Introduction
Secreted protein acidic and rich in cysteine
(SPARC), also termed as osteonectin or basement-membrane-40, is a
matrix-associated protein that elicits changes in cell shape,
inhibits cell-cycle progression and affects the synthesis of
extracellular matrix (ECM) (1).
The human SPARC gene was initially cloned from a human placenta
cDNA library (2). The final mature
SPARC protein has 286 amino acids with three distinct domains,
including an NH2-terminal acidic domain (NT), follistatin-like
domain (FS) and C terminus domain (EC). The NT domain, spanning the
first 52 amino acids (Ala1-Glu52), binds hydroxyapatite and calcium
ions. This is followed by FS, which comprises the next 85 amino
acids (Asn53-Pro137). This region contains several internal
disulfide bonds that stabilize two weakly interacting nodules. The
third domain, the EC, is 149 amino acids in length (Cys138-Ile286).
It contains two EF-hand motifs that bind calcium with high affinity
and is comprised almost entirely of β-helices.
SPARC binds fibrillar collagen and basal lamina
collagen IV (3) and is associated
with ECM assembly and fibrosis (4). SPARC has also been demonstrated to
regulate the activity of matrix metalloproteinases (MMPs), a family
of enzymes considered to be the primary mediators of ECM
proteolysis and turnover. SPARC has also been demonstrated to
modulate growth factor signaling mediated by cell surface
receptors, including vascular endothelial growth factor receptor,
basic fibroblast growth factor and transforming growth factor β1
(5). SPARC is also involved in
activating odontoblasts during tooth development (6). SPARC upregulation in endothelial
cells and fibroblasts may contribute to compensatory signaling for
controlling angiogenesis (7).
Numerous clinical studies have revealed a
correlation between SPARC expression, malignant progression and
patient survival (8,9). However, SPARC has demonstrated
seemingly contradictory effects on tumor progression in clinical
correlative studies and in animal models (10–13).
The capacity of SPARC to dictate the tumorigenic phenotype has been
attributed to its effects on the bioavailability and signaling of
integrins and growth factors/chemokines. These molecular pathways
contribute to a number of physiological events affecting malignant
progression, including ECM remodeling, angiogenesis, immune
modulation and metastasis (14–17).
Thus, comprehensive investigation regarding whether SPARC is
involved in various types of tumor formations is required.
In the present study, SPARC genes from humans,
chimpanzees, macaques, orangutans, dogs, cows, horses, mice, rats,
opossums, chickens, western clawed frog, zebrafish and fugu were
identified by comparative genomic analyses. Conserved transcription
factor-binding sites within promoter regions of human SPARC genes
were then searched. The expression data, functional relevant single
nucleotide polymorphisms (SNPs) and comparative proteomic analyses
were conducted. Furthermore, meta-analysis of the prognostic value
of SPARC genes in various types of cancer was also performed.
Materials and methods
Identification of novel SPARC genes in
vertebrate genomes and integrative genomic analyses
SPARC genes were searched in the genome sequences of
humans (Homo sapiens), chimpanzees (Pan troglodytes),
macaques (Macaca mulatta), orangutans (Pongo
pygmaeus), dogs (Canis familiaris), cows (Bos
taurus), horses (Equus caballus), mice (Mus
musculus), rats (Rattus norvegicus), opossums
(Monodelphis domestica), chickens (Gallus gallus),
frog (Xenopus tropicalis), zebrafish (Danio rerio)
and fugu (Takifugu rubripes) by the method described prior
to using the human SPARC (NM_003118.3) as queries. The assemblies
used were human NCBI 36, chimpanzee CHIMP2.1, macaque MMUL 1.0,
orangutan PPYG2, dog Canfam 2.0, cow Btau_4.0, horse Equ Cab 2,
mouse NCBI m37, rat RGSC 3.4, opossum monDom5, chicken WASHUC2,
frog JGI 4.1, zebrafish Zv8 and fugu FUGU 4.0. The identified
putative SPARC genes were BLASTed against the nr database of
GenBank to confirm that the best hits were SPARC genes (18–21).
Conserved transcription factor-binding sites within promoter
regions of the human SPARC gene was obtained from SABiosciences’
proprietary database which combines Text Mining Application and
data from the UCSC Genome Browser.
Comparative proteomic analyses of SPARC
proteins
The amino acid sequences of SPARC were deduced from
the identified SPARC genes and aligned using Clustal X 1.8 software
(22). The phylogenetic tree of
SPARC was obtained using maximum likelihood (ML; PHYML v2.4.4)
(23) and neighbor-joining (NJ;
MEGA 3.0) (24) methods, and the
reliability of the tree was evaluated by the bootstrap method with
1,000 replications. The program Codeml implemented in the PAML 3.14
b software package was used to investigate whether Ikaros proteins
are under positive selection (25). Six models of codon substitution,
one-ratio (M0), NearlyNeutral (M1a), PositiveSelection (M2a),
discrete (M3), β (M7) and β and ω (M8) were used in the analysis
(26).
Functional relevant SNP evaluation of the
human SPARC gene
Functional relevant SNPs of the human SPARC gene
were identified as previously described (18–21).
The SNPs were extracted from Ensembl (http://www.ensembl.org) and NCBI’s SNPdb (http://www.ncbi.nlm.nih.gov). The SNPs that were able
to disrupt exonic splicing enhancer/exonic splicing silencer
(ESE/ESS) motifs and cause missense mutations were also
identified.
In silico expression analyses of the
human SPARC gene
Expressed sequence tags (ESTs) derived from the
human SPARC gene were searched for using the BLAST programs as
previously described (27–30). The human SPARC gene (NM_003118) was
used as a query sequence for the BLAST programs. The expression
profiles for normal human tissues were obtained from GeneAnnot
(31) and ArrayExpress (32). Northern analysis of NCBI’s uniGene
dataset was also extracted (18–21).
Furthermore, the protein expression of SPARC was obtained from the
Systematic Protein Investigative Research Environment (SPIRE)
(33) and the Model Organism
Protein Expression Database (MOPED) (34).
Meta-analysis of the prognostic value of
the SPARC gene in cancer
A database named ‘PrognoScan’ has been developed
(35). This database is a large
collection of publicly available cancer microarray datasets with
clinical annotation, as well as a tool for assessing the biological
association between gene expression and prognosis. PrognoScan
employs the minimum P-value approach for grouping patients for
survival analysis. PrognoScan provides a powerful platform for
evaluating potential tumor markers and therapeutic targets and is
publicly accessible at http://gibk21.bio.kyutech.ac.jp/PrognoScan/index.html.
The human SPARC gene was inputted as queries and the data were
collected for analysis.
Results
Comparative proteomics of SPARC proteins
identified in vertebrate genomes
The SPARC genes were identified in the genome
sequences of humans, chimpanzees, macaques, orangutans, dogs, cows,
horses, mice, rats, opossums, chickens, western clawed frog,
zebrafish and fugu. The refined phylogenetic trees using the
identified SPARC proteins amino acid sequences by ML and NJ methods
were almost identical (Fig. 1).
The present study was unable to identify any site under positive
selection, with any of the six models, in the SPARC proteins.
Instead, the SPARC proteins were observed to be under purifying
selection (data not shown).
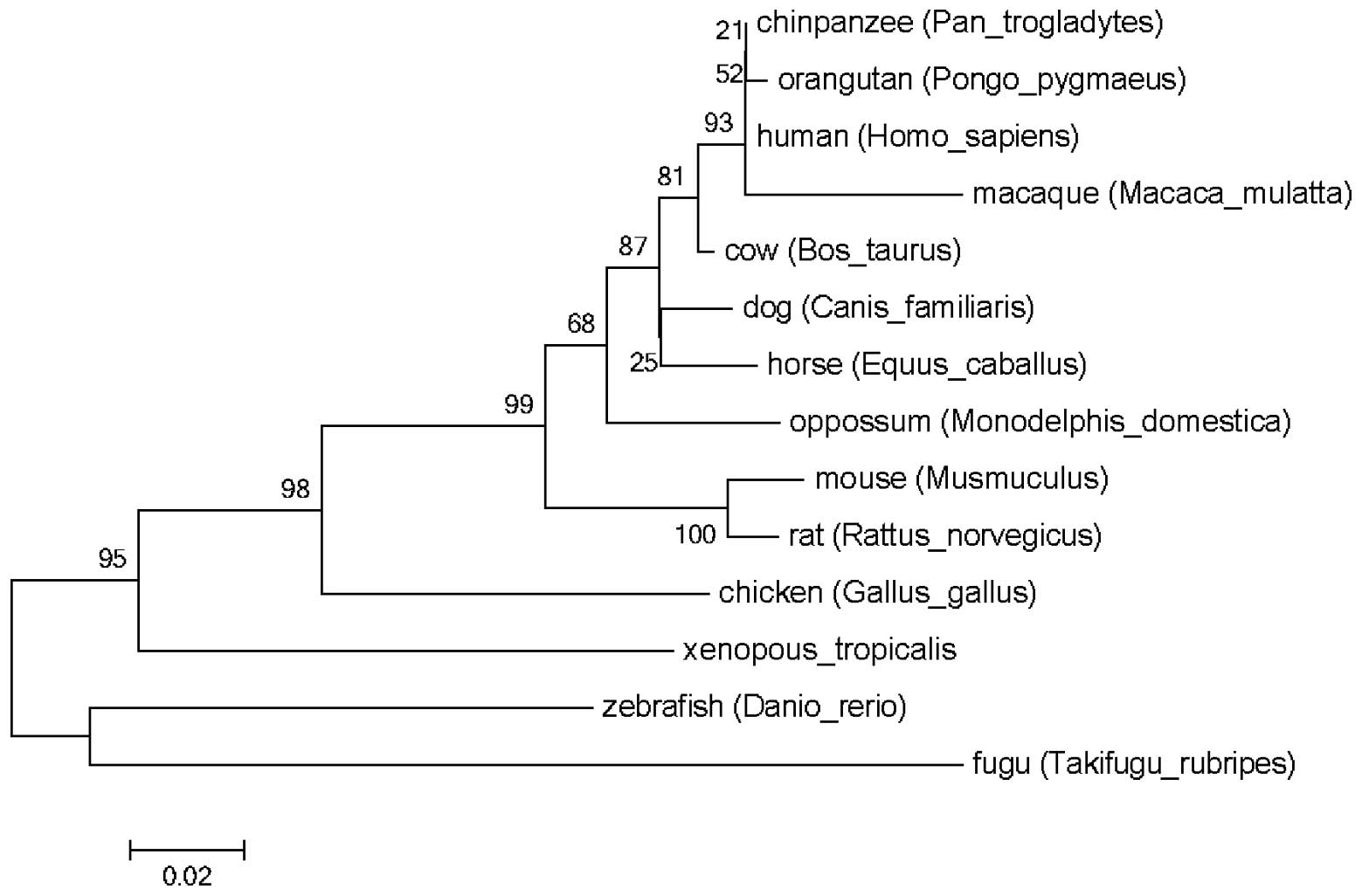 | Figure 1Phylogentic analysis of SPARC. SPARC
genes were identified in the genome sequences of humans,
chimpanzees, macaques, orangutans, dogs, cows, horses, mouses,
rats, opossums, chickens, western clawed frog, zebrafish and fugu.
The phylogenetic tree of the SPARC gene was obtained using maximum
likelihood and neighbor-joining methods. It appeared that the
primate SPARC gene clustered into one group, different from other
SPARC genes. SPARC, secreted protein acidic and rich in
cysteine. |
Expression profile of the human SPARC
gene
By searching for EST sequences, the human SPARC gene
was found to be expressed in the eye, placenta, fetal brain,
neuroblastoma, fetal liver and Lupski dorsal root ganglion. The
investigation of available microarray experiments and ‘virtual
Northern blot’ demonstrated a predominant expression of SPARC in
the bone marrow, whole blood, lymph node, thymus, brain,
cerebellum, retina, heart, smooth muscle, skeletal muscle, spinal
cord, intestine, colon, adipocyte, kidney, liver, pancreas,
thyroid, salivary gland, skin, ovary, uterus, placenta, cervix and
prostate. When searched in the PrognoScan database, the human SPARC
gene was also revealed to be expressed in bladder, blood, brain,
breast, colorectal, eye, lung, ovarian, prostate, renal, skin and
soft tissue cancer tissues. Among the protein expression databases
SPIRE and MOPED, SPARC protein was highly expressed in blood
plasma, blood monocyte, kidney HEK-293, liver and liver HuH-7
cancer cells; however, low levels of SPARC protein expression were
observed in blood erythroleukemia, blood neutrophil, blood
B-lymphocyte, blood T-lymph Jurkat, kidney urine, lung alveolar
lavage, pancreatic cancer and prostate cancer tissues.
Comparative genomics of the human SPARC
gene
Nkx2-5, Brachyury PPAR-α, AML1a and p53 regulatory
transcription factor binding sites were identified in the SPARC
gene upstream (promoter) region.
Functional relevant SNP evaluation of the
human SPARC gene
In total, 471 available SNPs were identified in the
human SPARC gene. Among these, 23 SNPs were functionally relevant,
including two available alleles, which disrupted an existing ESE
and 21 SNPs causing missense mutations (Table I).
 | Table IFunctional relevant SNP evaluation of
the human SPARC gene. |
Table I
Functional relevant SNP evaluation of
the human SPARC gene.
| SNP ID | Chr 5 position | Sequence | Type | Amino acid
change |
|---|
| rs707157 | 151047108(−) |
TGCGGA/G/TACTGG | Missense | ND/Y |
| rs1053296 | 151047111(−) | GCATGC/GGGGAC | Missense | R/D |
| rs11542492 | 151049293(−) | TGCCAC/TAAAGT | Missense | T/I |
| rs11542497 | 151054219(−) | CCTGCA/CTGATG | Missense | H/P |
| rs11542498 | 151054198(−) | GGTGGA/GAGAAA | Missense | E/G |
| rs41290587 | 151051255(+) | AGGGAT/CCTGTA | Missense | /N/S |
| rs7433231 | 151045923(+) | TTACCC/TGTCAA | Missense | R/G |
| rs113617771 | 151052711(+) | CTCTTC/TGGTTT | Missense | K/E |
| rs141567625 | 151045999(+) | TCGAAG/TTCCCG | Missense | E/D |
| rs142207246 | 151051184(+) | GCACAC/TGCACA | Missense | M/V |
| rs142378176 | 151051171(+) | TGGTGA/GGGTCC | Missense | P/L |
| rs142717464 | 151043728(+) | AAAAGC/TGGGTG | Missense | H/R |
| rs146500464 | 151052741(+) | TTCTCC/TTACTT | Missense | R/G |
| rs147557671 | 151051145(+) | AAACTC/TGCCAA | Missense | K/E |
| rs185684862 | 151047110(+) | AGTCCC/TGCATG | Missense | Q/R |
| rs188911380 | 151043660(+) | GATGCC/TGAAGC | Missense | S/G |
| rs199591638 | 151051252(+) | GGCAGG/TGATCT | Missense | H/P |
| rs199655940 | 151043747(+) | CTCCAC/TGGGGA | Missense | M/V |
| rs200777949 | 151047030(+) | TACCCA/GCAGCT | Missense | R/W |
| rs201797309 | 151049318(+) | AGAGTC/TGAAGG | Missense | N/D |
| rs201856432 | 151045950(+) | CTGGCC/TGAACT | Missense | S/G |
| rs2304049 | 151047100(+) | TTCTTG/CAGCCA | ESE | |
| rs11542495 | 151049274(−) | GAGGGC/TACCAA | ESE | |
Meta-analysis of the prognostic value of
the human SPARC gene in cancer
When provided with the gene, PrognoScan exhibits a
summary in table format of tests for the gene with columns for
dataset, cancer type, subtype, end point, cohort, contributor,
array type, probe ID, number of patient, optimal cut point, Pmin
and Pcor. Among the databases that detected the expression of the
SPARC gene, 27 out of 136 tests demonstrated an association between
microarray expression in the SPARC gene and cancer prognosis
(bladder cancer 0/4, blood cancer 5/18, brain cancer 0/8, breast
cancer 4/38, colorectal cancer 8/18, eye cancer 2/2, lung cancer
3/31, ovarian cancer 4/13, prostate cancer 1/1, renal cancer 0/1,
skin cancer 0/2 and soft tissue cancer 0/2) with a 5% significance
level (Table II, 36–50). Among
the five types of blood cancer, a higher expression of the SPARC
gene was associated with a poor survival rate and was found in
three acute myeloid leukemia (AML) cases (GSE12417-GPL96,
GSE12417-GPL96 and GSE12417-GPL570). However, in B-cell lymphoma
(GSE4475) and diffuse large B-cell lymphoma (DLBCL; TABM-346)
cases, a lower expression of the SPARC gene was associated with a
poor survival rate. Among the four types of breast cancer, a higher
expression of the SPARC gene was associated with a poor survival
rate and was identified in two cases (GSE11121 and E-TABM-158).
However, a lower expression of the SPARC gene was associated with a
poor survival rate in another two cases of breast cancer
(GSE3494-GPL96). The present study revealed that a higher
expression of the SPARC gene was associated with a poor survival
rate in all eight colorectal cancer cases. In lung cancer cases, a
higher expression of the SPARC gene was associated with a poor
survival rate in all three types of lung cancer, including
adenocarcinoma and non-small cell lung cancer (NSCLC). In addition,
a lower expression of the SPARC gene was associated with a poor
survival rate in four cases of ovarian cancer, two cases of eye
cancer and one case of prostate cancer.
 | Table IIDataset content from PrognoScan
demonstrated an association between microarray expression of SPARC
and cancer prognosis. |
Table II
Dataset content from PrognoScan
demonstrated an association between microarray expression of SPARC
and cancer prognosis.
| Database | Case type | Subtype | Patient number | End point | Cut point | P-value | Prognosis | Reference |
|---|
| GSE12417-GPL96 | Blood cancer | AML | 163 | Overall
survival | 0.86 | 0.04 | 2 | 66 |
| GSE12417-GPL96 | Blood cancer | AML | 163 | Overall
survival | 0.63 | 0.001 | 2 | 66 |
|
GSE12417-GPL570 | Blood cancer | AML | 79 | Overall
survival | 0.78 | 0.014 | 2 | 66 |
| GSE4475 | Blood cancer | B-cell
lymphoma | 158 | Overall
survival | 0.26 | 0.001 | 1 | 67 |
| TABM-346 | Blood cancer | DLBCL | 53 | Overall
survival | 0.38 | 0.033 | 1 | 68 |
| GSE11121 | Breast cancer | | 200 | Distant metastasis
free survival | 0.6 | 0.045 | 2 | 69 |
| E-TABM-158 | Breast cancer | | 117 | Disease specific
survival | 0.52 | 0.004 | 2 | 70 |
| GSE3494-GPL96 | Breast cancer | | 236 | Disease specific
survival | 0.27 | 0.00017 | 1 | 71 |
| GSE4922-GPL96 | Breast cancer | | 249 | Disease free
survival | 0.27 | 0.0054 | 1 | 72 |
| GSE17536 | Colorectal
cancer | | 177 | Disease specific
survival | 0.64 | 0.0001 | 2 | 73 |
| GSE17536 | Colorectal
cancer | | 177 | Overall
survival | 0.56 | 0.013 | 2 | 73 |
| GSE17536 | Colorectal
cancer | | 177 | Overall
survival | 0.64 | 0.007 | 2 | 73 |
| GSE17536 | Colorectal
cancer | | 177 | Disease free
survival | 0.58 | 0.01 | 2 | 73 |
| GSE17536 | Colorectal
cancer | | 177 | Disease free
survival | 0.66 | 0.0006 | 2 | 73 |
| GSE17536 | Colorectal
cancer | | 177 | Disease specific
survival | 0.56 | 0.0003 | 2 | 73 |
| GSE14333 | Colorectal
cancer | | 226 | Disease free
survival | 0.48 | 0.0063 | 2 | 74 |
| GSE14333 | Colorectal
cancer | | 226 | Disease free
survival | 0.56 | 0.0039 | 2 | 74 |
| GSE22138 | Eye cancer | Uveal melanoma | 63 | Distant metastasis
free survival | 0.56 | 0.0025 | 2 | 75 |
| GSE22138 | Eye cancer | Uveal melanoma | 63 | Distant metastasis
free survival | 0.59 | 0.0007 | 2 | 75 |
| GSE31210 | Lung cancer | Adenocarcinoma | 204 | Overall
survival | 0.83 | 0.048 | 2 | 76 |
| GSE31210 | Lung cancer | Adenocarcinoma | 204 | Relapse free
survival | 0.89 | 0.003 | 2 | 76 |
| GSE8894 | Lung cancer | NSCLC | 138 | Relapse free
survival | 0.36 | 0.044 | 2 | 77 |
| GSE9891 | Ovarian cancer | | 278 | Overall
survival | 0.67 | 0.011 | 2 | 78 |
| CSE9891 | Ovarian cancer | | 278 | Overall
survival | 0.59 | 0.006 | 2 | 78 |
| GSE26712 | Ovarian cancer | | 185 | Disease free
survival | 0.88 | 0.009 | 2 | 79 |
| GSE26712 | Ovarian cancer | | 185 | Overall
survival | 0.88 | 0.039 | 2 | 79 |
| GSE16560 | Prostate
cancer | | 281 | Overall
survival | 0.42 | 0.004 | 1 | 80 |
Discussion
SPARC, also known as osteonectin or BM-40, is a
matrix-associated protein that elicits changes in cell shape,
inhibits cell-cycle progression and affects the synthesis of the
extracellular matrix (ECM) (1). In
the present study, additional SPARC genes from 13 other vertebrate
genomes were identified and SPARC was found to exist in all types
of vertebrates, including fish, amphibians, birds and mammals.
Furthermore, all identified RON proteins contained NT, FS and EC
domains. The phylogenetic tree demonstrated that SPARC is separated
in the order fish, amphibians, birds and mammals. Primate SPARCs
are almost the identical and clustered together. From the alignment
and phylogenetic tree, mammalian SPARCs were observed to be
conserved among vertebrate genomes, suggesting that the function of
SPARC may be important physiologically for all the vertebrates in
the long evolutionary process. Furthermore, this process was under
purifying selection. It is in accordance with multiple biological
functions that have been ascribed to this protein, including its
involvement in tissue remodeling (51), morphogenesis (52,53)
and bone mineralization (54).
Matrix metalloproteinases (MMP-2, -3, -7 and -13),
plasmin and trypsin, have been demonstrated to cleave SPARC in
vitro, producing a KGHK-containing fragment (15,55,56–58).
The presence of the truncated form of the SPARC protein has been
reported in hepatocellular carcinoma samples (59,60)
and esophageal carcinoma (61). It
appeared that truncated SPARC may have an important pro-angiogenic
function in cancer. The present study identified 21 SNPs causing
missense mutations, which may affect the formation of the truncated
form of the SPARC protein. The effects of these SNPs on the
physiological and pathological functions of SPARC requires further
investigation.
SPARC was initially identified in bone and
endothelial cells (62,63). It is also highly expressed in
developing tissues, including the notochord (64), somites (65) and the embryonic skeleton (66), as well as in differentiating
chondrocytes (67), megakaryocytes
(68) and macrophages (69) at sites of tissue injury. The
systematic analysis of SPARC expression in normal tissues and
cancer samples has not been well studied. The present study
revealed that the human SPARC gene was expressed in numerous
tissues and organs, including the bone marrow, whole blood, lymph
node, thymus, brain, cerebellum, retina, heart, smooth muscle,
skeletal muscle, spinal cord, intestine, colon, adipocyte, kidney,
liver, pancreas, thyroid, salivary gland, skin, ovary, uterus,
placenta, cervix and prostate.
When searched in the PrognoScan database, the human
SPARC gene was also revealed to be expressed in bladder, blood,
breast, giloma, esophagus, colorectal, head and neck, ovarian, lung
and skin cancer tissues. SPARC is differentially expressed in
tumors and its surrounding stroma in various types of cancer in
comparison with the normal tissue, yet, its pattern of expression
is variable depending on the type of cancer. In total, 27 out of
136 tests demonstrated an association between microarray expression
of the SPARC gene and cancer prognosis (bladder cancer 0/4, blood
cancer 5/18, brain cancer 0/8, breast cancer 4/38, colorectal
cancer 8/18, eye cancer 2/2, lung cancer 3/31, ovarian cancer 4/13,
prostate cancer 1/1, renal cancer 0/1, skin cancer 0/2 and soft
tissue cancer 0/2) with a 5% significance level.
SPARC mRNA was significantly overexpressed in
pancreatic cancer; however, not in cancer of the papilla of Vater
(8). SPARC was demonstrated to be
associated with drug resistance in ovarian cancer (70). In addition, SPARC induced the
migration of glioblastoma cell lines (10) and the downregulation of SPARC
expression inhibited cell migration and invasion in malignant
gliomas (11). However, other
studies suggested that SPARC induced endoplasmic reticulum stress
leading to autophagy-mediated apoptosis in neuroblastoma (12), and RNA interference against SPARC
promoted the growth of malignant glioma cells (13). The aberrant methylation of SPARC
was identified in human laryngeal and hypopharyngeal carcinomas
(71). In addition, SPARC was
observed to be involved in the transformation of hamster oral
mucosa from precancerous lesions to squamous cell carcinoma
(72,73). SPARC protein expression was also
observed to be markedly induced in the supernatants of co-cultured
astrocytes (74). Despite
transcriptional silencing by aberrant hypermethylation of the
CpG-rich region in endometrial carcinoma, the SPARC protein
remained overexpressed (9).
Furthermore, microRNA-29a was able to suppress cell proliferation
by targeting SPARC in hepatocellular carcinoma (75).
This suggested that the expression of SPARC was
associated with the prognosis of numerous types of cancer,
including hematological and solid cancers. The underlying
mechanisms of SPARC involved in the process of these tumors
requires further investigation. It is important to note that the
association between the expression of SPARC and prognosis varied in
different types of cancer, even in the same cancer from different
databases. It implied that the function of SPARC in these tumors
may be multidimensional, functioning not just as a tumor suppressor
or oncogene.
Nkx2-5, Brachyury PPAR-α, AML1a and p53 regulatory
transcription factor binding sites were identified in the SPARC
gene upstream (promoter) region. Nkx2-5 encodes a
homeobox-containing transcription factor. This transcription factor
functions during heart formation and development. It was also
revealed that Nkx2-5 is the key transcription factor regulating its
genomic neighborhoods differently between the tumor types (76). The p53 gene is mutated in
approximately half of all types of human tumor. p53 is a
transcription factor and its activity gives rise to a variety of
cellular outcomes, most notably cell cycle arrest and apoptosis,
eliminating cancer-prone cells from the replicative pool (77–79).
These two tumor-related transcriptional factors may be involved in
the effect of SPARC on various types of tumor.
The present study demonstrated that the association
between the expression of SPARC and prognosis varied in different
types of cancer. However, the specific functions of SPARC in the
majority of tumors remain to be elucidated. Further studies are
required to focus on its different behaviors in different types of
cancer and its potential relative pathways, including MMPs.
Acknowledgements
This study was supported in part by the National
Natural Science Foundation of China (grant no. 81202077), the
Program for Development of Innovative Research Team in the First
Affiliated Hospital of NJMU (grant no. IRT-008) and a project
Funded by the Priority Academic Program Development of Jiangsu
Higher Education Institutions.
References
|
1
|
Bradshaw AD, Graves DC, Motamed K and Sage
EH: SPARC-null mice exhibit increased adiposity without significant
differences in overall body weight. Proc Natl Acad Sci USA.
100:6045–6050. 2003. View Article : Google Scholar
|
|
2
|
Schwartz RC, Young MF and Tsipouras P: Two
RFLPs in the 5′ end of the human osteonectin (ON) gene. Nucleic
Acids Res. 16:90761988.
|
|
3
|
Mayer U, Aumailley M, Mann K, Timpl R and
Engel J: Calcium-dependent binding of basement membrane protein
BM-40 (osteonectin, SPARC) to basement membrane collagen type IV.
Eur J Biochem. 198:141–150. 1991. View Article : Google Scholar
|
|
4
|
Bradshaw AD: The role of SPARC in
extracellular matrix assembly. J Cell Commun Signal. 3:239–246.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rivera LB and Brekken RA: SPARC promotes
pericyte recruitment via inhibition of endoglin-dependent TGF-beta1
activity. J Cell Biol. 193:1305–1319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi BD, Yun SH, Jeong SJ, et al:
Expression of thymosin beta4 in odontoblasts during mouse tooth
development. Int J Mol Med. 29:841–847. 2012.PubMed/NCBI
|
|
7
|
Nonogaki S, Campos HG, Butugan O, et al:
Markers of vascular differentiation, proliferation and tissue
remodeling in juvenile nasopharyngeal angiofibromas. Exp Ther Med.
1:921–926. 2010.
|
|
8
|
Prenzel KL, Warnecke-Eberz U, Xi H, et al:
Significant overexpression of SPARC/osteonectin mRNA in pancreatic
cancer compared to cancer of the papilla of Vater. Oncol Rep.
15:1397–1401. 2006.
|
|
9
|
Rodriguez-Jiménez FJ, Caldés T, Iniesta P,
Vidart JA, Garcia-Asenjo JL and Benito M: Overexpression of SPARC
protein contrasts with its transcriptional silencing by aberrant
hypermethylation of SPARC CpG-rich region in endometrial carcinoma.
Oncol Rep. 17:1301–1307. 2007.
|
|
10
|
Kunigal S, Gondi CS, Gujrati M, et al:
SPARC-induced migration of glioblastoma cell lines via uPA-uPAR
signaling and activation of small GTPase RhoA. Int J Oncol.
29:1349–1357. 2006.PubMed/NCBI
|
|
11
|
Seno T, Harada H, Kohno S, Teraoka M,
Inoue A and Ohnishi T: Downregulation of SPARC expression inhibits
cell migration and invasion in malignant gliomas. Int J Oncol.
34:707–715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sailaja GS, Bhoopathi P, Gorantla B, et
al: The secreted protein acidic and rich in cysteine (SPARC)
induces endoplasmic reticulum stress leading to autophagy-mediated
apoptosis in neuroblastoma. Int J Oncol. 42:188–196. 2013.
|
|
13
|
Liu H, Xu Y, Chen Y, et al: RNA
interference against SPARC promotes the growth of U-87MG human
malignant glioma cells. Oncol Lett. 2:985–990. 2011.PubMed/NCBI
|
|
14
|
Al Saleh S, Sharaf LH and Luqmani YA:
Signalling pathways involved in endocrine resistance in breast
cancer and associations with epithelial to mesenchymal transition
(Review). Int J Oncol. 38:1197–1217. 2011.
|
|
15
|
Zhang JL, Chen GW, Liu YC, et al: Secreted
protein acidic and rich in cysteine (SPARC) suppresses angiogenesis
by down-regulating the expression of VEGF and MMP-7 in gastric
cancer. PLoS One. 7:e446182012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arnold SA, Rivera LB, Carbon JG, et al:
Losartan slows pancreatic tumor progression and extends survival of
SPARC-null mice by abrogating aberrant TGFbeta activation. PLoS
One. 7:e313842012. View Article : Google Scholar
|
|
17
|
Said N, Frierson HF, Sanchez-Carbayo M,
Brekken RA and Theodorescu D: Loss of SPARC in bladder cancer
enhances carcinogenesis and progression. J Clin Invest.
123:751–766. 2013.PubMed/NCBI
|
|
18
|
Yang L, Luo Y and Wei J: Integrative
genomic analyses on Ikaros and its expression related to solid
cancer prognosis. Oncol Rep. 24:571–577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, Luo Y, Wei J and He S: Integrative
genomic analyses on IL28RA, the common receptor of
interferon-lambda1, -lambda2 and -lambda3. Int J Mol Med.
25:807–812. 2010.PubMed/NCBI
|
|
20
|
Yang L, Wei J and He S: Integrative
genomic analyses on interferon-lambdas and their roles in cancer
prediction. Int J Mol Med. 25:299–304. 2010.PubMed/NCBI
|
|
21
|
Yu H, Yuan J, Xiao C and Qin Y:
Integrative genomic analyses of recepteur d’origine nantais and its
prognostic value in cancer. Int J Mol Med. 31:1248–1254. 2013.
|
|
22
|
Thompson JD, Gibson TJ, Plewniak F,
Jeanmougin F and Higgins DG: The CLUSTAL_X windows interface:
flexible strategies for multiple sequence alignment aided by
quality analysis tools. Nucleic Acids Res. 25:4876–4882. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guindon S, Lethiec F, Duroux P and Gascuel
O: PHYML Online - a web server for fast maximum likelihood-based
phylogenetic inference. Nucleic Acids Res. 33:W557–W559. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kumar S, Tamura K and Nei M: MEGA3:
Integrated software for Molecular Evolutionary Genetics Analysis
and sequence alignment. Brief Bioinform. 5:150–163. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z: PAML: a program package for
phylogenetic analysis by maximum likelihood. Comput Appl Biosci.
13:555–556. 1997.PubMed/NCBI
|
|
26
|
Yang Z, Nielsen R, Goldman N and Pedersen
AM: Codon-substitution models for heterogeneous selection pressure
at amino acid sites. Genetics. 155:431–449. 2000.PubMed/NCBI
|
|
27
|
Katoh Y and Katoh M: Integrative genomic
analyses on GLI1: positive regulation of GLI1 by Hedgehog-GLI,
TGFbeta-Smads, and RTK-PI3K-AKT signals, and negative regulation of
GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol.
35:187–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Katoh M and Katoh M: Integrative genomic
analyses of WNT11: transcriptional mechanisms based on canonical
WNT signals and GATA transcription factors signaling. Int J Mol
Med. 24:247–251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Katoh M and Katoh M: Transcriptional
mechanisms of WNT5A based on NF-kappaB, Hedgehog, TGFbeta, and
Notch signaling cascades. Int J Mol Med. 23:763–769. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katoh M and Katoh M: Transcriptional
regulation of WNT2B based on the balance of Hedgehog, Notch, BMP
and WNT signals. Int J Oncol. 34:1411–1415. 2009.PubMed/NCBI
|
|
31
|
Chalifa-Caspi V, Yanai I, Ophir R, et al:
GeneAnnot: comprehensive two-way linking between oligonucleotide
array probesets and GeneCards genes. Bioinformatics. 20:1457–1458.
2004. View Article : Google Scholar
|
|
32
|
Parkinson H, Sarkans U, Shojatalab M, et
al: ArrayExpress - a public repository for microarray gene
expression data at the EBI. Nucleic Acids Res. 33:D553–D555. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kolker E, Higdon R, Morgan P, et al:
SPIRE: Systematic protein investigative research environment. J
Proteomics. 75:122–126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kolker E, Higdon R, Haynes W, et al:
MOPED: Model Organism Protein Expression Database. Nucleic Acids
Res. 40:D1093–D1099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: a new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Metzeler KH, Hummel M, Bloomfield CD, et
al: An 86-probe-set gene-expression signature predicts survival in
cytogenetically normal acute myeloid leukemia. Blood.
112:4193–4201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hummel M, Bentink S, Berger H, et al: A
biologic definition of Burkitt’s lymphoma from transcriptional and
genomic profiling. N Engl J Med. 354:2419–2430. 2006.
|
|
38
|
Jardin F, Jais JP, Molina TJ, et al:
Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct
gene expression signature and a poor prognosis under R-CHOP
treatment: a GELA study. Blood. 116:1092–1104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schmidt M, Böhm D, von Torne C, et al: The
humoral immune system has a key prognostic impact in node-negative
breast cancer. Cancer Res. 68:5405–5413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chin K, DeVries S, Fridlyand J, et al:
Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miller LD, Smeds J, George J, et al: An
expression signature for p53 status in human breast cancer predicts
mutation status, transcriptional effects, and patient survival.
Proc Natl Acad Sci USA. 102:13550–13555. 2005. View Article : Google Scholar
|
|
42
|
Ivshina AV, George J, Senko O, et al:
Genetic reclassification of histologic grade delineates new
clinical subtypes of breast cancer. Cancer Res. 66:10292–10301.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Smith JJ, Deane NG, Wu F, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
44
|
Jorissen RN, Gibbs P, Christie M, et al:
Metastasis-associated gene expression changes predict poor outcomes
in patients with Dukes stage B and C colorectal cancer. Clin Cancer
Res. 15:7642–7651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Laurent C, Valet F, Planque N, et al: High
PTP4A3 phosphatase expression correlates with metastatic risk in
uveal melanoma patients. Cancer Res. 71:666–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Okayama H, Kohno T, Ishii Y, et al:
Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee ES, Son DS, Kim SH, et al: Prediction
of recurrence-free survival in postoperative non-small cell lung
cancer patients by using an integrated model of clinical
information and gene expression. Clin Cancer Res. 14:7397–7404.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tothill RW, Tinker AV, George J, et al:
Novel molecular subtypes of serous and endometrioid ovarian cancer
linked to clinical outcome. Clin Cancer Res. 14:5198–5208. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bonome T, Levine DA, Shih J, et al: A gene
signature predicting for survival in suboptimally debulked patients
with ovarian cancer. Cancer Res. 68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sboner A, Demichelis F, Calza S, et al:
Molecular sampling of prostate cancer: a dilemma for predicting
disease progression. BMC Med Genomics. 3:82010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McCurdy SM, Dai Q, Zhang J, et al: SPARC
mediates early extracellular matrix remodeling following myocardial
infarction. Am J Physiol Heart Circ Physiol. 301:H497–H505. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cheng L, Sage EH and Yan Q: SPARC fusion
protein induces cellular adhesive signaling. PLoS One.
8:e532022013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nakamura K, Nakano S, Miyoshi T,
Yamanouchi K, Matsuwaki T and Nishihara M: Age-related resistance
of skeletal muscle-derived progenitor cells to SPARC may explain a
shift from myogenesis to adipogenesis. Aging (Albany NY). 4:40–48.
2012.PubMed/NCBI
|
|
54
|
Pataquiva-Mateus AY, Wu HC, Lucchesi C,
Ferraz MP, Monteiro FJ and Spector M: Supplementation of collagen
scaffolds with SPARC to facilitate mineralization. J Biomed Mater
Res B Appl Biomater. 100:862–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li B, Li F, Chi L, Zhang L and Zhu S: The
expression of SPARC in human intracranial aneurysms and its
relationship with MMP-2/-9. PLoS One. 8:e584902013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Seet LF, Tong L, Su R and Wong TT:
Involvement of SPARC and MMP-3 in the pathogenesis of human
pterygium. Invest Ophthalmol Vis Sci. 53:587–595. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Patterson J and Hubbell JA: SPARC-derived
protease substrates to enhance the plasmin sensitivity of
molecularly engineered PEG hydrogels. Biomaterials. 32:1301–1310.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Weaver MS, Sage EH and Yan Q: Absence of
SPARC in lens epithelial cells results in altered adhesion and
extracellular matrix production in vitro. J Cell Biochem.
97:423–432. 2006. View Article : Google Scholar
|
|
59
|
Lin ZY and Chuang WL: Genes responsible
for the characteristics of primary cultured invasive phenotype
hepatocellular carcinoma cells. Biomed Pharmacother. 66:454–458.
2012. View Article : Google Scholar
|
|
60
|
Zhang Y, Yang B, Du Z, et al: Aberrant
methylation of SPARC in human hepatocellular carcinoma and its
clinical implication. World J Gastroenterol. 18:2043–2052. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xue LY, Zou SM, Zheng S, et al:
Expressions of the gamma2 chain of laminin-5 and secreted protein
acidic and rich in cysteine in esophageal squamous cell carcinoma
and their relation to prognosis. Chin J Cancer. 30:69–78. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Termine JD, Kleinman HK, Whitson SW, Conn
KM, McGarvey ML and Martin GR: Osteonectin, a bone-specific protein
linking mineral to collagen. Cell. 26:99–105. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mason IJ, Murphy D, Münke M, Francke U,
Elliott RW and Hogan BL: Developmental and transformation-sensitive
expression of the Sparc gene on mouse chromosome 11. EMBO J.
5:1831–1837. 1986.PubMed/NCBI
|
|
64
|
Huynh MH, Sodek K, Lee H and Ringuette M:
Interaction between SPARC and tubulin in Xenopus. Cell Tissue Res.
317:313–317. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sage H, Decker J, Funk S and Chow M:
SPARC: a Ca2+-binding extracellular protein associated
with endothelial cell injury and proliferation. J Mol Cell Cardiol.
21(Suppl 1): 13–22. 1989.
|
|
66
|
Leboy PS, Shapiro IM, Uschmann BD, Oshima
O and Lin D: Gene expression in mineralizing chick epiphyseal
cartilage. J Biol Chem. 263:8515–8520. 1988.PubMed/NCBI
|
|
67
|
Aeschlimann D, Kaupp O and Paulsson M:
Transglutaminase-catalyzed matrix cross-linking in differentiating
cartilage: identification of osteonectin as a major glutaminyl
substrate. J Cell Biol. 129:881–892. 1995. View Article : Google Scholar
|
|
68
|
Breton-Gorius J, Clezardin P, Guichard J,
et al: Localization of platelet osteonectin at the internal face of
the alpha-granule membranes in platelets and megakaryocytes. Blood.
79:936–941. 1992.PubMed/NCBI
|
|
69
|
Komatsubara I, Murakami T, Kusachi S, et
al: Spatially and temporally different expression of osteonectin
and osteopontin in the infarct zone of experimentally induced
myocardial infarction in rats. Cardiovasc Pathol. 12:186–194. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Bioinformatic analysis of chemokine (C-C motif) ligand 21 and
SPARC-like protein 1 revealing their associations with drug
resistance in ovarian cancer. Int J Oncol. 42:1305–1316.
2013.PubMed/NCBI
|
|
71
|
He Q, Wei J, Zhang J, et al: Aberrant
methylation of secreted protein, acidic and rich in cysteine in
human laryngeal and hypopharyngeal carcinoma. Oncol Lett.
2:725–729. 2011.PubMed/NCBI
|
|
72
|
Chen D, Yang K, Mei J, Zhang G, Lv X and
Xiang L: Screening the pathogenic genes and pathways related to
DMBA (7,12-dimethylbenz[a]anthracene)-induced transformation of
hamster oral mucosa from precancerous lesions to squamous cell
carcinoma. Oncol Lett. 2:637–642. 2011.PubMed/NCBI
|
|
73
|
Suhr ML, Dysvik B, Bruland O, et al: Gene
expression profile of oral squamous cell carcinomas from Sri Lankan
betel quid users. Oncol Rep. 18:1061–1075. 2007.PubMed/NCBI
|
|
74
|
Gagliano N, Costa F, Cossetti C, et al:
Glioma-astrocyte interaction modifies the astrocyte phenotype in a
co-culture experimental model. Oncol Rep. 22:1349–1356. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhu XC, Dong QZ, Zhang XF, et al:
microRNA-29a suppresses cell proliferation by targeting SPARC in
hepatocellular carcinoma. Int J Mol Med. 30:1321–1326.
2012.PubMed/NCBI
|
|
76
|
Ylipää A, Yli-Harja O, Zhang W and Nykter
M: A systems biological approach to identify key transcription
factors and their genomic neighborhoods in human sarcomas. Chin J
Cancer. 30:27–40. 2011.PubMed/NCBI
|
|
77
|
Ren D, Wang M, Guo W, et al: Wild-type p53
suppresses the epithelial-mesenchymal transition and stemness in
PC-3 prostate cancer cells by modulating miR-145. Int J Oncol.
42:1473–1481. 2013.PubMed/NCBI
|
|
78
|
Abou-El-Ardat K, Derradji H, de Vos W, et
al: Response to low-dose X-irradiation is p53-dependent in a
papillary thyroid carcinoma model system. Int J Oncol.
39:1429–1441. 2011.PubMed/NCBI
|
|
79
|
Stefancikova L, Moulis M, Fabian P, et al:
Prognostic impact of p53 aberrations for R-CHOP-treated patients
with diffuse large B-cell lymphoma. Int J Oncol. 39:1413–1420.
2011.PubMed/NCBI
|















