Introduction
Hepatocellular carcinoma (HCC, also termed malignant
hepatoma) is one of the most common types of cancer and it is the
second most common cause of cancer-related mortality in China.
Common with other cancers, HCC is a complex disease that develops
when a mutation of the cellular machinery occurs, which results in
the cell replicating at a higher rate or avoiding apoptosis
(1). Due to the complicated
pathophysiology of HCC, it is difficult to treat this cancer
successfully and the prognosis is unsatisfactory. For example, only
10–20% of HCCs can be completely removed by surgery, which may lead
to recurrence of this disease within 3–6 months. However, the
molecular mechanisms involved in this biological process remain to
be fully understood. Thus further studies are required and may
contribute to the identification of key factors in HCC
progression.
Molecular changes in HCC are complex, including the
dysregulated genes, protein markers, non-coding RNAs and other
predictive biomarkers. Numerous dysregulated genes and potential
protein markers in HCC have been reported in the past decades. For
instance, the prospero homeobox protein (2) and high-mobility group A2 (3) were identified as critical factors
that regulate transcription factors relevant to
epithelial-mesenchymal transition, a significant event during
tumorigenesis, to promote HCC metastasis. Key dysregulated genes
involved in HCC have been identified by genomic and gene expression
analysis, including the oncogenes c-myc and N-ras and the tumor
suppressor genes tumor protein 53, retinoblastoma protein 1
(Rb1), cyclin-dependent kinase inhibitor 2A (CDKN2A)
and Axin 1. These genes were located on the regions of the
chromosome of genomic and epigenetic changes that were linked with
HCC development (4).
There is recent evidence indicating that microRNAs
(miRNAs; a type of non-coding RNA) have significant roles in
cancer, in particular in cancer development, progression and
metastasis, and as robust biomarkers for cancer prognosis. For
example, miR-129–2 methylation is highly accurate in distinguishing
HCC from cirrhosis patients and healthy individuals, implying its
potential utility as an early diagnostic marker for HCC (5). MiR-20a (6) and miR-195 (7) exhibit significant inhibitory roles in
HCC progression and may be potential therapeutic targets and
biomarkers for patients with HCC. It is noteworthy that certain
miRNAs have been revealed to target multiple genes involved in HCC
progression, including miR-221 and miR-222 that can target
p27, p57, B-cell lymphoma-2 (Bcl2)-modifying factor,
phosphatase and tensin homolog, metalloproteinase inhibitor 3,
DNA-damage-inducible transcript 4 protein and protein phosphatase
2, regulatory subunit Bα; and miR-122 can target cyclin G1,
disintegrin and metalloproteinase domain (ADAM)-containing protein
10, ADAM17, serum response factor, insulin-like growth
factor 1 receptor and Bcl-w (8).
These findings imply potential utility of miRNAs as therapeutic
targets and biomarkers for different pathophysiological processes
of HCC. Thus it is necessary to provide an overview of miRNA
expression and determine their target genes. In addition, the
development of computational prediction to identify potential miRNA
targets is convenient for the functional investigation of miRNAs
(9).
The present study aimed to identify featured target
genes of significant differentially-expressed miRNAs of HCC by
comparing normal and cancer tissue samples, and to analyze the
correlation of the target genes and HCC. Candidate target genes
identified by these approaches may provide the groundwork for a
combination therapy approach for HCC. However, further
investigation of their potential use in the treatment of HCC is
required.
Materials and methods
Microarray data
The miRNA profiling data was obtained from the study
by Wang et al (10), which
was found in the GEO (Gene Expression Omnibus) database (ID:
GSE31383) (11) and included nine
human healthy liver and 10 HCC samples. The miRNA profiling was
performed successfully using freshly frozen healthy liver and HCC
tissues obtained at the time of surgical treatment. The annotation
information of a total of 19 chips was available based on the
GPL10122 Platform (version 4; Luminex, New York, NY, USA) that was
designed by the bead-based technology.
Data pre-process and
differentially-expressed miRNA analysis
The microarray data in CEL format files were
converted into expression measures with the affy package
(Bioconductor) (12) in R language
(13,14) and standardized with the median
method. Next, the LIMMA package (Bioconductor) in R (15) was used to identify the miRNAs that
were differentially expressed between the nine human healthy liver
and the 10 HCC samples. Subsequently, the consequences were
adjusted for multiple testing with the Benjamin and Hochberg
(16) method in a multi-test
package. The miRNAs only with false discovery rate (FDR)<0.05
and |logFold Change|>1 were selected, and the
differentially-expressed miRNA that showed the greatest level of
upregulation (or downregulated) was selected for further
analysis.
Screening the target genes of the
differentially-expressed miRNA
In order to obtain the accurate target genes, the
two selected miRNAs were integrated into two miRNA databases:
miRecords (http://mirecords.biolead.org) (17) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) (18), in which the confirmed target genes
had been selected as functional genes of miRNAs.
The miRTarBase, a comprehensive database, contains a
manually curated collection of >1,300 experimentally supported
miRNA targets in several animal species, plants and viruses. The
miRecords, an integrated resource of interactions of experimentally
validated miRNA targets, consists of two components, the Validated
Targets component and the Predicated Targets component, and hosts
2,286 records of interactions between 548 miRNAs and 1,579 target
genes in nine animal species.
Network construction
The Search Tool for the Retrieval of the Interacting
Genes (STRING) database (19) was
used to search and construct the interaction network of the target
genes of the differentially-expressed miRNAs. Each score
represented a rough estimate of how likely a given association
describes a functional linkage between two genes. The pairs with a
combination score >0.5 were recorded. Finally, the interaction
network was analyzed, and the degree of each node was calculated
according to the experimental database and data mining of the
software.
Functional enrichment analysis
Functional enrichment analysis of the genes in the
interaction network was performed with software Gestalt (Leuven,
Belgium; Gene Set Analysis Toolkit) (20), an integrated system for analyzing
gene sets in various biological contexts, which incorporates
information of eight species, including humans, rats and mice, and
different public resources, including NCBI (21), Ensemble (22), KEGG (23) and GO (24). The function of algorithm based on
the hypergeometric distribution (25) and pathway enrichment analysis
(26) in this software identified
the correlated genes in the interaction network. FDR<0.05 was
set as the cut-off criterion.
Results
Screening differentially-expressed
miRNAs
To identify the specific differentially-expressed
miRNAs between human HCC tissues and healthy controls, publicly
available microarray datasets GSE31383 were obtained from the GEO
database. The miRNA expression profiling data were preprocessed
with the R affy package and were normalized by the median method
(Fig. 1).
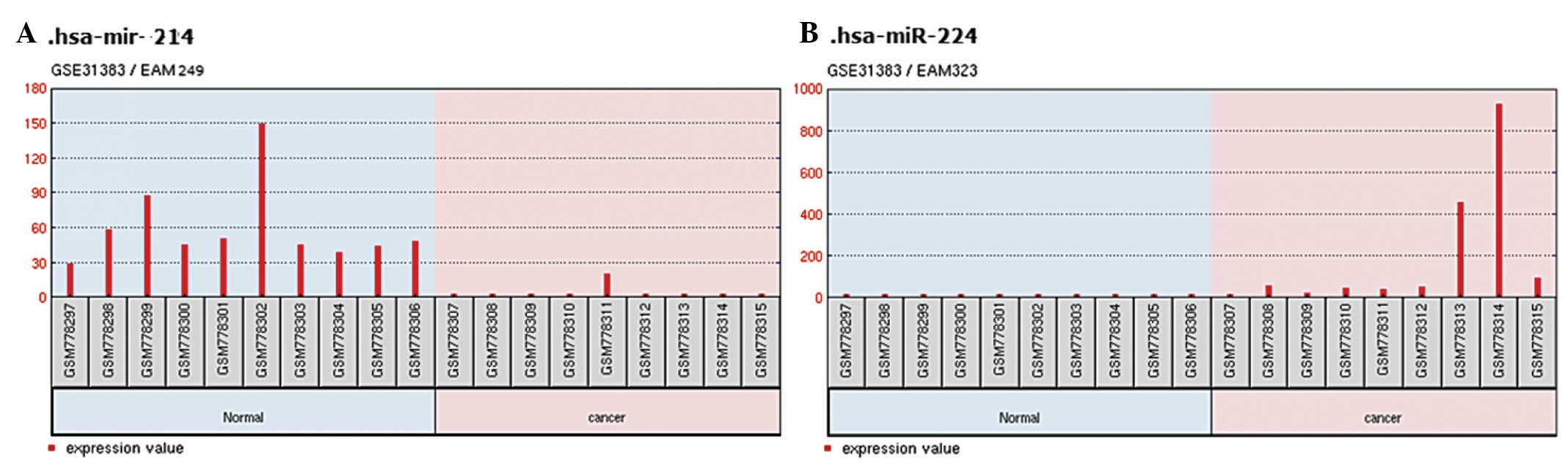
At an FDR<0.05 and |logFC|>1, a total
of 32 miRNAs was considered to be differentially-expressed between
the healthy controls and human HCC tissues, including 18
downregulated and 14 upregulated miRNAs (Table I). miRNA-224, exhibited the
greatest upregulation and thus, was selected from the 18
upregulated miRNAs. Similarly, miRNA-214 was selected from the
downregulated group (Fig. 2).
 | Table IDifferentially expressed miRNAs
(FDR<0.05, |logFC| >1). |
Table I
Differentially expressed miRNAs
(FDR<0.05, |logFC| >1).
| miRNA name | ID | FDR | logFC |
|---|
| hsa-miR-214 | EAM249 |
1.24×10−5 | −6.85536 |
| hsa-miR-503 | JLA186 |
6.19×10−11 | −6.53656 |
| hsa-mir-378 | JLA120 |
9.35×10−5 | −6.05122 |
| hsa-miR-200b | EAM305 |
1.20×10−3 | −5.9841 |
| hsa-miR-424 | JLA103 |
7.07×10−4 | −4.3832 |
| hsa-miR-375 | JLA106 |
2.54×10−3 | −4.2444 |
| hsa-miR-497 | JLA147 |
2.02×10−2 | −4.02889 |
| hsa-mir-30a | EAM280 |
6.36×10−3 | −3.75307 |
| hsa-miR-200c | JLA216 |
2.02×10−2 | −3.69177 |
| hsa-miR-130a | EAM159 |
6.36×10−3 | −3.26364 |
| hsa-miR-99a | EAM121 |
3.84×10−2 | −2.88995 |
| hsa-miR-145 | EAM212 |
1.44×10−3 | −2.75653 |
| hsa-miR-361-5p | JLA105 |
4.89×10−2 | −2.28546 |
| hsa-miR-451 | JLA5 |
3.69×10−2 | −2.15525 |
| hsa-miR-335 | EAM385 |
2.02×10−2 | −2.08815 |
| hsa-miR-101 | EAM311 |
1.90×10−2 | −1.69783 |
| hsa-miR-195 | EAM299 |
3.50×10−5 | −1.65589 |
| hsa-let-7c | EAM145 |
3.84×10−2 | −1.14039 |
| hsa-miR-146a | EAM139 |
1.54×10−2 | 1.38159 |
| hsa-miR-21 | EAM244 |
1.20×10−4 | 1.7009 |
| hsa-miR-7 | EAM109 |
2.81×10−2 | 2.55763 |
| hsa-miR-524-5p | JLA170 |
3.84×10−2 | 2.6693 |
| hsa-miR-10b | EAM190 |
7.07×10−4 | 3.39984 |
| hsa-miR-222 | EAM258 |
2.58×10−2 | 4.12295 |
| hsa-miR-520g | JLA176 |
3.69×10−2 | 4.81125 |
| hsa-mir-520c | JLA17 |
1.90×10−2 | 5.26842 |
| hsa-mir-519d | JLA29 |
2.02×10−2 | 5.50376 |
| hsa-miR-517c | JLA180 |
2.98×10−2 | 5.63958 |
| hsa-miR-516b | JLA27 |
2.02×10−2 | 5.72554 |
| hsa-miR-155 | EAM317 |
2.77×10−3 | 6.19272 |
| hsa-miR-517a | JLA26 |
2.76×10−2 | 6.72391 |
| hsa-miR-224 | EAM323 |
2.96×10−5 | 6.87628 |
By integrating the verified targets of miRNAs
downloaded from miRecords and miRTarBase, 14 target genes of
miRNA-224 and 8 target genes of miRNA-214 were obtained (Table II), respectively.
 | Table IITarget genes of the two significantly
differential expressed miRNAs. |
Table II
Target genes of the two significantly
differential expressed miRNAs.
| A, hsa-miR-224 |
|---|
|
|---|
| Target gene
name | Source | Test method |
|---|
| AP2M1 | miRTarBase | Luciferase reporter
assay |
| miRecord | Luciferase activity
assay |
| API-5 | miRTarBase | Western
blotting//Luciferase reporter assay//qPCR |
| miRecord | activity
assay//RT-PCR |
| CD40 | miRTarBase |
Microarray//qPCR |
| CDC42 | miRTarBase | Luciferase reporter
assay//qPCR//Western blotting |
| CXCR4 | miRTarBase | Luciferase reporter
assay//qPCR//Western blotting |
| EDNRA | miRTarBase | Northern
blot//qPCR//Western blotting |
| EYA4 | miRTarBase | Northern
blot//qPCR//Western blotting |
| FOSB | miRTarBase | Luciferase reporter
assay |
| KLK10 | miRTarBase | qPCR//Luciferase
reporter assay |
| miRecord | Luciferase | activity
assay//Western blotting |
| NCOA6 | miRTarBase | Luciferase reporter
assay |
| NIT1 | miRTarBase | Luciferase reporter
assay |
| PDGFRB | miRTarBase |
Microarray//Northern blotting |
| RAB9B | miRTarBase |
Microarray//Northern blotting |
| KLK1 | miRecord | ELISA |
|
| B, hsa-miR-214 |
|
| Target gene
name | Source | Test method |
|
| DAPK1 | miRTarBase |
Microarray//qPCR |
| EZH2 | miRTarBase | Luciferase reporter
assay//Northern blot |
| MAP2K3 | miRTarBase | GFP reporter
assay//Microarray//qPCR//Western blot |
| MAPK8 | miRTarBase | GFP reporter
assay//Microarray//qPCR//Western blot |
| PLXNB1 | miRTarBase |
Immunohistochemistry//qPCR//Western
blot |
| POU4F2 | miRTarBase | GFP reporter
assay//Northern blot//qPCR//Western blot |
| PTEN | miRTarBase | Western
blot//qPCR//Luciferase reporter assay |
| miRecord | RT-PCR |
| SRGAP1 | miRTarBase | Luciferase reporter
assay//qPCR//western blot |
Interaction network construction of
target genes
The target genes of miRNA-224 and miRNA-214 were
mapped using the software STRING and to predict their interactions.
By integrating these correlations, interaction networks between the
target genes and their interactive genes were constructed (Fig. 3). The core of the interaction
network of miRNA-224 targets (Fig.
3A) included CD40 [combined with CD40 ligand (LG), score
0.999], cell division cycle 42 (CDC42; combined with
Wiskott-Aldrich syndrome-like, score 0.999), chemokine (C-X-C
motif) receptor (CXCR) 4 (combined with protein kinase C, ζ (PRKCZ;
score 0.913), and platelet-derived growth factor receptor, β
polypeptide (PDGFRB; combined with phosphoinositide-3-kinase,
regulatory subunit 2, score 0.999) (27–29).
However, the network of miRNA-214 targets and their interactive
genes (Fig. 3B) revealed that
mitogen-activated protein kinase (MAPK) 8 was in the core and
program organized unit (POU) domain, class 4, transcription factor
2 (POU4F2) only interacted with cyclin D1 (CCND1).
Functional enrichment analysis of the
interaction network
In total, 15 and 13 function-clusters from the two
interaction networks were gained via the functional enrichment
analysis with software Gestalt (Fig.
4). For the miRNA-224 targets, the most significantly enriched
function-cluster of the total 15 categories was the cell signaling
cascade and others included phosphorus and phosphate metabolic
processes and the regulation of apoptosis.
As for the 13 categories of the miRNA-214 targets,
in which were in accordance with the 13 functional-clusters of the
15 categories in miRNA-224 targets, the interactive gene number of
each functional-cluster in miRNA-214 targets was larger than that
in miRNA-224 targets. In the common 13 functional-clusters in
miRNA-214 and in miRNA-224, the interactive genes of each target
were all correlated with the intracellular signaling cascade
(Fig. 4, circled in red).
Discussion
HCC, one of the most common types of tumor, results
in 662,000 mortalities per year worldwide and the prognosis of HCC
is poor (30). Therefore, there is
an urgent requirement to investigate the mechanism of HCC and to
develop an effective preventative strategy. Recently, in a hotspot
study of HCC, it has been demonstrated that miRNAs may represent
novel potential therapeutic targets and biomarkers for patients
with HCC (31,32).
In the present study, the gene expression profile
GSE31383 from GEO was used to analyze the differentially-expressed
miRNAs between HCC and healthy samples (10). As a result, two featured miRNAs
were obtained, the upregulated miRNA-224 and downregulated
miRNA-214, which were significantly differentially-expressed, as
well as 22 target genes, 14 targets of miRNA-224 and eight targets
of miRNA-214. These results were concurrent with a number of
previous studies, which demonstrated the important role of miR-224
and its targets in cell proliferation, migration, invasion and
anti-apoptosis of HCC by directly binding to interactive genes
(33). Target CD40 was
demonstrated to be the only upregulated molecule in the presence of
hepatitis B virus. CD40LG was the major signal that induced B cells
to efficiently present antigen to T cells. The CD40-CD40LG
interaction was crucial in the activation of antigen-presenting
cells and in the initiation of humoral and cellular immune
responses (34). Studies
demonstrated that CD40LG-mediated immune gene therapy for HCC was
an effective treatment as it activated the humoral and cellular
immune systems (35,36). In addition, PRKCZ has been proposed
to induce hepatocyte growth factor to regulate the CXCR4-CXC ligand
12, which has been demonstrated to mediate the metastasis of a
number of malignant tumors (37).
In the network analysis of the present study, it was initially
identified that PRKCZ directly combined with the target gene CXCR4
in HCC, which requires further analysis and experimental
verification (38). However, it
was noteworthy that the target gene nitrilase (NIT) 1 did not
combine with any other gene. Further functional analysis of NIT1 is
required and may be useful for the clarification of HCC
development.
miR-214 is likely to act as a tumor suppressor and
to have significant roles in inhibiting tumorigenesis of HCC by
suppressing the growth factors, such as EZH2, CTNNB1 and CDH1, in
human hepatoma (39). In the
network of eight target genes of miR-214, the majority of targets
were not in the core region and a number of interactive genes also
appeared in the network of miR-224. The target gene POU4F2 has been
demonstrated to function as a transcriptional factor in breast
cancer (40); however, no study
has been reported with regard to the interaction of POU4F2 and
CCND1.
Finally, the functional enrichment analysis of
interactive genes of the two target groups were enriched to 15 and
13 function-clusters respectively. In each cluster, the most
significant function was the intracellular signaling cascade. The
initiation of intracellular signaling cascades may occur via
cell-substratum interactions, triggering subcellular events, as
reported previously (41). The
interactive genes correlated with this function included the tumor
necrosis factor (TNF) receptor-associated factor,
phosphatidylinositol 3-kinase (PI3K) and MAPK, which had been
demonstrated to function in the regulation of inflammation,
antiviral responses and apoptosis of HCC (42,43).
Clinical trials of target genes combined with inhibitors, such as
p16 and p27, have been conducted for decades and have revealed
positive outcomes for treating HCC (44). Researchers reported that activation
of PI3K/AKT and MAPK pathway through a PDGFRB-dependent feedback
loop compromised the antitumor activity of rapamycin in HCC, and
blockade of this feedback loop by sorafenib was an attractive
approach to improve the antitumor effect of rapamycin, particularly
in preventing or treating HCC recurrence following liver
transplantation (27). However,
the present study confirmed anticancer effects of a synthesized
novel PI3Kα inhibitor on HCC cells and the inhibitor exhibited
anticancer activities (45). These
results demonstrated the potential function of miRNAs and their
targets in the treatment of HCC.
In conclusion, miR-224 and miR-214 were identified
by comparing normal and HCC tissue samples and the targeted genes
regulated by miRNAs and their interaction network indicated that
they were correlated with the intracellular signaling cascade, a
significant pathway of signaling transduction. The target genes and
their associated interactive genes, are significant in cellular
responses, including proliferation regulation, gene expression,
differentiation, mitosis, cell survival and apoptosis, and are
likely to be presented as novel potential therapeutic targets and
biomarkers of HCC. Further analyses are required to unravel the
involvement HCC development.
Acknowledgements
This study was supported by the Natural Science
Foundation Project of CQ CSTC (grant no. cstc2011jjA10001).
References
|
1
|
Chen CJ, Yang HI, Su J, et al: Risk of
hepatocellular carcinoma across a biological gradient of serum
hepatitis B virus DNA level. JAMA. 295:65–73. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Y, Zhang JB, Qin Y, et al: PROX1
promotes hepatocellular carcinoma metastasis by way of
up-regulating hypoxia-inducible factor 1α expression and protein
stability. Hepatology. 58:692–705. 2013.PubMed/NCBI
|
|
3
|
Luo Y, Li W and Liao H: HMGA2 induces
epithelial-to-mesenchymal transition in human hepatocellular
carcinoma cells. Oncol Lett. 5:1353–1356. 2013.PubMed/NCBI
|
|
4
|
Zhao H, Wang J, Han Y, et al: ARID2: a new
tumor suppressor gene in hepatocellular carcinoma. Oncotarget.
2:886–891. 2011.PubMed/NCBI
|
|
5
|
Lu CY, Lin KY, Tien MT, Wu CT, Uen YH and
Tseng TL: Frequent DNA methylation of MiR-129–2 and its potential
clinical implication in hepatocellular carcinoma. Genes Chromosomes
Cancer. 52:636–643. 2013.
|
|
6
|
Shrivastava S, Petrone J, Steele R, Lauer
GM, Bisceglie AM and Ray RB: Up-regulation of circulating miR-20a
is correlated with hepatitis C virus-mediated liver disease
progression. Hepatology. 58:863–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding J, Huang S, Wang Y, et al:
Genome-wide screening revealed that miR-195 targets the TNF-α/NF-κB
pathway by downregulating IκB kinase alpha and TAB3 in
hepatocellular carcinoma. Hepatology. 58:654–666. 2013.
|
|
8
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barbato C, Arisi I, Frizzo ME, Brandi R,
Da Sacco L and Masotti A: Computational challenges in miRNA target
predictions: to be or not to be a true target? J Biomed Biotechnol.
2009:8030692009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang PR, Xu M, Toffanin S, Li Y, Llovet JM
and Russell DW: Induction of hepatocellular carcinoma by in vivo
gene targeting. Proc Natl Acad Sci USA. 109:11264–11269. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barrett T, Troup DB, Wilhite SE, et al:
NCBI GEO: mining tens of millions of expression profiles - database
and tools update. Nucleic Acids Res. 35(Suppl 1): D760–D765. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gautier L, Irizarry R, Cope L and Bolstad
B: Description of Affy. http://www.bioconductor.org/packages/release/bioc/vignettes/affy/inst/doc/affy.pdf.
Accessed April 15, 2014
|
|
13
|
Fujita A, Sato JR, de Rodrigues LO,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Troyanskaya O, Cantor M, Sherlock G, et
al: Missing value estimation methods for DNA microarrays.
Bioinformatics. 17:520–525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R
and Dudoit S: Springer; New York, NY: pp. 397–420. 2005, View Article : Google Scholar
|
|
16
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: an integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009.PubMed/NCBI
|
|
18
|
Hsu SD, Lin FM, Wu WY, et al: miRTarBase:
a database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Franceschini A, Kuhn M, et
al: The STRING database in 2011: functional interaction networks of
proteins, globally integrated and scored. Nucleic Acids Res.
39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zwanzig R: Ensemble method in the theory
of irreversibility. J Chem Phys. 33:13381960. View Article : Google Scholar
|
|
23
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: two different approaches for Gene Ontology
analysis. BMC Proc. 3(Suppl 4): S102009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Exton H: Handbook of Hypergeometric
Integrals: Theory, Applications, Tables, Computer Programs. Ellis
Horwood; Chichester, England: 1978
|
|
26
|
Weng L, Macciardi F, Subramanian A, et al:
SNP-based pathway enrichment analysis for genome-wide association
studies. BMC Bioinformatics. 12:992011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li QL, Gu FM, Wang Z, et al: Activation of
PI3K/AKT and MAPK pathway through a PDGFRβ-dependent feedback loop
is involved in rapamycin resistance in hepatocellular carcinoma.
PLoS One. 7:e333792012.
|
|
28
|
Stengel K and Zheng Y: Cdc42 in oncogenic
transformation, invasion, and tumorigenesis. Cell Signal.
23:1415–1423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Manu KA, Shanmugam MK, Ong TH, et al:
Emodin suppresses migration and invasion through the modulation of
CXCR4 expression in an orthotopic model of human hepatocellular
carcinoma. PLoS One. 8:e570152013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shi YH, Ding ZB, Zhou J, et al: Prognostic
significance of Beclin 1-dependent apoptotic activity in
hepatocellular carcinoma. Autophagy. 5:380–382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Llovet JM and Bruix J: Molecular targeted
therapies in hepatocellular carcinoma. Hepatology. 48:1312–1327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu AX, Sahani DV, Duda DG, et al:
Efficacy, safety, and potential biomarkers of sunitinib monotherapy
in advanced hepatocellular carcinoma: a phase II study. J Clin
Oncol. 27:3027–3035. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Takahashi S, Tasaka A, Yoshima T,
Ochi H and Chayama K: Involvement of microRNA-224 in cell
proliferation, migration, invasion, and anti-apoptosis in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:565–575.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang YF, Ma J, He Y, Zhang YH, Xu Y and
Gong GZ: Cationic liposome-mediated transfection of CD40 ligand
gene inhibits hepatic tumor growth of hepatocellular carcinoma in
mice. J Zhejiang Univ Sci B. 10:7–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang Y, Chen Y, Ni B, Yang D, Guo S and Wu
Y: Up-regulation of the expression of costimulatory molecule CD40
in hepatocytes by hepatitis B virus X antigen. Biochem Biophys Res
Commun. 384:12–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan Y, Ma X, Li X and Yi J: A novel
immunotherapy to hepatocellular carcinoma: CD40-activated B
lymphocytes transfected with AFPmRNA. Med Hypotheses. 73:835–837.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang S, Ouyang N, Lin L, et al:
HGF-induced PKCζ activation increases functional CXCR4 expression
in human breast cancer cells. PLoS One. 7:e291242012.
|
|
38
|
Liu Y, Wang B, Wang J, et al:
Down-regulation of PKCzeta expression inhibits chemotaxis signal
transduction in human lung cancer cells. Lung Cancer. 63:210–218.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xia H, Ooi LL and Hui KM: MiR-214 targets
β-catenin pathway to suppress invasion, stem-like traits and
recurrence of human hepatocellular carcinoma. PLoS One.
7:e442062012.
|
|
40
|
Fujita R, Ounzain S, Wang AC, Heads RJ and
Budhram-Mahadeo VS: Hsp-27 induction requires POU4F2/Brn-3b TF in
doxorubicin-treated breast cancer cells, whereas phosphorylation
alters its cellular localisation following drug treatment. Cell
Stress Chaperones. 16:427–439. 2011. View Article : Google Scholar
|
|
41
|
Golemis EA and Chernoff J: Analysis and
manipulation of intracellular signaling cascades. Methods.
32:347–348. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang L, Jiang G, Yao F, et al: Growth
inhibition and apoptosis induced by osthole, a natural coumarin, in
hepatocellular carcinoma. PLoS One. 7:e378652012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang DM, Liu JS, Deng LJ, et al:
Arenobufagin, a natural bufadienolide from toad venom, induces
apoptosis and autophagy in human hepatocellular carcinoma cells
through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis.
34:1331–1342. 2013. View Article : Google Scholar
|
|
44
|
Matsuda Y, Wakai T, Kubota M, et al:
Clinical significance of cell cycle inhibitors in hepatocellular
carcinoma. Med Mol Morphol. 46:185–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yun SM, Lee JH, Jung KH, et al: Induction
of apoptosis and suppression of angiogenesis of hepatocellular
carcinoma by HS-159, a novel phosphatidylinositol 3-kinase
inhibitor. Int J Oncol. 43:201–109. 2013.PubMed/NCBI
|