Introduction
Colorectal carcinoma is the third most common type
of malignant tumor worldwide, and resulted in ~400,000 deaths in
2010 (1). Although treatments have
greatly improved in recent years, the five-year survival rate of
advanced colorectal carcinoma has not significantly improved. Thus
far, the pathogenesis of colorectal carcinoma has not been clearly
characterized.
The microenvironment of tumors has increasingly been
recognized as having an important role in the occurrence and
development of cancer (2).
Chemokines and their receptors can induce migration, chemotaxis and
rearrangement of the cytoskeleton in the target cell, and therefore
promote multiple physiological functions of cells, including cell
growth, development, differentiation and apoptosis (3–5). It
has been reported that chemokine receptors and their functions
differ between tumor cells and normal cells, suggesting that
dysregulation of chemokines may be involved in the development of
malignancy (6,7). It has been demonstrated that the
expression levels of the receptor CCR6 are associated with the
metastasis of hepatocellular carcinoma (8) and lymphatic metastasis in colorectal
carcinoma (9). In addition,
studies conducted by Hojo et al (10) indicated that high CXCL16 expression
levels are associated with good prognosis in colorectal carcinoma
(10).
Using the Expressed Sequence Tags database, Hromas
et al (11) described a
novel CXC chemokine located in human kidney and breast tissue,
termed BRAK, which is now termed CXCL14 (11). CXCL14 is highly concentrated in
normal cells, but is mostly absent from cancer cells (12,13).
Due to its ELR-motif, CXCL14 exhibits multiple functions, including
an inhibitory effect on angiogenesis and chemotactic effect on
natural killer cells, B-cells, macrophages, monocytes and immature
dendritic cells (14–18). The identification of CXCL14 led to
clinical research into other diseases, including obesity (19,20),
bacterial infections (21) and
immune system disorders (22,23).
In addition to breast cancer tissues, studies have demonstrated low
or absent CXCL14 expression in other tissues, including lung
cancer, head and neck squamous cell carcinoma, hepatocellular
carcinoma and gastric cancer (23–28).
However, in prostate and pancreatic cancer, CXCL14 is generally
expressed at a high level (29–31).
Different studies have reported varied expression levels within one
tissue type; Zeng et al (32) observed that CXCL14 expression
levels were higher in colorectal carcinoma tissue than in normal
tissues, while Cao et al (33) produced an opposite result. As a
homeostasis-associated chemokine, CXCL14 may function in the
development of colorectal carcinoma by blocking the chemotaxis of
immune cells. However, its role in the genesis and development of
colorectal carcinoma as either an anti-oncogene or an oncogene is
not entirely clear. In the present study, the expression of CXCL14
and its clinical significance in colorectal carcinoma tissues were
investigated, and the direct effect of CXCL14 on colorectal
carcinoma cells was evaluated by upregulating CXCL14
expression.
Materials and methods
Collection and processing of tissue
specimens
Tumor samples were collected from 40 patients with
colorectal carcinoma that were diagnosed by endoscopic biopsy and
surgically treated in Taizhou First People’s Hospital (Taizhou,
China) between December 2008 and April 2009. The diagnoses of
colorectal cancer in all patients were confirmed after surgery by
histopathological examination according to World Health
Organization standards (34). The
clinicopathological features of the patients are listed in Table I. None of the patients had received
preoperative radiotherapy or chemotherapy and all patients received
strict chemotherapy following surgery according to the colorectal
carcinoma treatment guidelines stipulated by National Comprehensive
Cancer Network (35). Tumor
samples were paired with normal tissues that had been resected from
a location 2 cm away from the edge of the tumor. Each sample was
divided into two further samples. One was cryopreserved in liquid
nitrogen within 30 min of removal for RNA extraction, and the other
was formalin-fixed and embedded in paraffin for sectioning and
immunohistochemical analysis. Informed written consent was obtained
from each patient and the study was approved by the Human Research
Ethics Committee of Taizhou First People’s Hospital.
 | Table IClinicopathological features and
CXCL14 protein expression of 40 specimens. |
Table I
Clinicopathological features and
CXCL14 protein expression of 40 specimens.
| Clinical
feature | n | Relative protein
expression | P-value |
|---|
| Gender | | | >0.05 |
| Male | 30 | 0.183
(0.007–1.151) | |
| Female | 10 | 0.157
(0.005–0.035) | |
| Age (years) | | | >0.05 |
| <60 | 6 | 0.217
(0.028–0.096) | |
| ≥60 | 34 | 0.170
(0.006–1.164) | |
| Tumor size
(cm) | | | >0.05 |
| <5 | 20 |
0.178(0.002–1.56) | |
| ≥5 | 20 | 0.176
(0.001–0.107) | |
| Lymph node
metastasis | | | <0.05 |
| Present | 22 | 0.146
(0.006–0.061) | |
| Absent | 18 | 0.215
(0.004–0.120) | |
| Tumor
infiltration | | | >0.05 |
| Serosal layer | 16 | 0.176
(0.001–0.106) | |
| Serous outer | 20 | 0.173
(0.003–0.161) | |
| Subserosa | 2 | 0.243 | |
| Muscular
layer | 2 | 0.152 | |
| Tumor location | | | <0.05 |
| Rectum | 20 | 0.146 (0.
006–1.283) | |
| Colon | 20 | 0.206
(0.009–0.047) | |
| Anatomical
stagea | | | <0.05 |
| I, II | 18 |
0.215(0.004–1.200) | |
| III, IV | 22 | 0.146
(0.005–0.061) | |
| CEA (μg/l) | | | >0.05 |
| ≤5 | 18 |
0.200(0.001–1.351) | |
| >5 | 22 |
0.158(0.004–0.124) | |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA extraction was conducted using TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according
to the manufacturer’s instructions. In order to remove any DNA, the
extracted RNA was processed using DNase I (Takara Bio, Inc., Shiga,
Japan) with phenol-chloroform extraction (Kangwei Biological
Company, Beijing, China). The purified RNA was dissolved in
diethylpyrocarbonate water (Takara Bio, Inc.) and its concentration
was measured with an ultraviolet spectrophotometer (DU640; Beckman
Coulter, Miami, FL, USA), prior to storage at −80°C. Extracted RNA
(1 μg) was taken from each sample for reverse transcription using
Oligo dT primers (ReverTra Ace qPCR RT kit, Toyobo, Osaka, Japan).
The reaction conditions were as follows: 42°C for 60 min, 75°C for
15 min, followed by −20°C at the end of the reaction. qPCR was
conducted based on a 15 μl reaction system. Table II lists the primer sequences used
in the analysis. The quantitative fluorescent PCR reaction system
consisted of the following: 1 μl reverse transcription product, 1X
SYBR-Green I Mastermix (Toyobo), and 0.5 μmol/l each of the
specific forward and reverse primers. The reaction conditions were
as follows: 40 cycles of 95°C for 2 min, 95°C for 15 sec and 60°C
for 1 min. qPCR was conducted using the CFX96 Real-Time PCR
Detection system (Bio-Rad, Hercules, CA, USA). All samples were run
in triplicate to increase reliability of results. Based on the Ct
values of the specimens using GAPDH as the normalizer gene, the
relative quantitative method of qPCR was adopted.
N=2−ΔΔCt was considered to represent the CXCL14 relative
expression level in specimens, where,
ΔΔCt=(CtCXCL14−CtGAPDH) tumor −
(CtCXCL14−CtGAPDH) normal.
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Gene | Direction | Primer
sequence | Product length
(bp) |
|---|
| CXCL14 | F |
5′-AGCCAAAGTACCCGCACTG-3′ | 156 |
| R |
5′-AGACCCTGCGCTTCTCGTTC-3′ | |
| GAPDH | F |
5′-CAGGGCTGCTTTTAACTCTGGTAA-3′ | 101 |
| R |
5′-GGGTGGAATCATATTGGAACATGT-3′ | |
Immunohistochemical detection
Formalin-fixed specimens embedded in paraffin were
sectioned into 4-μm samples. The slides were processed with 0.1%
poly-L-lysine and tissues were dewaxed using dimethyl benzene
followed by immersion in distilled water. Endogenous peroxidase
activity was inhibited by incubation in a 0.3% hydrogen peroxide
bath for 10 min followed by three washes with 0.01 mol/l
phosphate-buffered saline (PBS; pH 7.4). Slides were then immersed
in citrate antigen retrieval buffer (Zhongshan Golden Bridge
Biotechnology, Beijing, China) for 1.5 min. Once the slides were
cooled to room temperature, they were washed with PBS three
additional times, followed by blocking in sheep serum for 2 h. The
sections were incubated in primary anti-CXCL14 antibody (Abcam,
Cambridge, MA, USA) diluted to 1:500 in PBS, in a humidified
chamber at 4°C overnight. Sections were then incubated in
horseradish peroxidase (HRP)-conjugated secondary goat anti-mouse
antibodies (MaiXin Bio, Fuzhou, China) at 37°C for 30 min. The
color reaction was developed using a 3,3′-diaminobenzidine kit
(Zhongshan Golden Bridge Biotechnology) according to the
manufacturer’s instructions. Slides were then counterstained using
hematoxylin, dehydrated and sealed with neutral gum. In the
negative control group, PBS was used in place of the primary
antibody. As CXCL14 is located in the plasma, cells stained with
brown cytoplasm were considered as positive for CXCL14. Image-Pro
Plus, version 6.0 image processing software (Media Cybernetics,
Rockville, MD, USA) was used to calculate the mean optical density
for further statistical analyses.
Construction of overexpressed CXCL14
lentivirus
In order to facilitate the CXCL14 expression
analysis, the study adopted the pLenti6.3_MCS_IRES-EGFP lentivirus
expression vector (Invitrogen Life Technologies) with an internal
ribosome entry site (IRES). The full-length sequence of CXCL14
(accession no. BC003513.1) was synthesized by Invitrogen and cloned
into the plasmid of pLenti6.3_MCS_IRES-EGFP using Nhel and
Ascl (Toyobo) double enzyme digestion. The
pLenti6.3_CXCL14_IRES-EGFP recombinant plasmid was then identified
by CXCL14 gene-specific PCR and sequencing analysis. The plasmid
was extracted using a PureLink HiPure Plasmid Maxiprep kit
(Invitrogen Life Technologies) in preparation for the lentivirus
package. To obtain the lentivirus, 293T cells (ATCC, Manassas, VA,
USA) at logarithmic phases were transformed with the packaged
plasmids of pLP1, pLP2, pLP/VSVG and pLenti6.3_CXCL14_IRES-EGFP
lentivirus expression plasmid at a ratio of 3:1 according to
manufacturer’s instructions of Lipofectamine 2000 (Invitrogen Life
Technologies). After 48 h, the cell culture supernatant was
collected and centrifuged at 3,000 × g for 10 min to remove the
residual cells and fragments. The virus supernatant was then
ultracentrifugated at 50,000 × g for 2 h, and the concentration was
resuspended in Opti-MEM solution (Invitrogen, Life Technologies).
Following titer determination, the virus solution was stored at
−80°C.
Western blot analysis
HT29 colorectal carcinoma cells (ATCC) were
transfected by CXCL14 lentivirus with a multiplicity of infection
(MOI) of 20, and total protein was extracted with a protein
extraction kit (Shanghai Biyuntian Biological Co., Ltd, Shanghai,
China) following 48 h of infection. Extracted protein (20 μg) was
then separated by 10% SDS-PAGE gel electrophoresis and transferred
onto polyvinylidene fluoride membranes (Millipore, Billerica, MA,
USA). After blocking with 5% non-fat milk for 2 h, the membranes
were then incubated with the CXCL14 monoclonal antibody (1:100,000)
at 4°C overnight. Following three cycles of washing with
Tris-buffered saline and Tween-20, HRP-conjugated goat anti-mouse
antibodies (Beyotime Biotech, Shanghai, China) were incubated for 2
h at room temperature. A BeyoECL Plus (Beyotime Biotech) detection
system was used for analysis of the final chemiluminescence
reaction. The samples of HT29 colorectal carcinoma cell lines
transfected with the control lentivirus were regarded as negative
controls and β-actin served as an internal reference gene.
Cell proliferation and activity
detection
HT29 colorectal carcinoma cell lines were cultured
in a 96-well plate and were transfected with CXCL14 lentivirus at
an MOI of 20. Following 48-h culture, the activity of the cells was
detected with a cell counting kit-8 assay kit (Beyotime Biotech).
The absorbency was measured with a spectrophotometer (BioTek,
Winooski, VT, USA) at a wavelength of at 450 nm according to the
manufacturer’s instructions, and cell viability was calculated.
Cell cycle detection by flow cytometry
(FCM)
The transfected HT29 cells were digested with 0.25%
pancreatic enzymes, then centrifuged at 800 × g for 10 min.
Following washing, the cells were infiltrated and fixed with 70%
precooled ethyl alcohol at 4°C overnight. After washing twice with
PBS, the cells were digested by RNase-A (Toyobo) at 37°C for 30
min. The cells were stained with propidium iodide in an ice bath in
the dark for 30 min, then filtered. Flow cytometry (BD Biosciences,
San Jose, CA, USA) was utilized for analytic detection (Beckman
Coulter, Brea, CA, USA).
Statistical analysis
The Wilcoxon signed-rank test was adopted for the
comparison of paired samples; the Mann-Whitney U test was applied
for the comparison of two groups of independent samples; the
Kruskal-Wallis H test was applied for the comparison of three or
more groups of independent samples; the χ2 test was
adopted for the comparison of inter-group rates; and the
association between CXCL14 protein expression and prognosis was
analyzed with a Kaplan-Meier survival curve. P<0.05 was
considered to represent a statistically significant difference. All
the analyses were conducted using SPSS, version 19.0 (IBM SPSS,
Armonk, NY, USA).
Results
Expression of CXCL14 in colorectal
carcinoma tissues
The expression levels of CXCL14 mRNA in colorectal
carcinoma were analyzed using RT-qPCR, with GAPDH as an internal
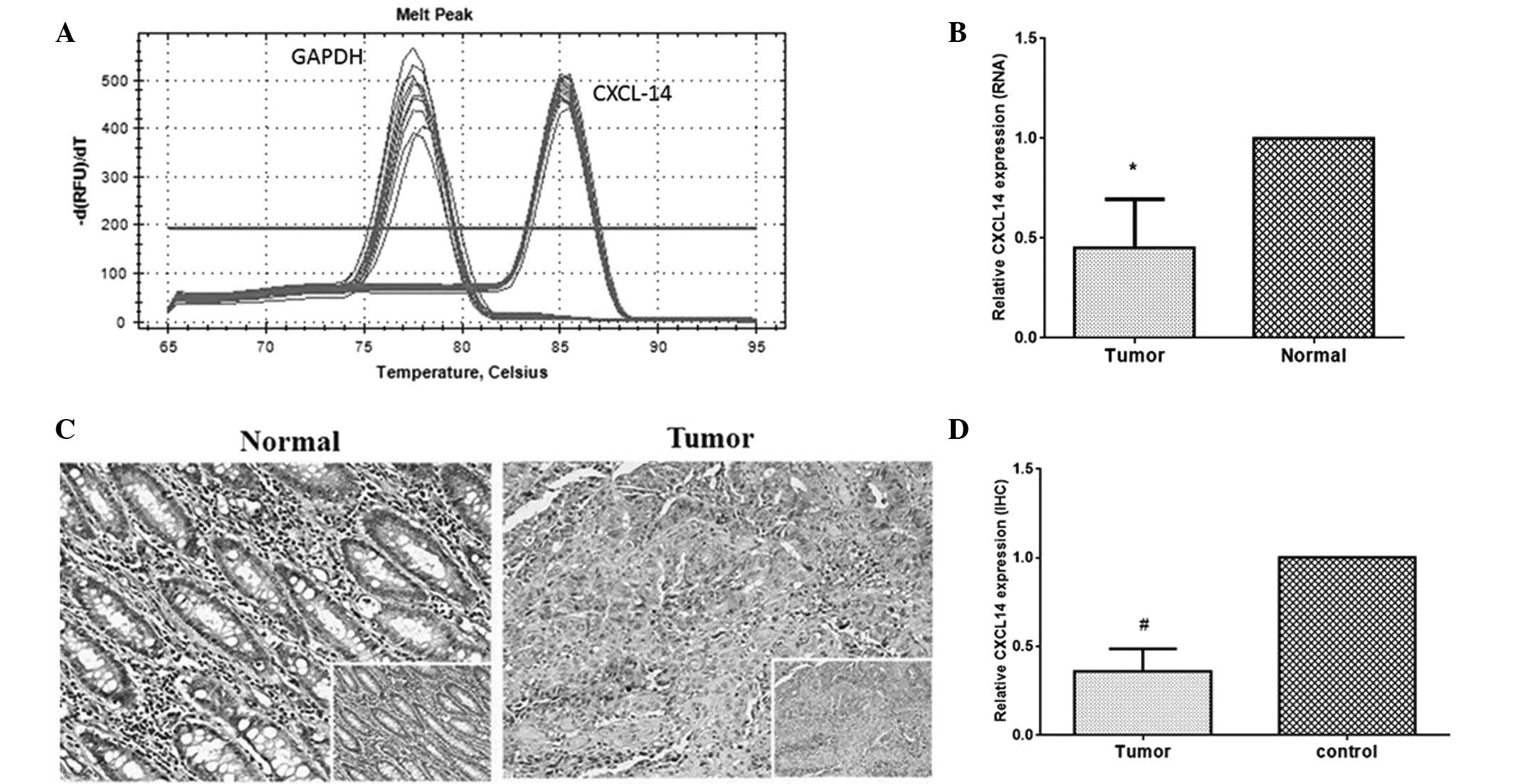
reference. The melting curves of CXCL14 and GAPDH were sharply
defined curves with a narrow peak, indicating that the established
PCR method effectively amplified the target genes (Fig. 1A). As seen in Fig. 1B, the relative level of CXCL14
expression in tumor specimens was 0.502, which was significantly
lower than the level in paired normal tissues (P<0.01). The
CXCL14 protein expression level was further analyzed by
immunohistochemical analysis. The expression of CXCL14 protein was
observed in the cytoplasm (Fig.
1C). In normal tissues, there was strong positive staining,
while the tumor tissues expressed CXCL14 at a low level, if at all.
Semi-quantitative analyses of immunohistochemical results indicated
that the expression level of CXCL14 in colorectal carcinoma tissue
was lower than that in normal tissues (Fig. 1D, P<0.01). The mean optical
densities of CXCL14 in tumor specimens and normal cells were 0.5411
and 0.1769, respectively.
Correlation between CXCL14 expression and
the clinicopathological features of colorectal carcinoma
Further analysis of the correlation between CXCL14
expression and the clinicopathological features of colorectal
carcinoma indicated that the expression of CXCL14 protein is
significantly correlated with tumor location, lymphatic metastasis
and clinicopathological stages (P<0.05; Table II). The relative expression level
of CXCL14 in colorectal carcinoma tumor tissue with lymphatic
metastasis was 0.146 (0.006~0.061), which is significantly lower
than the level in tumor tissue without lymphatic metastasis [0.215
(0.004~0.120)] (P<0.05). The relative expression levels in
colorectal carcinoma tissues at pathological stages III and IV were
significantly lower than those of specimens at pathological stages
I and II (P<0.05), whose relative expression levels were 0.146
(0.005~0.061) and 0.215(0.004~1.200), respectively. No significant
correlation was identified between the expression levels of CXCL14
and age, gender, infiltration degree or tumor marker (P>0.05).
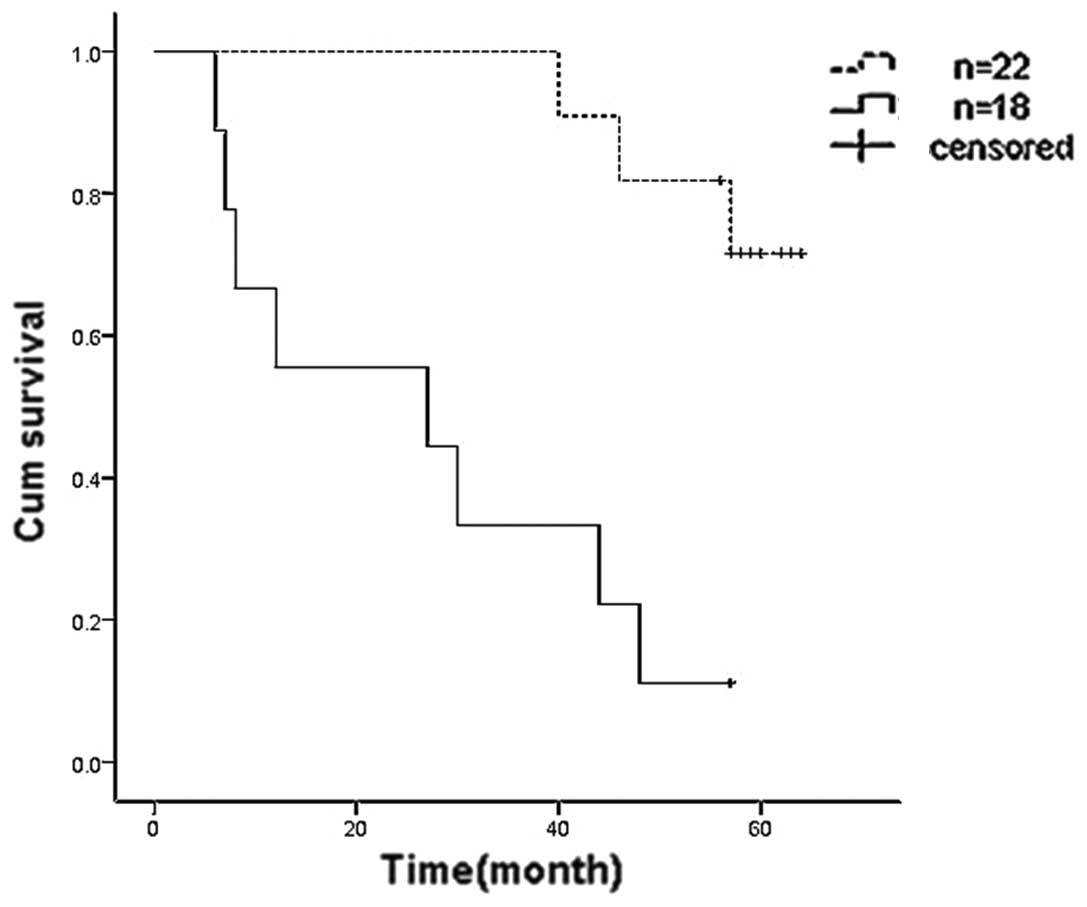
According to the relative values of the mean optical density of
CXCL14 proteins in paired tumor and normal tissues, specimens with
a relative value ≥1/3 were set as the high expression group and
specimens with a relative value <1/3 were denoted as the low
expression group. Kaplan-Meier survival analysis demonstrated that
the five-year survival rate of the high expression group was 72.7%,
while in the low expression group it was 11.1% (Fig. 2). Therefore, CXCL14 may be an
independent factor that positively affects prognosis.
CXCL14 overexpression influences
proliferation and changes in cell cycle distributions of HT29
colorectal carcinoma cells
To analyze the effect of CXCL14 on colorectal
carcinoma cells, a lentivirus that overexpressed CXCL14 was
constructed. The PCR product (~336 bp) was obtained from the
constructed plasmid of recombinant pLenti6.3_CXCL14_IRES-EGFP.
Nhel and Ascl double enzyme digestions also revealed
that the CXCL14 gene had been successfully cloned into the vectors
(Fig. 3A). DNA sequencing further
verified the successful construction of a recombinant plasmid. The
CXCL14-overexpressing lentivirus was further packaged by infecting
293T cells for 48 h. The virus titer was detected through the
enhanced green fluorescent protein (EGFP) carried by the virus. As
demonstrated in Fig. 3B, the cells
expressing EGFP were detected in cells infected with
2×10−8 ml virus and the titer of the virus was
calculated as 1.2×109 TU/ml. To confirm the expression
of CXCL14, HT29 colorectal carcinoma cells were transfected with
the CXCL14 lentivirus for 48 h followed by CXCL14 protein detection
with western blotting. Compared with the lentivirus control, CXCL14
expression increased markedly in the cells transfected with the
CXCL14 lentivirus (Fig. 3C).
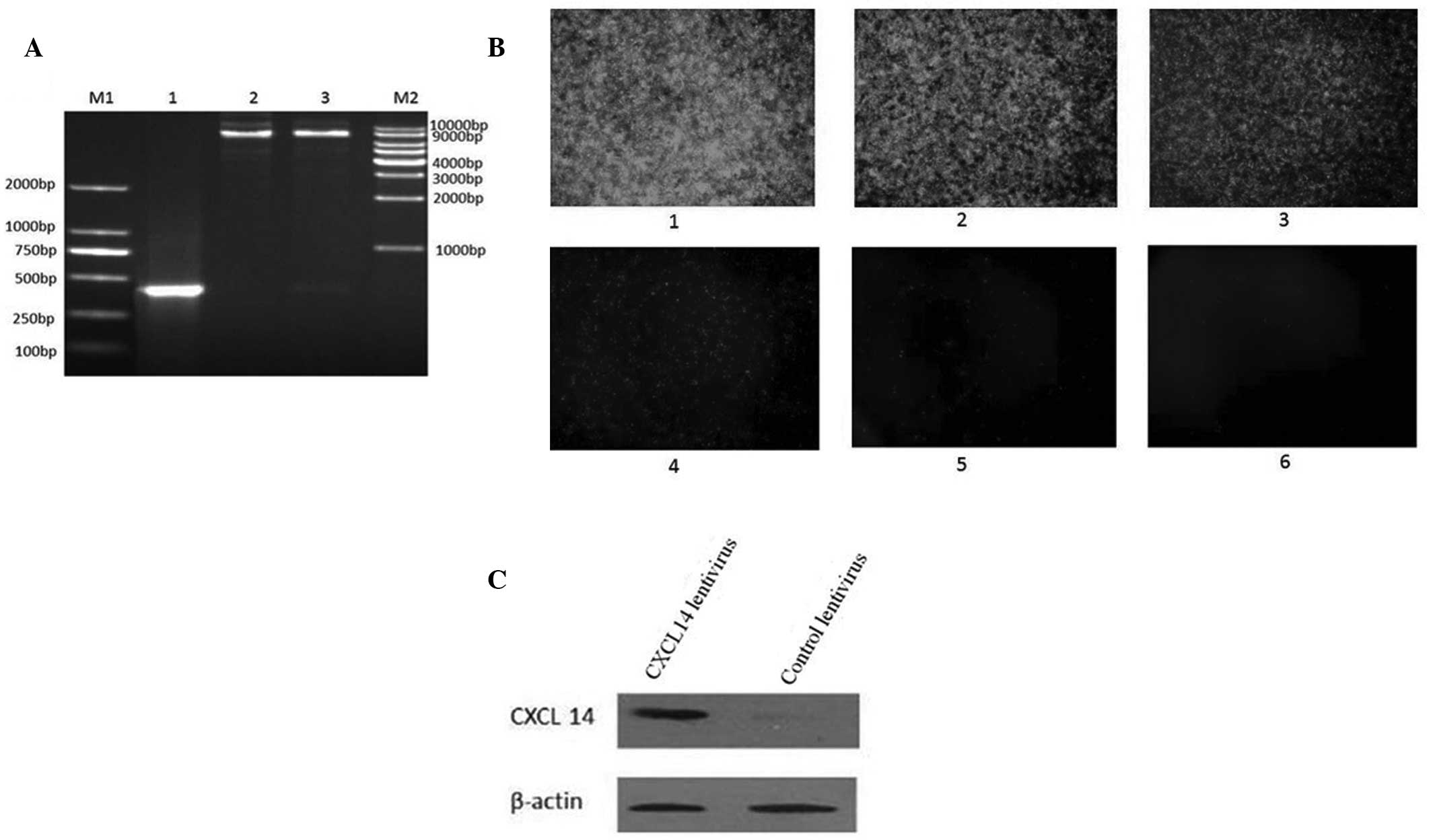 | Figure 3(A) Identification of PLenti6.3
_CXCL14_IRES-EGFP recombinant plasmid. M1, 250 bp DNA ladder
marker; lane 1, pLenti6.3_CXCL14_IRES-EGFP recombinant plasmid;
lane 2, plasmid digested by NheI; lane 3, plasmid degested
by NheI and AscI; M2, 1 kbp DNA ladder marker. (B)
Titer of CXCL14-overexpressing lentivirus by GFP detection: 1,
2×10−3 ml virus infection; 2, 2×10−4 ml virus
liquid; 3, 2×10−5 ml virus infection; 4,
2×10−6 ml virus infection; 5, 2×10−7 ml virus
infection; and 6, 2×10−8 ml virus infection
(magnification, ×40). (C) Western blot displaying overexpression of
CXCL14. GFP, green fluorescent protein. |
Further investigation of the effect of
CXCL14 overexpression in HT29 cells
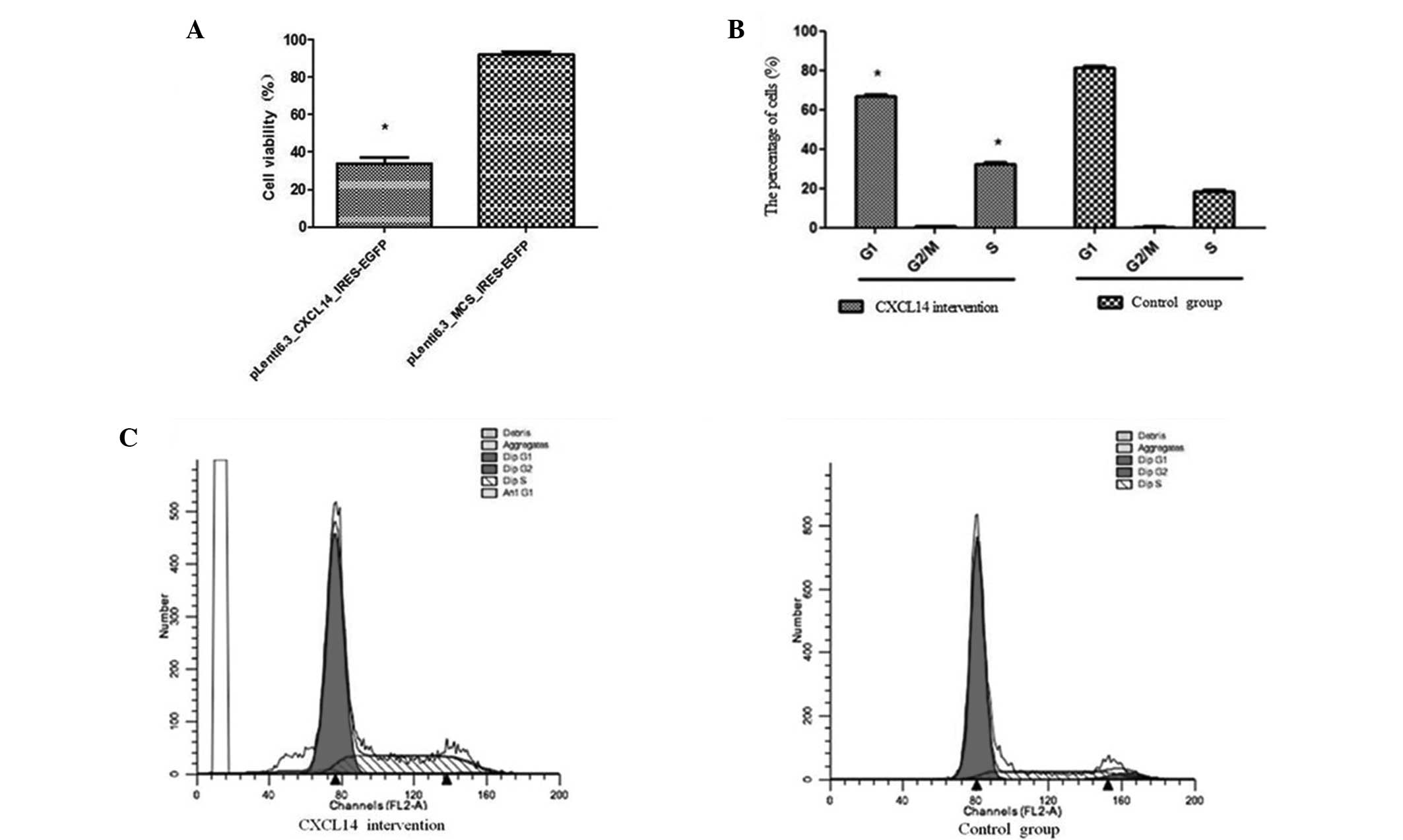
Ectopic expression of CXCL14 led to a significant
reduction in the viability of the HT29 cells, compared with the
group transfected with the control lentivirus (Fig. 4A; P<0.05). The cell viabilities
in the CXCL14-overexpression and control groups were 96.47±2.53%
and 36.56±5.47%, respectively. Further FCM analysis indicated that
the CXCL14-overexpression group presented a lower percentage of
cells in the G1 stage (67.46±0.92%) compared with the
control group (82.34±0.75%) (P=0.02); and a significantly higher
percentage of cells in the S stage (36.47±0.59) compared with the
control group (21.97±0.64%) (P=0.01) (Fig. 4B). No significant difference
between the two groups was observed in the percentages of cells in
the G2/M stage. These results suggest that the
inhibition of HT29 cell proliferation by CXCL14 overexpression was
mainly due to cell-cycle arrest at the G0/G1
stage (Fig. 4B and C).
Discussion
Chemokines can be sorted into inflammatory and
self-stable sub-functional families (36). Chemokines are important in
chemotaxis, i.e. the adhesion and migration of leukocytes, in
addition to being influential in pro-inflammatory reactions
(37,38). Chemokines can also control the
migration of immune cells from lymphoid to non-lymphoid tissues in
a stable condition, and therefore function in immunological
surveillance (39,40). It has been reported that cells in
tumor microenvironments are able to secrete several chemokines to
regulate basic biological processes in tumor cells and normal
cells, such as angiogenesis and specific immune activation of host
and tumor cell proliferation (41). Therefore, chemokines are essential
for the genesis, development and metastasis of tumors (41,42).
CXCL14, consisting of 77 amino acids, is a novel
member of the chemokine family and is crucial in maintaining a
self-stable physiological function. When stimulated by
lipopolysaccharide or Prostaglandin E2, peripheral monocytes have
been indicated to be chemoattracted to sites of high CXCL14
expression (12,39). Therefore, CXCL14 may be associated
with the homeostasis of monocyte differentiation into
macrophagocytes in tissues (12,13,17).
To analyze the association between CXCL14 expression
and colorectal carcinoma in the current study, the mRNA and protein
expression levels of CXCL14 in colorectal carcinoma and adjacent
normal tissues were detected using RT-qPCR and
immunohistochemistry. Consistent with the results reported by a
number of previous tumor-related studies (23–28),
the present study detected that the expression levels of CXCL14 in
colorectal carcinoma tissues were significantly lower than those in
normal tissues. The current study did contradict the findings of
Zeng et al (32), but was
consistent with the results of Cao et al (33). In order to diminish the occurrence
of experimental errors, a parallel immunohistochemical analysis was
conducted using the method described by Zeng et al (32). The results from the parallel study
were concurrent and indicated repeatedly that the expression levels
of CXCL14 in colorectal carcinoma were reduced compared with levels
in normal tissues. This further supports the hypothesis that the
lack of CXCL14 expression in colorectal carcinoma may result in
local immune deficiencies (including attenuated immune
surveillance, immune evasion, weakened presentation of antigens and
a disordered internal immune environment) caused by low
permeability of immune cells in tissues (14). An analysis of the correlation of
clinical features indicated that the expression level of CXCL14 in
colorectal carcinoma tissue with lymphoid metastasis was
significantly lower than that of tumor tissue without lymphoid
metastasis; the relative expression levels of CXCL14 in colorectal
carcinoma tissue at pathological stages III and IV were lower than
those of specimens at stages I and II. Additionally, the expression
levels of CXCL14 in tumor tissues were significantly correlated
with the prognosis of colorectal carcinoma. These findings suggest
that downregulation of CXCL14 expression may contribute to the more
aggressive phenotype of colorectal carcinoma.
In addition to anticancer immune mechanisms, CXCL14
may suppress tumor vasculature by inhibiting the chemotaxis of
vascular smooth muscle cells and the formation of microvascular
systems (16,19,21),
and therefore suppress the metabolism and growth of a tumor. In
addition, CXCL14 may also influence the proliferation, invasion and
migration of tumor cells via auto/paracrine pathways (24,43,44).
It is controversial whether CXCL14 expression levels are elevated
in prostate and pancreatic cancer (29–31).
However, endogenously expressed CXCL14 has been demonstrated to
promote the growth and invasiveness of breast and pancreatic cancer
cells (31,45).
In order to analyze the direct effect of CXCL14 on
colorectal carcinoma cells in the present study,
pLenti6.3_CXCL14_IRES-EGFP vectors containing an IRES sequence and
enhanced green fluorescent protein (EGFP) were constructed to
produce lentiviral overexpression of CXCL14. This method enabled
rapid analysis of CXCL14 overexpression through the detection of
EGFP. The successful construction of CXCL14-overexpressing
lentivirus was confirmed by immunoblotting using CXCL14-specific
monoclonal antibodies of cells transfected with the CXCL14
lentivirus. When HT29 cells were transfected with the CXCL14
lentivirus, the proliferation of HT29 cells was significantly
inhibited. FCM analysis indicated that the overexpression of CXCL14
in HR29 cells promoted cell cycle arrest in the G1
stage. The results of the current study were consistent with the
results reported by Wang et al (28) in hepatocellular carcinoma
cells.
In conclusion, the present study further suggests
that the downregulated expression levels of CXCL14 protein and mRNA
in colorectal carcinoma tissues influence the local immune response
of the tumor. In addition, the CXCL14 gene serves as a potential
tumor-inhibiting gene in colorectal carcinoma. This
characterization of the expression levels of CXCL14 in tumor
tissues may provide a novel clinical auxiliary index for future
treatment and prognosis evaluation of colorectal carcinoma.
Acknowledgements
The present study was supported by grants from the
Department of Education, Zhejiang (grant no. Y201327980); the
Taizhou Science and Technology Bureau (grant no. 130lky40); and the
Zhejiang Science and Technology Bureau (grant nos. 2012C33126 and
2012C37080). These sponsors provided the funding for the
experiments and the collection of specimens.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Devaud C, John LB, Westwood JA, Darcy PK
and Kershaw MH: Immune modulation of the tumor microenvironment for
enhancing cancer immunotherapy. Oncoimmunology. 2:e259612013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allen SJ, Crown SE and Handel TM:
Chemokine: receptor structure, interactions, and antagonism. Annu
Rev Immunol. 25:787–820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mellado M, Rodríguez-Frade JM, Mañes S and
Martínez AC: Chemokine signaling and functional responses: the role
of receptor dimerization and TK pathway activation. Annu Rev
Immunol. 19:397–421. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sallusto F, Mackay CR and Lanzavecchia A:
The role of chemokine receptors in primary, effector, and memory
immune responses. Annu Rev Immunol. 18:593–620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruffini PA, Morandi P, Cabioglu N,
Altundag K and Cristofanilli M: Manipulating the
chemokine-chemokine receptor network to treat cancer. Cancer.
109:2392–2404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mantovani A, Savino B, Locati M, Zammataro
L, Allavena P and Bonecchi R: The chemokine system in cancer
biology and therapy. Cytokine Growth Factor Rev. 21:27–39. 2010.
View Article : Google Scholar
|
|
8
|
Uchida H, Iwashita Y, Sasaki A, Shibata K,
Matsumoto T, Ohta M and Kitano S: Chemokine receptor CCR6 as a
prognostic factor after hepatic resection for hepatocellular
carcinoma. J Gastroenterol Hepatol. 21:161–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghadjar P, Coupland SE, Na IK, Noutsias M,
Letsch A, Stroux A, Bauer S, Buhr HJ, Thiel E, Scheibenbogen C and
Keilholz U: Chemokine receptor CCR6 expression level and liver
metastases in colorectal cancer. J Clin Oncol. 24:1910–1916. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hojo S, Koizumi K, Tsuneyama K, Arita Y,
Cui Z, Shinohara K, Minami T, et al: High-level expression of
chemokine CXCL16 by tumor cells correlates with a good prognosis
and increased tumor-infiltrating lymphocytes in colorectal cancer.
Cancer Res. 67:4725–4731. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hromas R, Broxmeyer HE, Kim C, Nakshatri
H, Christopherson K II, Azam M and Hou YH: Cloning of BRAK, a novel
divergent CXC chemokine preferentially expressed in normal versus
malignant cells. Biochem Biophys Res Commun. 255:703–706. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frederick MJ, Henderson Y, Xu X, Deavers
MT, Sahin AA, Wu H, Lewis DE, El-Naggar AK and Clayman GL: In vivo
expression of the novel CXC chemokine BRAK in normal and cancerous
human tissue. Am J Pathol. 156:1937–1950. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meuter S and Moser B: Constitutive
expression of CXCL14 in healthy human and murine epithelial
tissues. Cytokine. 44:248–255. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Starnes T, Rasila KK, Robertson MJ, Brahmi
Z, Dahl R, Christopherson K and Hromas R: The chemokine CXCL14
(BRAK) stimulates activated NK cell migration: implications for the
downregulation of CXCL14 in malignancy. Exp Hematol. 34:1101–1105.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Juremalm M and Nilsson G: Chemokine
receptor expression by mast cells. Chem Immunol Allergy.
87:130–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shellenberger TD, Wang M, Gujrati M,
Jayakumar A, Strieter RM, Burdick MD, Ioannides CG, Efferson CL,
El-Naggar AK, Roberts D, et al: BRAK/CXCL14 is a potent inhibitor
of angiogenesis and a chemotactic factor for immature dendritic
cells. Cancer Res. 64:8262–8270. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sleeman MA, Fraser JK, Murison JG, Kelly
SL, Prestidge RL, Palmer DJ, Watson JD and Kumble KD: B cell- and
monocyte-activating chemokine (BMAC), a novel non-ELR
alpha-chemokine. Int Immunol. 12:677–689. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kurth I, Willimann K, Schaerli P, Hunziker
T, Clark-Lewis I and Moser B: Monocyte selectivity and tissue
localization suggests a role for breast and kidney-expressed
chemokine (BRAK) in macrophage development. J Exp Med. 194:855–861.
2001. View Article : Google Scholar
|
|
19
|
Hara T and Nakayama Y: CXCL14 and insulin
action. Vitam Horm. 80:107–123. 2009. View Article : Google Scholar
|
|
20
|
Tanegashima K, Okamoto S, Nakayama Y, Taya
C, Shitara H, Ishii R, Yonekawa H, Minokoshi Y and Hara T: CXCL14
deficiency in mice attenuates obesity and inhibits feeding behavior
in a novel environment. PLoS One. 5:e103212010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maerki C, Meuter S, Liebi M, Muhlemann K,
Frederick MJ, Yawalkar N, Moser B and Wolf M: Potent and
broad-spectrum antimicrobial activity of CXCL14 suggests an
immediate role in skin infections. J Immunol. 182:507–514. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lindberg J, af Klint E, Catrina AI,
Nilsson P, Klareskog L, Ulfgren AK and Lundeberg J: Effect of
infliximab on mRNA expression profiles in synovial tissue of
rheumatoid arthritis patients. Arthritis Res Ther. 8:R1792006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hara T and Tanegashima K: Pleiotropic
functions of the CXC-type chemokine CXCL14 in mammals. J Biochem.
151:469–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tessema M, Klinge DM, Yingling CM, Do K,
Van Neste L and Belinsky SA: Re-expression of CXCL14, a common
target for epigenetic silencing in lung cancer, induces tumor
necrosis. Oncogene. 29:5159–5170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gu XL, Ou ZL, Lin FJ, Yang XL, Luo JM,
Shen ZZ and Shao ZM: Expression of CXCL14 and its anticancer role
in breast cancer. Breast Cancer Res Treat. 135:725–735. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ozawa S, Kato Y, Ito S, Komori R, Shiiki
N, Tsukinoki K, Ozono S, Maehata Y, et al: Restoration of
BRAK/CXCL14 gene expression by gefitinib is associated with
antitumor efficacy of the drug in head and neck squamous cell
carcinoma. Cancer Sci. 100:2202–2209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu C, Lin F, Zhu G, Xue X, Ding Y, Zhao Z,
Zhang L and Shen X: Abnormal hypermethylation of promoter region
downregulates chemokine CXC ligand 14 expression in gastric cancer.
Int J Oncol. 43:1487–1494. 2013.PubMed/NCBI
|
|
28
|
Wang W, Huang P, Zhang L, Wei J, Xie Q,
Sun Q, Zhou X, Xie H, Zhou L and Zheng S: Antitumor efficacy of
C-X-C motif chemokine ligand 14 in hepatocellular carcinoma in
vitro and in vivo. Cancer Sci. 104:1523–1531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song EY, Shurin MR, Tourkova IL, Gutkin DW
and Shurin GV: Epigenetic mechanisms of promigratory chemokine
CXCL14 regulation in human prostate cancer cells. Cancer Res.
70:4394–4401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Augsten M, Hagglof C, Olsson E, Stolz C,
Tsagozis P, Levchenko T, Frederick MJ, Borg A, Micke P, Egevad L
and Ostman A: CXCL14 is an autocrine growth factor for fibroblasts
and acts as a multi-modal stimulator of prostate tumor growth. Proc
Natl Acad Sci USA. 106:3414–3419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wente MN, Mayer C, Gaida MM, Michalski CW,
Giese T, Bergmann F, Giese NA, Büchler MW and Friess H: CXCL14
expression and potential function in pancreatic cancer. Cancer
Lett. 259:209–217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeng J, Yang X, Cheng L, Liu R, Lei Y,
Dong D, Li F, Lau QC, et al: Chemokine CXCL14 is associated with
prognosis in patients with colorectal carcinoma after curative
resection. J Transl Med. 11:62013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao B, Yang Y, Pan Y, Jia Y, Brock MV,
Herman JG and Guo M: Epigenetic silencing of CXCL14 induced
colorectal cancer migration and invasion. Discov Med. 16:137–147.
2013.PubMed/NCBI
|
|
34
|
Winawer SJ, Krabshuis J, Lambert R,
O’Brien M and Fried M; World Gastroenterology Organization
Guidelines Committee. Cascade colorectal cancer screening
guidelines: a global conceptual model. J Chin Gastroenterol.
45:297–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Benson AB 3rd, Bekaii-Saab T, Chan E, Chen
YJ, Choti MA, Cooper HS, et al: Localized colon cancer, version
3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc
Netw. 11:519–528. 2013.PubMed/NCBI
|
|
36
|
Zlotnik A and Yoshie O: Chemokines: a new
classification system and their role in immunity. Immunity.
12:121–127. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Campanelli AP, Brodskyn CI, Boaventura V,
Silva C, Roselino AM, Costa J, Saldanha AC, de Freitas LA, et al:
Chemokines and chemokine receptors coordinate the inflammatory
immune response in human cutaneous leishmaniasis. Hum Immunol.
71:1220–1227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wong MM and Fish EN: Chemokines:
attractive mediators of the immune response. Semin Immunol.
15:5–14. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Moser B, Wolf M, Walz A and Loetscher P:
Chemokines: multiple levels of leukocyte migration control. Trends
Immunol. 25:75–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Krieg C and Boyman O: The role of
chemokines in cancer immune surveillance by the adaptive immune
system. Semin Cancer Biol. 19:76–83. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Balkwill FR: The chemokine system and
cancer. J Pathol. 226:148–157. 2012. View Article : Google Scholar
|
|
42
|
Zlotnik A, Burkhardt AM and Homey B:
Homeostatic chemokine receptors and organ-specific metastasis. Nat
Rev Immunol. 11:597–606. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shurin GV, Ferris RL, Tourkova IL, Perez
L, Lokshin A, Balkir L, Collins B, Chatta GS and Shurin MR: Loss of
new chemokine CXCL14 in tumor tissue is associated with low
infiltration by dendritic cells (DC), while restoration of human
CXCL14 expression in tumor cells causes attraction of DC both in
vitro and in vivo. J Immunol. 174:5490–5498. 2005. View Article : Google Scholar
|
|
44
|
Izukuri K, Suzuki K, Yajima N, Ozawa S,
Ito S, Kubota E and Hata R: Chemokine CXCL14/BRAK transgenic mice
suppress growth of carcinoma cell transplants [corrected].
Transgenic Res. 19:1109–1117. 2010.PubMed/NCBI
|
|
45
|
Allinen M, Beroukhim R, Cai L, Brennan C,
Lahti-Domenici J, Huang H, et al: Molecular characterization of the
tumor microenvironment in breast cancer. Cancer Cell. 6:17–32.
2004. View Article : Google Scholar : PubMed/NCBI
|