Introduction
Hepatitis C is currently the dominant cause of
chronic liver disease, cirrhosis and hepatocellular carcinoma
(HCC). It is estimated that ~170 million individuals worldwide are
infected with HCV, with a global prevalence of ~3% (1,2). In
total, there are >3.5 million new cases of hepatitis C virus
(HCV) infection per year (3). In
China, there are ~38 million individuals who are infected with HCV,
with a prevalence of ~3.2%. Endemic strains of HCV have been
identified that persist in specific locations (4). There are 11 HCV genotypes with >70
subtypes, among which, four HCV genotypes (genotypes 1, 2, 3 and 6)
are prevalent in the Chinese population (5). The distribution of HCV genotypes is
significantly regional. HCV genotype 1 is globally spread, and
found particularly within Europe and North America (6). Genotype 2 is located in West Africa,
while genotype 6 is found in South East Asia (7). Recently, genotype 3A has been found
to dominantly infect intravenous drug-user populations in Europe
(4). However, the geographical
distribution of HCV genotypes in the Chinese population has rarely
been investigated.
An acute HCV infection can induce host innate and
adaptive immune responses (8,9). In
total, 15–25% of HCV-infected patients successfully eliminate the
virus, whereas the majority of patients develop chronic liver
disease, cirrhosis and HCC (10).
Various host factors can affect treatment-induced
control and the spontaneous clearance of HCV infection. HCV
clearance is associated with polymorphisms in the region of the
interleukin-28B (IL-28B) gene, which indicates that interferon
(IFN)-λ3, the gene product, is vital in the immune response to HCV
(11). Spontaneous HCV clearance
occurs in 15–25% of cases (12),
which may be affected by a number of factors, including gender,
ethnicity, jaundice and co-infections. Numerous studies have
demonstrated that host genetic polymorphisms may lead to
differences in host immune function and therefore affect the
clinical outcome of HCV infection. Host genetic variation in the
IL-28B gene was shown to markedly predict viral clearance and
sustained virological response (SVR) rates in patients with HCV
infection (13–15). The region on chromosome 19 that is
associated with HCV treatment response contains multiple single
nucleotide polymorphisms (SNPs) in linkage disequilibrium around
the IL-28B gene (16–18) In the past years, genetic studies
have identified several SNPs in and near IL-28B that are associated
with viral clearance. Sequence analysis of the IL-28B region
indicated two SNPs (rs8103142 and rs28416813) as candidate causal
variants (16), but little is
known about other SNPs associated with IL-28B. Two good responder
alleles, rs12979860 and rs8099917, have been found to be associated
with natural viral clearance and low levels of mRNA IL-28B in
peripheral mononuclear cells (17,18).
However, IL-28B gene expression in liver samples appears not to be
regulated by the IL-28B genotypes, and studies on the association
between IFN-λ mRNA and IL-28B genotypes lead to conflicting
results. Therefore, there is an urgent requirement to clarify
detailed epidemiological information with regard to the SNPs of
IL-28B correlated with the future prospects and treatment of HCV
patients. Furthermore, the current data on other SNPs in IL-28B are
poorly understood and require further study.
The aim of the present study was to analyze the
epidemiology of Chinese hepatitis C patients through i) the host
genotypes and SNPs associated with IL-28B, ii) the HCV genotypes in
different regions and iii) the possible association between
IL-28B-related genetic variations and HCV genotypes in hepatitis
C.
Patients and methods
Patients population
A total of 1,014 patients infected with HCV (554
males and 460 females; mean age, 45 years) were randomly recruited
from 28 hospitals in varying regions in China (Table I). The registration ID of the
present study is NCT01293279 (www.clinicaltrials.gov). The study was approved by the
ethical committees of all the hospitals that patients were
recruited from in the present study. Written informed consent was
obtained from each patient. All the patients had been confirmed as
positive for anti-HCV antibody and serum-HCV RNA. All the hospitals
were divided territorially into North, Northeast, Southwest, South,
Central, Northwest and East China (Fig. 1) (19). The study included 92 patients
infected with HCV in North China, 91 in the Southwest, 104 in the
South, 224 in Central China, 162 in the Northwest, 252 in the East
and 89 in the Northeast. All the patients with HCV infection did
not receive any treatment, and the age, gender and regions were
taken into consideration.
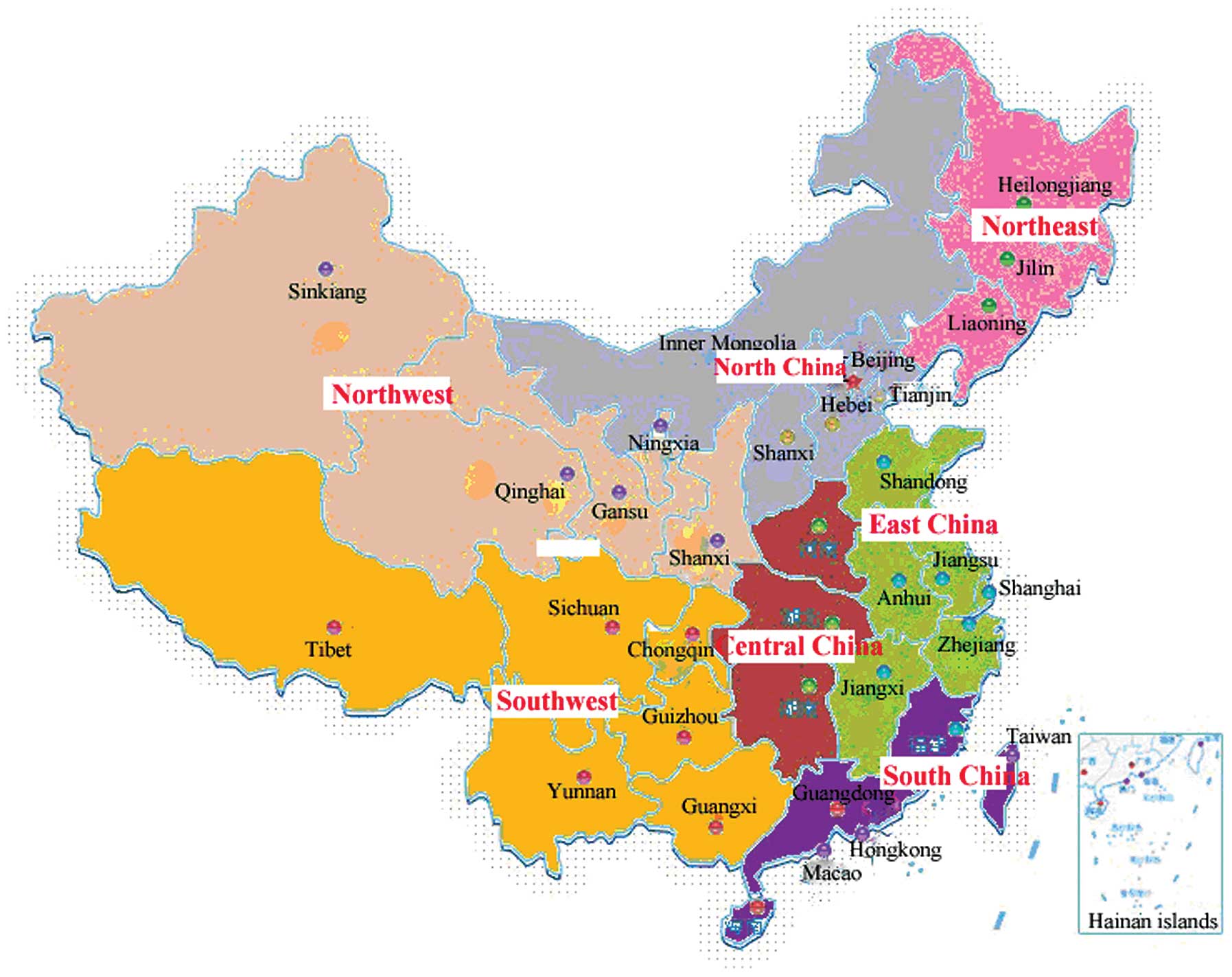 | Figure 1Regional distribution of HCV patients
recruited. In total, 28 hospitals were divided territorially into
North, Northeast, Southwest, South, Central, Northwest and East
China. The study included 92 HCV-infected patients in North China,
91 in the Southwest, 104 in the South, 224 in Central China, 162 in
the Northwest, 252 in the East and 89 in the Northeast (19). HCV, hepatitis C virus. |
 | Table IRegional information of patients
infected with HCV in China. |
Table I
Regional information of patients
infected with HCV in China.
| Regions of China | Hospital | No. of patients |
|---|
| North | Peking University
People’s Hospital | 59 |
| Beijing Friendship
Hospital | 33 |
| Northeast | Shengjing Hospital of
China Medical Hospital | 34 |
| The First Hospital of
Jilin University | 34 |
| The Second Affiliated
Hospital of Haerbin Medical University | 21 |
| Southwest | West China Hospital,
Suchuan | 22 |
| The First Affiliated
Hospital of Kunming Medical College | 40 |
| Southwest
Hospital | 29 |
| South | The First Affiliated
Hospital of Guangxi Medical University | 40 |
| Nanfang Hopital | 14 |
| The Third Affiliated
Hospital of Sun Yat-sen University | 50 |
| Central | Henan Provincial
People’s Hospital | 95 |
| The First Affiliated
Hospital of Zhengzhou University | 28 |
| People’s Hospital of
Hubei Wuhan University | 38 |
| Affiliated Tongji
Hospital of Tongji Medical College of Huazhong University of
Science and Technology | 32 |
| The Second Xiangya
Hospital of Central South University | 31 |
| Northwest | Tangdu Hospital
40 | |
| The First Affiliated
Hospital of Shanxi Medical University | 37 |
| The First Affiliated
Hospital of Lanzhou University | 58 |
| Ningxia People’s
Hospital | 27 |
| East | The First Affiliated
Hospital of Nanchang University | 37 |
| The First Affiliated
Hospital of Anhui Medical University | 19 |
| The Second Hospital
of Shangdong University | 41 |
| The First Affiliated
Hospital of Medical College Zhejiang University | 33 |
| The First
Affiliated Hospital of Fujian Medical University | 11 |
| Shanghai Ruijin
Hospital | 52 |
| The First
Affiliated People’s Hospital of Shanghai Jiaotong University | 2 |
| Jiangsu Province
Hospital | 57 |
Genomic DNA and RNA extraction
Genomic DNA and total RNA from liver biopsies were
extracted using the Qiagen RNeasy Mini kit (Qiagen, Inc., Valencia,
CA, USA) (20).
HCV genotyping
HCV genotyping was conducted using the INNO-LiPA HCV
II assay (Innogenetics, Zwijnaarde, Belgium), which is based on
hybridization of probes with the 5′ non-coding region of HCV.
According to the HCV gene sequences published in GenBank (AF333324,
D16435, KF035127 and KC844040), the HCV 5′ non-coding region
(nt299-1) was selected, and one pair of specific primers were
designed for nested polymerase chain reaction (PCR). The sequences
of the PCR primers were as follows: Sense: −299
5′-CCCTGTGAGGAACTWCTGTCTTCACGC-3′ (−299~ −273); and antisense:
5′-GGTGCACGGTCTACGAGACCT-3′ (−1~−21) (W is A or T). The probes used
for the detection of the different serum types are also listed as
follows: Genotype 1: −170 AATTGCCAGGACGACC; genotype 2: −83
TAGCGTTGGGTTGCGA; genotype 3: −170 AATCGCTGGGGTGACC; and genotype
6: −81 AGTAGCGTTGGGTTGC. The HCV 5′ non-coding region of the HCV
genome was amplified using nested PCR. Briefly, 5 μl cDNA was
amplified for >40 cycles in a total volume of 50 μl, each
consisting of 1 min at 95°C, 1 min at 55°C and 1 min at 72°C. From
the second round of amplification, 1 μl product was amplified again
with the same primers for another 40 cycles. The nested PCR
products were subjected to electrophoresis in a 2% agarose gel. The
probes specific for the different types of HCV were added with a
poly(dT) tail at their 3′ end. Primer (20 pmol) was incubated for 1
h at 37°C in 25 μl buffer (3.2 mM-dTTP, 25 mM-Tris-HC1 pH 7.5, 0.1
M-sodium cacodylate, 1 mM-CoCl2, 0.1 M-dithiothreitol
and 60 U terminal deoxynucleotidyl transferase). The reaction was
stopped by adding 2.5 μl EDTA (0.5 M, pH 8.0) and diluted with 20X
SSC. This solution was applied on a nitrocellulose membrane, and
was fixed to the membrane by baking at 80°C for 2 h, and then was
hybridized with equal volumes of the nested PCR products.
Assay for HCV RNA
Serum was obtained from the HCV-infected patients,
and the total RNA was isolated using an RNeasy Mini kit and
RNase-Free DNase Set (Qiagen, Hilden, Germany). The HCV RNA content
was detected by Abbott RealTime HCV (Abbott Laboratories, Des
Plaines, IL, USA).
IL-28B genotyping
Genotyping for the IL-28B gene was performed by the
iPLEX system [MassARRAY® Real-Time Transactional Memory
(RTTM) software for SNP genotyping; iPLEX Gold; Sequenom, San
Diego, CA, USA]. The DNA was blind coded and tested using a
384-SpectroCHIP® microarray (Sequenom). A
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometer was used for data acquisitions from the SpectroCHIP
microarray. The results were analyzed using Sequenom MassARRAY RTTM
software. The region on around the IL-28B gene on chromosome 19
contains multiple SNPs in linkage disequilibrium. The present study
focused on the genotyping of thirteen SNPs associated with IL-28B:
rs8013142, rs28416813, rs10853728, rs7248668, rs8105790,
rs11881222, rs12979860, rs12980275, rs4803219, rs8099917,
rs8109886, rs4803223 and rs10853727.
Statistical analysis
The Hardy-Weinberg disequilibrium test was performed
for each SNP genotype associated with IL-28B. The gene frequencies
were compared between groups using the χ2 test with
Yates correction or Fisher’s exact test. Group means were presented
as the mean ± standard deviation, and were compared by analysis of
variance and Student’s t-test. The serum HCV RNA levels were
expressed as the original values. The HCV RNA levels were divided
into high, medium and low level groups. The statistical analyses
were performed with the SPSS 12.0 statistical package (SPSS, Inc.,
Chicago, IL, USA). All statistical analyses were based on two-sided
hypothesis tests, with P<0.05 used to indicate a statistically
significant difference.
Results
Baseline characteristics
A total of 1,014 HCV-infected patients were randomly
recruited from 28 hospitals in the 7 regions of China for the
present study (Fig. 1 and Table I). The overall gender ratio of male
to female was 1.204 (554/460). The HCV-infected population was
divided according to their age into four groups: The old-aged
adults (>60 years old), the middle-aged adults (45–59 years
old), the young adults (18–44 years old) and the young (<18
years old). HCV genotypes 1, 2, 3 and 6 were included in the study.
Furthermore, HCV RNA expression was detected and divided into high
(RNA≥1×107), medium
(1×105≤RNA<1×107) and low
(RNA<1×105) expression.
Geographical distribution of HCV
genotypes
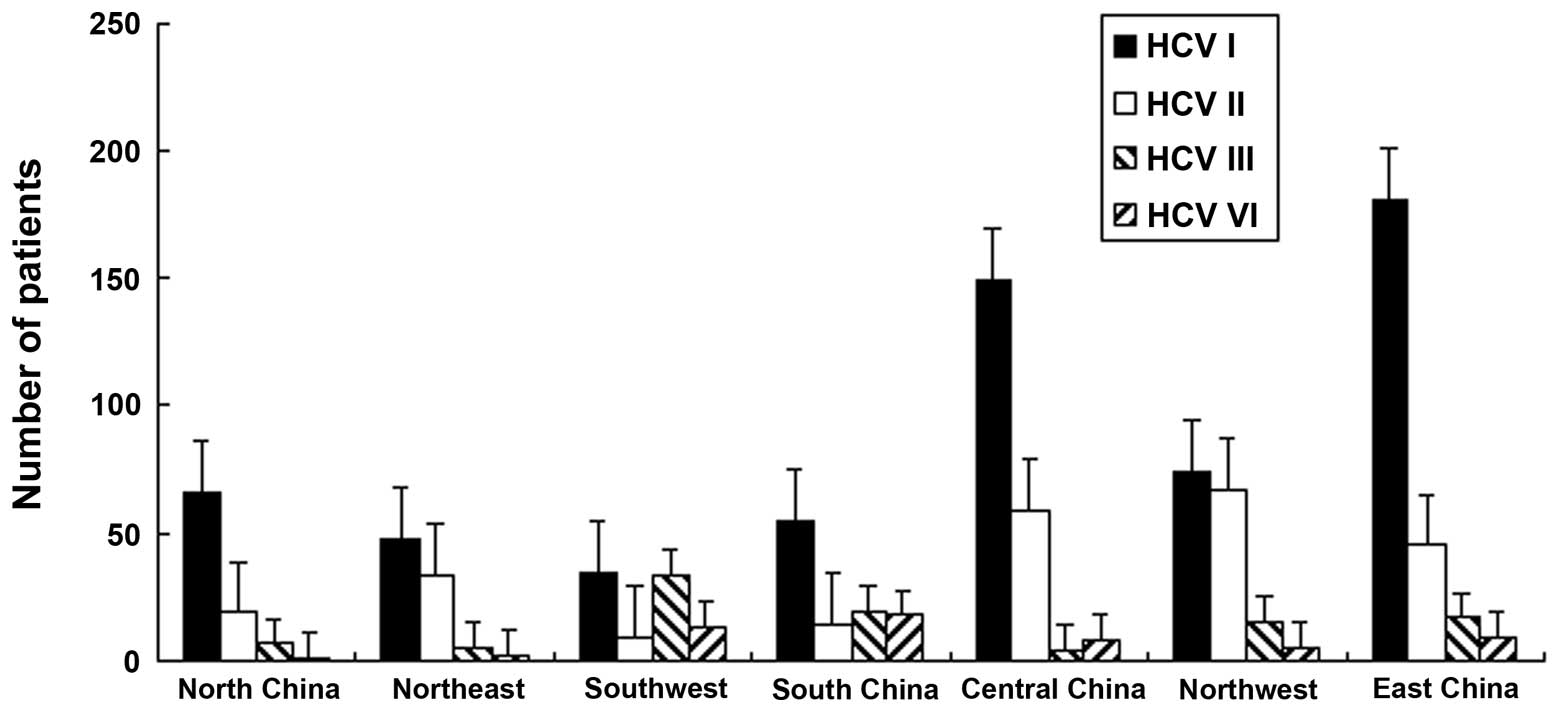
The HCV genotypes in Chinese patients were
investigated. The data revealed that HCV genotype 1 was distributed
more extensively compared with other genotypes in all regions, with
the exception of South and Northwest China (P<0.01). In South
China, the distribution of HCV genotypes 1 and 3 was higher
compared with other genotypes, with percentages of 38 and 37%,
respectively. The percentages of HCV genotypes 1 and 2 in the
Northwest region were 45.6 and 41%, respectively. In all regions,
with the exception of Southwest (15%) and South (17%) China, the
percentage of genotype 6 was recorded to be the lowest, (Fig. 2).
Distribution of HCV-infected patients in
different genders and ages in China
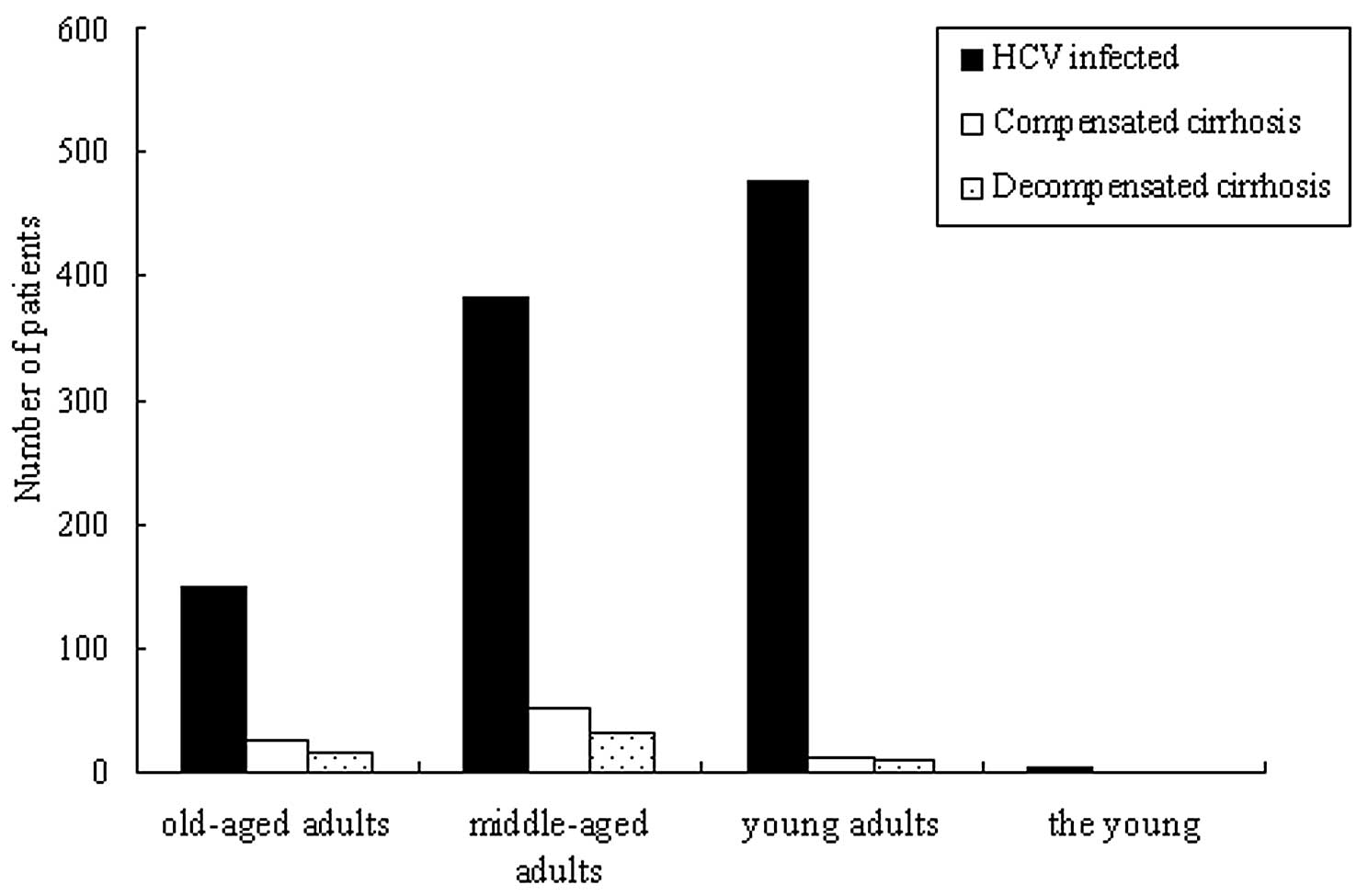
In all the recruited HCV-infected patients, the
old-aged adults, middle-aged adults, young adults and the young
accounted for 14.8 (150/1,014), 37.7 (382/1,014), 47.3 (480/1,014)
and 0.2% (2/1,014) of cases, respectively. The patients with
compensated and decompensated cirrhosis accounted for 8.8
(89/1,014) and 5.3% (54/1,014) of the cases, respectively (Fig. 3). The old-aged, middle-aged and
young adults accounted for 27, 60 and 13% of cases, respectively,
in the compensated cirrhosis group, and for 28, 57 and 15% of
cases, respectively, in the decompensated cirrhosis group (Fig. 3). The distribution of the young in
the purely HCV-infected patients was significantly higher compared
with that in the compensated and decompensated cirrhosis groups.
However, in the compensated and decompensated cirrhosis groups, the
distribution rate of the middle-aged adults was higher than the
other groups (P<0.01).
In the middle-aged adults group, the number of male
patients was higher compared with the number of females in North
and South China, which was the opposite of the results found in the
other regions. In particular, in Eastern China, the number of
female patients was significantly higher compared with the number
of males (P<0.01). However, in the young adult group, there were
far more males compared with females in the majority of the
regions, with the exception of North China (P<0.01) (Table II).
 | Table IIGender differences in HCV patients of
different ages and regions in China. |
Table II
Gender differences in HCV patients of
different ages and regions in China.
| Age, % |
|---|
|
|
|---|
| Regions and
gender | Old-aged
adults | Middle-age
adults | Young adults | The young |
|---|
| North |
| Male | 12.09 | 24.18 | 17.58 | 1.10 |
| Female | 18.68 | 17.58 | 8.79 | 0.00 |
| Northeast |
| Male | 8.99 | 19.10 | 19.10 | 0.00 |
| Female | 10.11 | 24.72a | 17.98 | 0.00 |
| Southwest |
| Male | 4.49 | 7.87 | 46.07 | 0.00 |
| Female | 5.62 | 11.24a | 24.72 | 0.00 |
| South |
| Male | 3.85 | 13.46 | 47.12 | 0.00 |
| Female | 6.73 | 11.54 | 17.31 | 0.00 |
| Central |
| Male | 4.02 | 12.50 | 30.36 | 0.00 |
| Female | 3.57 | 29.91a | 19.64 | 0.00 |
| Northwest |
| Male | 5.56 | 17.90 | 38.27 | 0.00 |
| Female | 4.94 | 20.37 | 12.96 | 0.00 |
| East |
| Male | 10.71 | 17.46 | 25.79 | 0.40 |
| Female | 9.13 | 24.21a | 12.30 | 0.00 |
Gender differences of HCV genotypes in
the varying regions
In all regions, no gender difference was found in
HCV genotype 1 (P<0.05). However, in HCV genotype 2, there were
significantly less males compared with females in North and
Southwest China (P<0.01). In HCV genotype 3, the males were
extensively distributed in all the regions, with the exception of
the Northeast. In HCV genotype 6, the infected rate in males was
significantly higher than that in females in South China
(P<0.01) (Table III).
 | Table IIIGender difference of geographical HCV
genotypes in China. |
Table III
Gender difference of geographical HCV
genotypes in China.
| HCV genotypes,
% |
|---|
|
|
|---|
| Regions and
gender | 1 | 2 | 3 | 6 |
|---|
| North |
| Male | 55.00 | 31.58 | 100.00a | 0.00 |
| Female | 45.00 | 68.42a | 0.00 | 0.00 |
| Northeast |
| Male | 45.65 | 54.84 | 33.33 | 50.00 |
| Female | 54.35 | 45.16 | 66.67 | 50.00 |
| Southwest |
| Male | 52.94 | 62.50 | 66.67 | 53.85 |
| Female | 47.06 | 37.50a | 33.33 | 46.15 |
| South |
| Male | 62.00a | 50.00 | 81.82 | 76.00 |
| Female | 38.00 | 50.00 | 18.18 | 24.00a |
| Central |
| Male | 44.93 | 46.43 | 75.00 | 55.56a |
| Female | 55.07a | 53.57 | 25.00 | 44.44 |
| Northwest |
| Male | 56.16a | 56.06 | 100.00 | 100.00 |
| Female | 43.84 | 43.94 | 0.00 | 0.00 |
| East |
| Male | 55.43a | 47.75 | 75.00 | 44.44 |
| Female | 44.57 | 52.25 | 25.00 | 55.56 |
HCV RNA expression among HCV
genotypes
The HCV RNA in all the genotypes was highly
expressed, with the exception of genotype 1b+2. There was no
difference in HCV RNA expression among HCV genotypes 1, 2 and 3.
However, the number of patients with high RNA expression in
genotype 6 was significantly higher compared with that in the other
genotypes (P<0.01). In genotype 1b+2 patients, the number of
patients with high RNA expression was lower compared with the
patients with medium and low levels of RNA expression, but no
statistical significance was found due to the small number of
specimens (Fig. 4).
Distribution of SNPs associated with
IL-28B
A total of 13 SNPs associated with IL-28B
(rs8013142, rs28416813, rs10853728, rs7248668, rs8105790,
rs11881222, rs12979860, rs12980275, rs4803219, rs8099917,
rs8109886, rs4803223 and rs10853727) were investigated in 1,014
HCV-infected patients in the present study. The distribution of
host genotypes in each SNP were detected (Table IV). The data show that the
percentage of genotype C/T in rs8013142 (98.13%), G/C in rs28416813
(77.71%), C/C in rs10853728, rs12979860, rs4803129 and rs8109886
(66.07, 84.71, 84.32 and 81.56%, respectively), G/G in rs7248668
(85.21%), T/T in rs8105790, rs8099917 and rs10853727 (71.40, 85.40
and 98.52%, respectively), and A/A in rs11881222, rs12980275 and
rs4803223 (83.73, 81.36 and 84.32%, respectively) were
significantly higher compared with the other two allelic genes.
Notably, there were significant differences in the genotype
percentages of all SNPs between HCV genotypes 1 and 2 in the
majority of SNPs (with the exception of rs8013142, rs8099917,
rs8109886 and rs10853727). Genotypes rs28416813 (C/C), rs4803219
(T/T) and rs10853727 (C/C) were not found in any of the recruited
HCV patients (Table IV).
 | Table IVDistribution of SNP genotypes
associated with IL-28B in HCV genotypes 1 and 2. |
Table IV
Distribution of SNP genotypes
associated with IL-28B in HCV genotypes 1 and 2.
| SNPs | HCV infected | HCV genotype 1 | HCV genotype 2 |
|---|
| rs8013142, %
(n) |
| C/T | 98.13 (995)a | 99.66 (587) | 99.23 (258) |
| T/T | 0.20 (2) | 0.00 (0) | 0.38 (1) |
| C/C | 1.67 (17) | 0.34 (2) | 0.38 (1) |
| Gene frequency,
% |
| T:C | 49/51 | 50/50 | 50/50 |
| rs28416813, %
(n) |
| G/C | 77.71 (788)a | 79.80 (470) | 88.46 (230)b |
| G/G | 22.29 (226) | 20.20 (119) | 11.54 (30) |
| Gene frequency,
% |
| G:C | 62/38 | 60/40 | 58/42 |
| rs10853728, %
(n) |
| C/G | 30.47 (309) | 34.63 (204) | 27.69 (72) |
| G/G | 3.46 (35) | 3.06 (18) | 3.85 (10) |
| C/C | 66.07 (670)a | 62.31 (367) | 68.46 (178)b |
| Gene frequency,
% |
| G:C | 19/81 | 20/80 | 18/82 |
| rs7248668, %
(n) |
| G/A | 14.60 (148) | 17.66 (104) | 11.15 (29) |
| G/G | 85.21 (864)a | 82.17 (484) | 88.46 (230)b |
| A/A | 0.20 (2) | 0.17 (1) | 0.38 (1) |
| Gene frequency,
% |
| G:A | 93/7 | 91/7 | 94/6 |
| rs8105790, %
(n) |
| T/C | 28.4 (288) | 33.62 (198) | 22.31 (58) |
| T/T | 71.4 (724)a | 66.38 (391) | 76.54 (200)b |
| C/C | 0.2 (2) | 0.00 (0) | 0.77 (2) |
| Gene frequency,
% |
| T:C | 86/14 | 83/17 | 88/12 |
| rs11881222, %
(n) |
| G/A | 16.17 (16) | 19.86 (117) | 10.77 (28) |
| A/A | 83.73 (849)a | 80.14 (472) | 88.46 (230)b |
| G/G | 0.20 (2) | 0.00 (0) | 0.77 (2) |
| Gene frequency,
% |
| G:A | 8/92 | 10/90 | 6/94 |
| rs12979860, %
(n) |
| C/T | 15.09 (153) | 18.17 (107) | 11.15 (29) |
| C/C | 84.71 (859)a | 81.66 (481) | 88.46 (230)b |
| T/T | 0.20 (2) | 0.17 (1) | 0.38 (1) |
| Gene frequency,
% |
| T:C | 8/92 | 9/91 | 6/94 |
| rs12980275, %
(n) |
| G/A | 15.68 (159) | 19.52 (115) | 10.77 (28) |
| G/G | 2.96 (30) | 0.00 (0) | 1.15 (3) |
| A/A | 81.36 (825) | 80.48 (474) | 88.08 (229)b |
| Gene frequency,
% |
| G:A | 11/89 | 10/90 | 7/93 |
| rs4803219, %
(n) |
| C/T | 15.68 (159) | 19.86 (117) | 11.54 (30) |
| C/C | 84.32 (855)a | 80.14 (472) | 88.46 (230)b |
| Gene frequency,
% |
| T:C | 8/92 | 10/90 | 6/94 |
| rs8099917, %
(n) |
| G/T | 14.40 (146) | 17.83 (105) | 11.60 (29) |
| T/T | 85.40 (866)a | 82.00 (483) | 88.00 (220) |
| G/G | 0.20 (2) | 0.17 (1) | 0.40 (1) |
| Gene frequency,
% |
| G:T | 7/93 | 9/91 | 6/94 |
| rs8109886, %
(n) |
| C/A | 18.24 (185) | 21.73 (128) | 15.77 (41) |
| C/C | 81.56 (827)a | 78.10 (460) | 83.85 (218)b |
| A/A | 2.00 (2) | 0.17 (1) | 0.38 (1) |
| Gene frequency,
% |
| C:A | 91/9 | 89/11 | 92/8 |
| rs4803223, %
(n) |
| G/A | 15.38 (156) | 18.17 (107) | 11.92 (31) |
| A/A | 84.32 (855)a | 81.83 (482) | 88.08 (229)b |
| G/G | 0.30 (3) | 0.00 (0) | 0.00 (0) |
| Gene frequency,
% |
| G:A | 8/92 | 9/91 | 6/94 |
| rs10853727, %
(n) |
| T/C | 1.48 (15) | 1.36 (8) | 0.00 (0) |
| T/T | 98.52 (999)a | 98.64 (581) | 100.00 (260) |
| Gene frequency,
% |
| T:C | 99/1 | 99/1 | 100/0 |
Overall, no geographical differences in SNPs existed
in the Chinese HCV-infected population. However, SPSS analysis
revealed that several correlations exist between the SNPs genotypes
and HCV (P<0.01).
Discussion
As a major cause of chronic liver disease globally,
the prevalence of HCV infection exhibits significant geographical
variations. Distinct epidemiological characteristics and
differences in methodologies are reflected by these variations.
Therefore, in the present study, all HCV samples, originally
obtained from 28 different hospitals around China, were analyzed in
the same center.
The geographical differences in HCV genotypes have
long since been discovered, and underlie variations when conducting
epidemiological surveys, etiological diagnoses, clinical treatment
and vaccine development in a specific region (21). For example, the HCV genotype 1 is
predominant in Japan and South Asia. Genotype 2 is prevalent in
Taiwan and genotypes 1, 2 and 6 are prevalent in Thailand. The
prevalence of genotype 3 is higher in Europe compared with Africa
and Asia (22).
In the present study, HCV genotype 1b was shown to
be extensively distributed in North (71.8), Northeast (53.9), South
(52), Central (66.5) and East (71.8%) China. HCV genotypes 1b and 3
were the main genotypes in Southwest China. HCV genotypes 1b and 2a
were the prevalent genotypes in Northeast China. The geographical
distribution of HCV genotypes has been investigated by a variety of
studies in China, however the results are paradoxical. A number of
studies show that HCV genotype 1b is the most widely distributed
genotype (23), and is most
predominant in the North (Beijing, 56.8%), East (Shanghai, 69.1%)
and Southwest (Chongqing, 32.9%), while genotype 2a is most
predominant in the Northwest (Wuwei, 59.1%) (24–27).
The differences in the distribution of HCV genotypes may arise from
the varying detection methods and small sample size used among the
literature. HCV genotype 6 was relatively uncommon throughout all
regions, while in the Southwest it is the most predominant (Yunnan
Province, 47%) (28). In the
present study, the distribution of HCV genotypes varied and
demonstrated geographical properties in China. HCV genotype
distribution may be caused by a source of infection, presence of
ethnic groups and individual differences. However, the mechanism of
HCV genotype variation and the relevance with host genovariation
remain unknown and require further investigation.
HCV genotypes are considered to be a major
determinant of the response to treatment in HCV infection (29,30).
Furthermore, increasing studies clearly indicate that host genetics
are also critical factors affecting the response to treatment
(31). HCV clearance is associated
with polymorphisms in the IL-28B gene region, indicating a vital
role for the IFN-λ3 gene product in the immune response to HCV
(11). The present study initially
investigated 13 SNPs associated with IL-28B in a range of regions
and HCV genotypes in the Chinese population. It has previously been
clarified that host genetics play an essential role in clearing
acute hepatitis C infection and achieving an SVR (32). Variants in the minor alleles,
rs8099917 and rs12979860, are associated with SVR and natural viral
clearance (33). In the present
study of Chinese HCV-infected populations, the rate of T/T in
rs8099917 and C/C in rs12979860 was significantly higher compared
with the other two allelic genes (P<0.01) and was correlated
with HCV genotype variation. Other SNPs associated with IL-28B were
also investigated in the present study. Notably, the genotype
percentages of all the SNPs were significantly different between
HCV genotypes 1 and 2, with the exception of rs8013142, rs8099917,
rs8109886 and rs10853727. The SPSS analysis data revealed that
there was significant relevance between the host and HCV genotypes
(P<0.01). This indicated that a certain number of interactions
were occurring between the host and HCV genotypes in the Chinese
HCV-infected population. The molecular mechanism of these
associations require elucidating in further studies. The present
study found that these susceptible SNPs correlated with HCV
infection, which may be useful for use in epidemiological surveys
and etiological diagnoses of HCV infection.
In conclusion, the present study investigated the
geographical distribution of HCV genotypes and SNPs associated with
IL-28B in Chinese patients infected with HCV, and found a
geographical distribution in the HCV genotypes, and a correlation
between HCV genotypes and several IL-28B SNPs. The study indicated
that these variants may be associated with spontaneous and
treatment-induced HCV clearance.
Acknowledgements
This study was supported by grants from the National
Science and Technology Major Project for Infectious Disease Control
during the 11th Five-Year Plan Period (grant nos. 2008ZX10002-012
and 2008ZX10002-013) and the 12th Five-Year Plan Period (grant no.
2012ZX10002003), and by grants from the Natural Science Foundation
of Gansu Province (grant no. 0803RJZA057), Fundamental Research
Funds for the Central Universities (lzujbky-2009-150) and
Bristol-Myers Squibb. Operational support and statistical analyses
were provided by Research Pharmaceutical Services, Beijing,
China.
References
|
1
|
Martins T, Narciso-Schiavon JL and
Schiavon LL: Epidemiology of hepatitis C virus infection. Rev Assoc
Med Bras. 57:107–112. 2011.(In Portuguese).
|
|
2
|
Alter MJ: Epidemiology of hepatitis C
virus infection. World J Gastroenterol. 13:2436–2441. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lauer GM and Walker BD: Hepatitis C virus
infection. N Engl J Med. 345:41–52. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klenerman P and Gupta PK: Hepatitis C
virus: current concepts and future challenges. QJM. 105:29–32.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liew M, Erali M, Page S, Hillyard D and
Wittwer C: Hepatitis C genotyping by denaturing high-performance
liquid chromatography. J Clin Microbiol. 42:158–163. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Simmonds P: Genetic diversity and
evolution of hepatitis C virus-15 years on. J Gen Virol.
85:3173–3188. 2004.PubMed/NCBI
|
|
7
|
Pybus OG, Barnes E, Taggart R, et al:
Genetic history of hepatitis C virus in East Asia. J Virol.
83:1071–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thio CL, Thomas DL and Carrington M:
Chronic viral hepatitis and the human genome. Hepatology.
31:819–827. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thio CL: Host genetic factors and
antiviral immune responses to hepatitis C virus. Clin Liver Dis.
12:713–726. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seeff LB: Natural history of chronic
hepatitis C. Hepatology. 36:S35–S46. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Osaki R, Nishimura T, Shioya M, et al:
Interleukin-28B genotypes determine response to
pegylated-interferon plus ribavirin therapy in patients with
hepatitis C virus infection. Mol Med Rep. 5:525–528.
2012.PubMed/NCBI
|
|
12
|
Thomas DL, Astemborski J, Rai RM, et al:
The natural history of hepatitis C virus infection: host, viral,
and environmental factors. JAMA. 284:450–456. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rauch A, Kutalik Z and Descombes P; Swiss
HIV Cohort Study. Genetic variation in IL28B is associated with
chronic hepatitis C and treatment failure: a genome-wide
association study. Gastroenterology. 138:1338–1345. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grebely J, Petoumenos K and Hellard M;
ATAHC Study Group. Potential role for interleukin-28B genotype in
treatment decision-making in recent hepatitis C virus infection.
Hepatology. 52:1216–1224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomas DL, Thio CL, Martin MP, et al:
Genetic variation in IL28B and spontaneous clearance of hepatitis C
virus. Nature. 461:798–801. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ge D, Fellay J, Thompson AJ, et al:
Genetic variation in IL-28B predicts hepatitis C treatment-induced
viral clearance. Nature. 461:399–401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suppiah V, Moldovan M, Ahlenstiel G, et
al: IL28B is associated with response to chronic hepatitis C
interferon-alpha and ribavirin therapy. Nat Genet. 41:1100–1104.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanaka Y, Nishida N, Sugiyama M, et al:
Genome-wide association of IL-28B with response to pegylated
interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat
Genet. 41:1105–1109. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mao XR, Zhang LT, Chen H, et al: Possible
factors affecting thyroid dysfunction in hepatitis C virus-infected
untreated patients. Exp Ther Med. (In Press).
|
|
20
|
Bouwknegt M, Lodder-Verschoor F, van der
Poel WH, Rutjes SA and de Roda Husman AM: Hepatitis E virus RNA in
commercial porcine livers in The Netherlands. J Food Prot.
70:2889–2895. 2007.PubMed/NCBI
|
|
21
|
Viazov S, Zibert A, Ramakrishnan K, et al:
Typing of hepatitis C virus isolates by DNA enzyme immunoassay. J
Virol Methods. 48:81–91. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seeff LB and Hoofnagle JH: Appendix: The
National Institutes of Health Consensus Development Conference
Management of Hepatitis C 2002. Clin Liver Dis. 7:261–287.
2003.PubMed/NCBI
|
|
23
|
Chen YD, Liu MY, Yu WL, Li JQ, Peng M, Dai
Q, Liu X and Zhou ZQ: Hepatitis C virus infections and genotypes in
China. Hepatobiliary Pancreat Dis Int. 1:194–201. 2002.PubMed/NCBI
|
|
24
|
Rao H, Wei L, Lopez-Talavera JC, Shang J,
Chen H, Li J, Xie Q, Gao Z, Wang L, Wei J, et al: Distribution and
clinical correlates of viral and host genotypes in Chinese patients
with chronic hepatitis C virus infection. J Gastroenterol Hepatol.
29:545–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong ZX, Zhou HJ, Wang JH, Xiang XG,
Zhuang Y, Guo SM, Gui HL, Zhao GD, Tang WL, Wang H and Xie Q:
Distribution of hepatitis C virus genotypes in Chinese patients
with chronic hepatitis C: correlation with patients’
characteristics and clinical parameters. J Dig Dis. 13:564–570.
2012.
|
|
26
|
Yan Z, Fan K, Wang Y, Fan Y, Tan Z and
Deng G: Changing pattern of clinical epidemiology on hepatitis C
virus infection in Southwest China. Hepat Mon. 12:196–204. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li D, Long Y, Wang T, Xiao D, Zhang J, Guo
Z, Wang B and Yan Y: Epidemiology of hepatitis C virus infection in
highly endemic HBV areas in China. PLoS One. 8:e548152013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Z, Yao Y, Wu W, Feng R, Wu Z, Cun W
and Dong S: Hepatitis C virus genotype diversity among intravenous
drug users in Yunnan Province, Southwestern China. PLoS One.
8:e825982013. View Article : Google Scholar
|
|
29
|
Sibbing B and Nattermann J: Hepatitis C
virus infection and genetic susceptibility to therapy. J
Gastrointestin Liver Dis. 20:397–406. 2011.PubMed/NCBI
|
|
30
|
Zeuzem S, Berg T, Moeller B, et al: Expert
opinion on the treatment of patients with chronic hepatitis C. J
Viral Hepat. 16:75–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Asselah T, Bièche I, Paradis V, et al:
Genetics, genomics, and proteomics: implications for the diagnosis
and the treatment of chronic hepatitis C. Semin Liver Dis.
27:13–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pearlman BL: The IL-28 genotype: how it
will affect the care of patients with hepatitis C virus infection.
Curr Gastroenterol Rep. 13:78–86. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Antaki N, Bibert S, Kebbewar K, Asaad F,
Baroudi O, Alideeb S, Hadad M, Abboud D, Sabah H, Bochud PY and
Negro F: IL28B polymorphisms predict response to therapy among
chronic hepatitis C patients with HCV genotype 4. J Viral Hepat.
20:59–64. 2013. View Article : Google Scholar : PubMed/NCBI
|