not shown). Primers flanking >51
TRs were designed using the Primer 5.0 software (Premier Biosoft
International, Palo Alto, CA, USA) and synthesized by Sangon
Company (Shanghai, China). The polymorphism of PCR fragments
amplified with primers of 51 TRs was analyzed by agarose
electrophoresis and nine VNTR loci (TR27, TR51, TR41, TR43, TR5,
TR40, TR44, TR24 and TR49) were verified to be polymorphic
(Fig. 1). The nine VNTRS of 19
CMCC strains except ATCC9150 were sequenced. The repeat numbers for
each locus corresponding to 20 CMCC and AKU_12601 are summarized in
Table II.
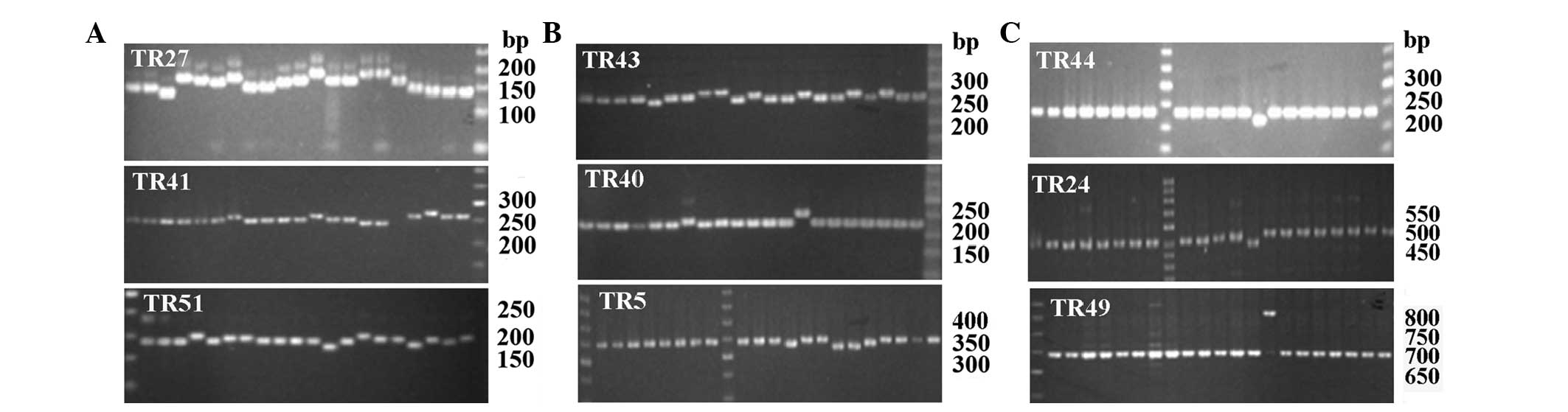 | Figure 1The polymorphisms of nine VNTR loci
TR27, TR51, TR41, TR43, TR5, TR40, TR44, TR24 and TR49 analyzed by
agarose gel electrophoresis, capillary electrophoresis and
sequencing for PCR products amplified from 20 CMCC strains and one
Yuxi isolate YN07044. (A) The agarose gel electrophoresis for PCR
products of nine VNTR loci. Lanes: left to right, ATCC9150, 50078,
50001, 50502, 50506, 50501, 50505, 50154, 50434, 50101, 50507,
50509,50672, 50674, 50084, 50433, 50508, 50701, 50002, 50504,
YN07044. DNA Marker, 50bp DNA Ladder Marker. (B) The representative
electropherogram from pooled capillary electrophoresis runs of
FAM-labeled or HEX-labeled primers of nine VNTRs. The PCR products
for TR27, TR41, TR51, TR43, TR40 and TR5 were amplified from
YN07044, demonstrating 143, 259, 194, 276, 217 and 338 bp,
respectively. The PCR products for TR44 and TR24 were amplified
from 50504 with 254 and 527 bp respectively. (C) The precise
numbers of TR40 in three representative sequences amplified from
ATCC9150 (3), 50154 (4), and 50674 (6) were analyzed by sequencing. CMCC,
Chinese Medical culture Collection Center; SPA, Salmonella
enterica serotype paratyphi A; VNTR, variable number of tandem
repeats. |
 | Table IICharacteristics of 9 VNTR loci for 20
CMCC SPA strains and AKU_12601. |
Table II
Characteristics of 9 VNTR loci for 20
CMCC SPA strains and AKU_12601.
| VNTR locus | Primer sequence
(5′-3′) | Repeat model | Repeat number |
|---|
| TR27 | F:
GGAAAGACTGGCGAACAAAT | | |
| R:
TCGCCAATACCATGAGTACG | TACTGG | 9–16 |
| TR51 | F:
CCATGGCTGCAGTTAATTTCT | | |
| R:
TGATACGCTTTTGACGTTGC | ACCATG | 1–11 |
| TR41 | F:
TGGGAAACTTATCTTCGA | | |
| R:
TAATCAGTCTGGCCTGTG | ACATCTCCT | 1–6 |
| TR43 | F:
TACTGCTTTCGCCATCGG | | |
| R:
ATAATCCGGGTAAAGACC | CCGTTAACCG | 3–5 |
| TR5 | F:
GCATACACCGCAGCACTC | | |
| R:
TTCCTTTCCCTGCTTATTTGTC | TAGCAGGTAA | 2–4 |
| TR40 | F:
CGGGTGATTCTGTTATCT | | |
| R:
ATAGTGTTACGCACCTCA | TTTTTTAAG | 3–6 |
| TR44 | F:
CAGAAGCAGTTCCACCACCT | | |
| R:
CATTTCACATCGCCGACTTT |
GCAGGAGCTGGTGGGCGA | 1–3 |
| TR24 | F:
GCTGAAGAAGCGGCAAAAC | | |
| R:
GTACCGCTATCTTTCGATGGC | 45bpa | 7–8 |
| TR49 | F:
GCTTGCAGCTAAATGGAT | | |
| R:
ATCTGACGAAAGCGGAAC | 232bpb | 2–3 |
PCR and agarose electrophoresis
analysis
All selected loci were amplified from the genomic
DNA of the 20 CMCC SPA strains by PCR as described previously
(23). Briefly, 1 μl bacterial
lysate was amplified by a thermal cycler PTC-200 DNA Engine (MJ
Search Partners, Inc., Lake Forest, IL, USA) in a 25 μl final
reaction volume containing 0.1 μmol/ml dNTPs, 0.2 μmol/ml primers,
0.5 U Taq DNA polymerase (Takara Bio, Inc., Shiga, Japan)
under the following conditions: 10 min at 95°C, followed by 30
cycles of three temperatures (15 sec at 95°C, 1 min at 55~60°C, 1
min at 72°C) and then 10 min at 72°C. A total of 5 μl of the PCR
products were separated in 1.5% agarose gels in 1X TAE buffer
(AppliChem Inc., St. Louis, MO, USA) at a voltage of 6 V/cm for ~3
h. The gels were stained in ethidium bromide for visualization
under UV light and were photographed on a Gel Doc 2000 system
(Bio-Rad, Hercules, CA, USA). The 50 bp (base pair) DNA Ladder
Marker (Takara Bio, Inc.) was loaded in all of the gels to
facilitate determining the size of the DNA fragments. To ensure the
accuracy of agarose electrophoresis and to compare the results
between multiple gels, the PCR products of ATCC9150 in each locus
were obtained as a positive control. The TRs were identified to be
polymorphic if large differences between their PCR fragments in the
agarose gel electrophoresis were observed. The PCR products were
purified with the QIAquick PCR Purification kit (Qiagen, Hilden,
Germany) following the manufacturer’s instructions.
MLVA typing and data analysis
In order to confirm that any length polymorphism of
fragment was due to variations in the VNTR copy number (24), the purified PCR products amplified
from 195 Yuxi isolate and 20 CMCC strains were sequenced by the
Sangon Company. The numbers of repeats in each allele were analyzed
by BioNumerics version 6.0 (Applied Maths, Austin, TX, USA)
(25), and the numerical profile
for each locus was created according to the copies of VNTR
(14,26). The dendogram for MLVA profiles was
drawn using the categorical coefficient and the alignment of
unweighted pair group method using arithmetic averages (27). A minimum spanning tree (MST) was
constructed using the categorical coefficient (10,23,28).
The priority rule for constructing MST was set so that the
genotypes that had the highest number of single-locus variants
would be linked first (23).
Results
MLVA genotyping and phylogenetic analysis
for 215 SPA isolates
The dendogram for the MLVA types distribution
demonstrates that 23 MLVA types (MLVA 1-23) were identified in the
215 SPA isolates and were grouped into six distinct cluster groups
A, B, C, D, E and F. All of the Yuxi isolates were exclusively
grouped into cluster C with nine MLVA genotypes (MLVA 10-18;
Fig. 2). The 20 CMCC isolates were
grouped in cluster A B, D, E and F with 14 MLVA types (MLVA 1-9 and
MLVA 19-23; Fig. 2). There are two
key observations to note among the 20 CMCC strains. Firstly, the
same MLVA type of SPA emerged in different countries. For example,
50501, 50502 and 50506 (Dalian, China) and 50001 (Denmark) were
typed as MLVA7, ATCC9150, 50002 (Denmark) and 50701 (Czech
Republic) were typed as MLVA19, and 50507 (Lanzhou, China) and
50101 (Former Soviet Union) were typed as MLVA6, although they were
from different countries. Secondly, there existed different MLVA
types in the same region or country. For instance, one strain from
Dalian, China (50505) was identified to be MLVA8, which is
different from the other three Dalian strains of MLVA7 (50501,
50502 and 50506). Despite the fact that 50672 and 50674 were from
Poland their MLVA types were MLVA9 and MLVA23, respectively. The
same phenomena were observed in two strains from Bulgaria 50433
(MLVA2) and 50434 (MLVA5).
 | Figure 2MLVA type distribution of 215 SPA
isolates using categorical coefficient and unweighted pair group
method using arithmetic average. The 215 SPA isolates, including
195 Yuxi isolates and 20 CMCC strains were subtyped into 23 MLVA
types (MLVA1~23) and grouped into six distinct clusters (A, B, C,
D, E and F). All of the 195 Yuxi isolates were grouped in the C
cluster with nine MLVA genotypes (MLVA10~18). The 20 CMCC strains
were grouped into A, B, D, E and F clusters with 14 MLVA types
(MLVA 1-9 and MLVA 19-23). MLVA, multiple-locus variable number of
tandem repeats analysis; SPA, Salmonella enterica serotype
paratyphi A; CMCC, Chinese Medical Culture Collection Center. |
The genetic correlations among the 215 isolates were
determined based on the MLVA profiles using the MST algorithm
(10,23,28).
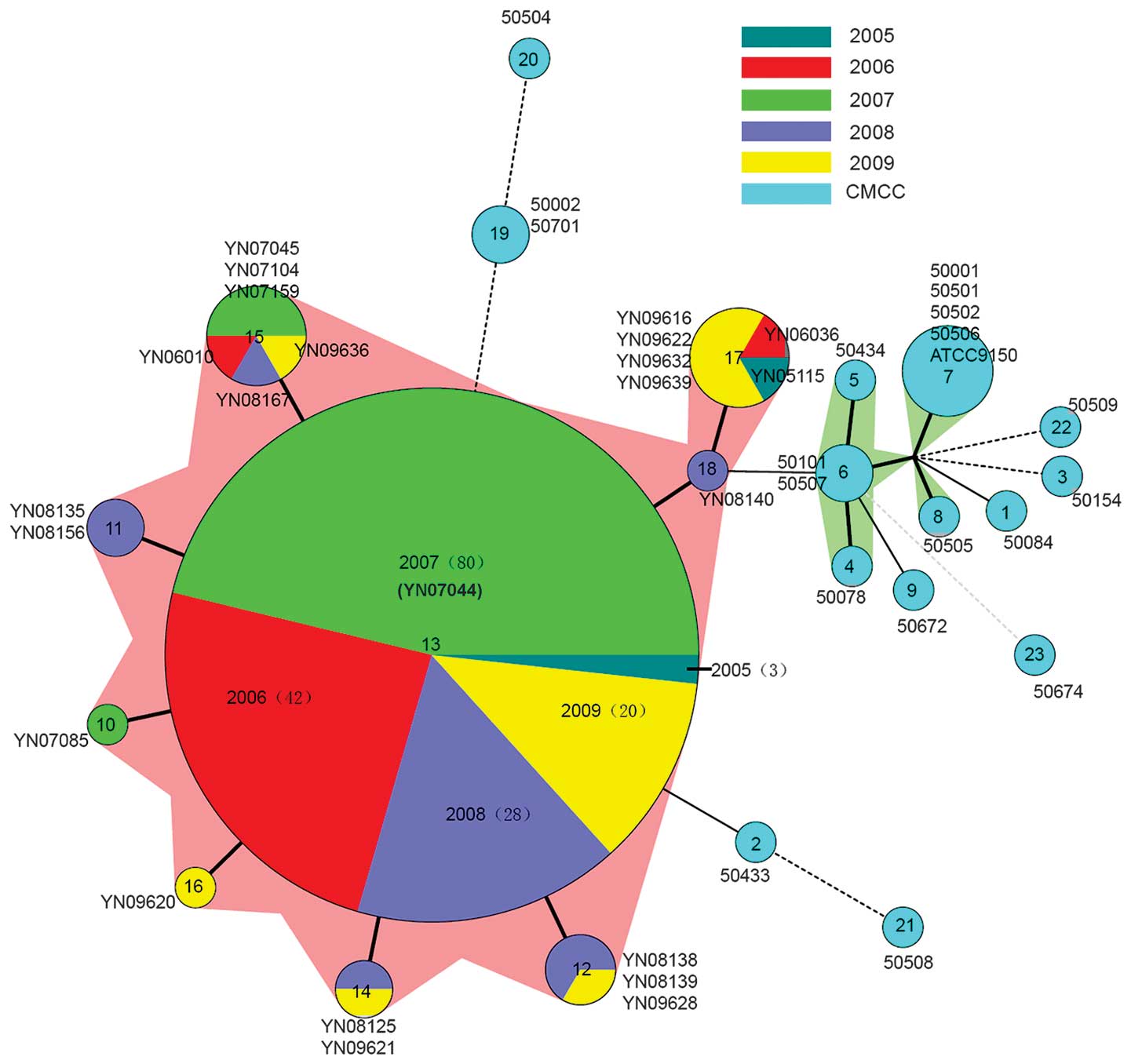
As demonstrated in Fig. 3, MST
offers a more detailed view of the diversity of the 215 isolates
and highlights the closer subtypes that differ by few allelic
changes (23). Isolates with the
same MLVA profiles were clustered in a circle. A total of 173 Yuxi
isolates form the MLVA13 circle surrounded by 22 other isolates
with 8 MLVA types. It indicates that the 195 Yuxi isolates are
closely related with each other. Although the 195 Yuxi isolates are
distinct from the 20 CMCC strains, they are relatively close to
50433 (Bulgaria) with MLVA2, and also close to 50002 (Denmark), and
50701 (Czech) with MLVA19. YN08140 (Yuxi) with MLVA18 is closely
related to 50101 (Former Soviet Union) and 50507 (Lanzhou, China)
with MLVA6, which varied in only two VNTR loci (Fig. 2).
Epidemiology of SPA in Yuxi
Although 195 Yuxi SPA isolates distribute in nine
genotypes (MLVA 10-18), they express only one or two VNTR loci that
are different from each other (Fig.
2). The MST demonstrated that they are closely related with
each other and separated from the 20 CMCC strains (Fig. 3). Table III reveals the MLVA type
distribution of 195 Yuxi isolates collected between 2005 and 2009.
In all, MLVA13 accounted for 88.7% (173/195) of the Yuxi isolates.
Among the 48 outbreak isolates in 2007, MLVA13 accounted for 91.7%
(44/48). Outside of the 2007 outbreak, MLVA13 accounted for 87.8%
(129/147) of Yuxi sporadic isolates.
 | Table IIIThe MLVA types distribution of 195
SPA Yuxi isolates between 2005 and 2009. |
Table III
The MLVA types distribution of 195
SPA Yuxi isolates between 2005 and 2009.
|
MLVA
type |
|---|
|
|
|---|
| Year | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | Total |
|---|
| 2005 | | | | 3 | | | | 1 | | 4 |
| 2006 | | | | 42 | | 1 | | 1 | | 44 |
| 2007 | 1 | | | 80 | | 3 | | | | 84 |
| 2008 | | 2 | 2 | 28 | 1 | 1 | | | 1 | 35 |
| 2009 | | | 1 | 20 | 1 | 1 | 1 | 4 | | 28 |
| Total | 1 | 2 | 3 | 173 | 2 | 6 | 1 | 6 | 1 | 195 |
Discussion
A reliable method for subtyping bacterial isolates
is a prerequisite for the identification of sources and
transmission routes of an infectious disease (29). There is no doubt that PFGE is
currently the gold-standard technique for subtyping numerous
bacteria, including Salmonella serotypes with reproducible
patterns and high resolution and is widely used by the CDC PulseNet
surveillance program worldwide (30). However, the widespread use of PFGE
is limited in the CDC of numerous Chinese cities and counties by
the lack of specifically trained personnel, sophisticated and
expensive equipment and precise standard protocols (31). Conversely, MLVA, which is based on
the evaluation of differences in the number of TRs, is a quick,
cheap and simple method for the molecular typing of bacteria
(20). In the present study, a
MLVA with nine VNTR markers was developed, which exhibited a wide
range of variability for subtyping 215 SPA isolates into 23 MLVA
types. The phylogenetic association among the 20 CMCC SPA strains
with various backgrounds was elucidated clearly with 14 MLVA types.
Clonal groups among the 195 Yuxi isolates in the different years
were discerned with nine other MLVA types. The outbreak-related
isolate was identified to be MLVA13 in 2007. Eight novel SPA
isolates separated from patients in 2010 were examined with the
MLVA method developed in the present study, and it was identified
that six of the isolates were MLVA13 while one was MLVA14 and
another was MLVA16. These results indicate that the VNTR markers
identified in the present study are applicable to subtype SPA.
Yuxi, a medium-sized city (15,285 km2)
with 2,095,532 residents distributed into two districts (Hongta and
Eshan) and six counties (Chengjiang, Tonghai, Jiangchuan, Huaning,
Xinping and Yuanjiang) has been one of the most severely endemic
areas of paratyphoid fever in China since 1999. There was a
progressive increase in the number of SPA cases in Yuxi between
2005 and 2009. The results of MLVA typing for Yuxi isolates
indicate that the MLVA13 isolate was the epidemic clone in Yuxi in
outbreaks and sporadic cases. Consistent with the sources and
transmission routes of enteric fever (32,33),
contaminated water and food are major sources of SPA in Yuxi. It
was identified that contaminated well water in a vegetable market
of Hongta was the direct factor leading to the 2007 outbreak of
SPA. More than 90% of patients in the 2007 outbreak were
retrospectively investigated to have purchased vegetables from the
Hongta vegetable market near the infected well, where the vendors
watered the vegetables using the well water. Subsequently, the SPA
isolates were separated from the water in the well. The sources of
SPA from the well water were further confirmed by the result of
MLVA typing for SPA in the present study, demonstrating that MLVA13
SPA were the major clones isolated from the well water, vegetables
and patients during the epidemic. In Asia, SPA may also be
transmitted by consumption of contaminated foods from street
vendors (4). The contaminated
foods sold by street vendors may be important vectors of the SPA
sporadic isolates in Yuxi as it is highly common in Yuxi to eat at
street vendors with poor sanitary conditions. From the patients who
had eaten at street vendors, a variety of MLVA types were separated
with the majority being the MLVA13 type of SPA. The incidence rate
of enteric fever has decreased significantly and remained at a low
level following 2010 with the strengthened surveillance of stock
sold in the markets and by street vendors.
A total of 20 SPA isolates have been collected from
different countries and regions during different periods by the
CMCC thus far. To investigate the Yuxi SPA isolates, the MLVA type
of 20 CMCC SPA strains was analyzed, and revealed a large diversity
with 14 MLVA types which are unrelated to the 195 Yuxi
isolates.
Acknowledgements
This study was supported by grants from the
Foundation of Provincial Education Department of Hunan Province,
China (grant no. 05B046), and the Cooperative Foundation of Yuxi
Science and Technology Committee, Yunnan, China (grant no.
HZ200706).
References
|
1
|
Smith KP, George J, Cadle KM, Kumar S,
Aragon SJ, Hernandez RL, et al: Elucidation of antimicrobial
susceptibility profiles and genotyping of Salmonella
enterica isolates from clinical cases of Salmonellosis in New
Mexico in 2008. World J Microbiol Biotechnol. 26:1025–1031.
2010.PubMed/NCBI
|
|
2
|
Maskey AP, Day JN, Phung QT, Thwaites GE,
Campbell JI, Zimmerman M, et al: Salmonella enterica serovar
Paratyphi A and S. enterica serovar Typhi cause
indistinguishable clinical syndromes in Kathmandu, Nepal. Clin
Infect Dis. 42:1247–1253. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sheikh A, Charles RC, Rollins SM, Harris
JB, Bhuiyan MS, Khanam F, et al: Analysis of Salmonella
enterica serotype paratyphi A gene expression in the blood of
bacteremic patients in Bangladesh. PLoS Negl Trop Dis.
4:e9082010.
|
|
4
|
Fangtham M and Wilde H: Emergence of
Salmonella paratyphi A as a major cause of enteric fever:
need for early detection, preventive measures, and effective
vaccines. J Travel Med. 15:344–350. 2008.
|
|
5
|
Dong BQ, Yang J, Wang XY, Gong J, von
Seidlein L, Wang ML, et al: Trends and disease burden of enteric
fever in Guangxi province, China, 1994–2004. Bull World Health
Organ. 88:689–669. 2010.PubMed/NCBI
|
|
6
|
Wang SK, Chu CJ, Sun PS, Shan DS, Kong FL,
Liu HY, et al: Study on blood cultures and bacteria counts in the
blood of paratyphoid fever A patients. Eur J Clin Microbiol Infect
Dis. 28:1259–1261. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gaind R, Paglietti B, Murgia M, Dawar R,
Uzzau S, Cappuccinelli P, et al: Molecular characterization of
ciprofloxacin-resistant Salmonella enterica serovar Typhi
and Paratyphi A causing enteric fever in India. J Antimicrob
Chemother. 58:1139–1144. 2006.PubMed/NCBI
|
|
8
|
Gal-Mor O, Suez J, Elhadad D, Porwollik S,
Leshem E, Valinsky L, et al: Molecular and cellular
characterization of a Salmonella enterica serovar Paratyphi
A outbreak strain and the human immune response to infection. Clin
Vaccine Immunol. 19:146–156. 2012.
|
|
9
|
Elberse KE, Nunes S, Sá-Leão R, van der
Heide HG and Schouls LM: Multiple-locus variable number tandem
repeat analysis for Streptococcus pneumoniae: comparison
with PFGE and MLST. PLoS One. 6:e196682011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tien YY, Ushijima H, Mizuguchi M, Liang SY
and Chiou CS: Use of multilocus variable-number tandem repeat
analysis in molecular subtyping of Salmonella enterica
serovar Typhi isolates. J Med Microbiol. 61:223–232. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
U’Ren JM, Schupp JM, Pearson T, Hornstra
H, Friedman CL, Smith KL, et al: Tandem repeat regions within the
Burkholderia pseudomallei genome and their application for
high resolution genotyping. BMC Microbiol. 7:232007.
|
|
12
|
Heck M: Multilocus variable number of
tandem repeats analysis (MLVA) - a reliable tool for rapid
investigation of Salmonella typhimurium outbreaks. Euro
Surveill. 14pii: 19177. 2009.PubMed/NCBI
|
|
13
|
Kruy SL, van Cuyck H and Koeck JL:
Multilocus variable number tandem repeat analysis for Salmonella
enterica subspecies. Eur J Clin Microbiol Infect Dis.
30:465–473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tien YY, Wang YW, Tung SK, Liang SY and
Chiou CS: Comparison of multilocus variable-number tandem repeat
analysis and pulsed-field gel electrophoresis in molecular
subtyping of Salmonella enterica serovars Paratyphi A. Diagn
Microbiol Infect Dis. 69:1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McClelland M, Sanderson KE, Clifton SW,
Latreille P, Porwollik S, Sabo A, et al: Comparison of genome
degradation in Paratyphi A and Typhi, human-restricted serovars of
Salmonella enterica that cause typhoid. Nat Genet.
36:1268–1274. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Killgore GE and Kato H: Use of arbitrary
primer PCR to type Clostridium difficile and comparison of results
with those by immunoblot typing. J Clin Microbiol. 32:1591–1603.
1994.PubMed/NCBI
|
|
17
|
Queipo-Ortuño MI, De Dios Colmenero J,
Macias M, Bravo MJ and Morata P: Preparation of bacterial DNA
template by boiling and effect of immunoglobulin G as an inhibitor
in real-time PCR for serum samples from patients with brucellosis.
Clin Vaccine Immunol. 15:293–296. 2008.PubMed/NCBI
|
|
18
|
Jiang Y, Liu HC, Zheng HJ, Tang B, Dou XF,
Zhao XQ, et al: Evaluation of four candidate VNTR Loci for
genotyping 225 Chinese clinical Mycobacterium tuberculosis
complex strains. Biomed Environ Sci. 25:82–90. 2012.PubMed/NCBI
|
|
19
|
Benson G: Tandem repeats finder: a program
to analyze DNA sequences. Nucleic Acids Res. 27:573–580. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haguenoer E, Baty G, Pourcel C, Lartigue
MF, Domelier AS, Rosenau A, et al: A multi locus variable number of
tandem repeat analysis (MLVA) scheme for Streptococcus
agalactiae genotyping. BMC Microbiol. 11:1712011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mouton L, Nong G, Preston JF and Ebert D:
Variable-number tandem repeats as molecular markers for biotypes of
Pasteuria ramosa in Daphnia spp. Appl Environ
Microbiol. 73:3715–3718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O’Dushlaine CT and Shields DC: Tools for
the identification of variable and potentially variable tandem
repeats. BMC Genomics. 7:2902007.PubMed/NCBI
|
|
23
|
Ramisse V, Houssu P, Hernandez E, Denoeud
F, Hilaire V, Lisanti O, et al: Variable number of tandem repeats
in Salmonella enterica subsp. enterica for typing purposes.
J Clin Microbiol. 42:5722–5730. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, Lee MA, Ooi EE, Mavis Y, Tan AL and
Quek HH: Molecular typing of Salmonella enterica serovar
typhi isolates from various countries in Asia by a multiplex PCR
assay on variable-number tandem repeats. J Clin Microbiol.
41:4388–4394. 2003.PubMed/NCBI
|
|
25
|
Guo C, Liao Y, Li Y, Duan J, Guo Y, Wu Y,
et al: Genotyping analysis of Helicobacter pylori using
multiple-locus variable-number tandem-repeats analysis in five
regions of China and Japan. BMC Microbiol. 11:1972011.PubMed/NCBI
|
|
26
|
Yazdankhah SP and Lindstedt BA: Variable
number tandem repeat typing of bacteria. Methods Mol Biol.
396:395–405. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lavania M, Katoch K, Sharma R, Sharma P,
Das R, Gupta AK, et al: Molecular typing of Mycobacterium
leprae strains from northern India using short tandem repeats.
Indian J Med Res. 133:618–626. 2011.
|
|
28
|
Witonski D, Stefanova R, Ranganathan A,
Schutze GE, Eisenach KD and Cave MD: Variable-number tandem repeats
that are useful in genotyping isolates of Salmonella
enterica subsp. enterica serovars Typhimurium and Newport. J
Clin Microbiol. 44:3849–3854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nielsen EM, Engberg J, Fussing V, Petersen
L, Brogren CH and On SL: Evaluation of phenotypic and genotypic
methods for subtyping Campylobacter jejuni isolates from
humans, poultry, and cattle. J Clin Microbiol. 38:3800–3810.
2000.PubMed/NCBI
|
|
30
|
Harbottle H, White DG, McDermott PF,
Walker RD and Zhao S: Comparison of multilocus sequence typing,
pulsed-field gel electrophoresis, and antimicrobial susceptibility
typing for characterization of Salmonella enterica serotype
Newport isolates. J Clin Microbiol. 44:2449–2457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Montesinos I, Salido E, Delgado T, Cuervo
M and Sierra A: Epidemiologic genotyping of methicillin-resistant
Staphylococcus aureus by pulsed-field gel electrophoresis at
a university hospital and comparison with antibiotyping and protein
A and coagulase gene polymorphisms. J Clin Microbiol. 40:2119–2125.
2002.PubMed/NCBI
|
|
32
|
Korbsrisate S, Sarasombath S, Janyapoon K,
Ekpo P and Pongsunk S: Immunological detection of Salmonella
paratyphi A in raw prawns. Appl Environ Microbiol. 60:4612–4613.
1994.
|
|
33
|
Basnyat B, Maskey AP, Zimmerman MD and
Murdoch DR: Enteric (typhoid) fever in travelers. Clin Infect Dis.
41:1467–1472. 2005. View
Article : Google Scholar : PubMed/NCBI
|